Abstract
We sought a genotype-phenotype association: between single-nucleotide polymorphisms (SNPs) in olfactory receptor (OR) genes from the two largest OR gene clusters and odor-triggered nonallergic vasomotor rhinitis (nVMR). In the initial pedigree screen, using transmission disequilibrium test (TDT) analysis, six SNPs showed “significant” p-values between 0.0449 and 0.0043. In a second case-control population, the previously identified six SNPs did not re-emerge, whereas four new SNPs showed p-values between 0.0490 and 0.0001. Combining both studies, none of the SNPs in the TDT analysis survived the Bonferroni correction. In the population study, one SNP showed an empirical p-value of 0.0066 by shuffling cases and controls with 105 replicates; however, the p-value for this SNP was 0.83 in the pedigree study. This study emphasizes that underpowered studies having p-values between <0.05 and 0.0001 should be regarded as inconclusive and require further replication before concluding the study is “informative.” However, we believe that our hypothesis that an association between OR genotypes and the nVMR phenotype remains feasible. Future studies using either a genomewide association study of all OR gene-pseudogene regions throughout the genome—at the current recommended density of 2.5 to 5 kb per tag SNP—or studies incorporating microarray analyses of the entire “OR genome” in well-characterized nVMR patients are required.
Keywords: vasomotor rhinitis, olfactory receptor genes, genotype-phenotype association study, transmission disequilibrium test, case-control study, multiple-testing, Bonferroni correction, idiopathic environmental intolerance
Introduction
A number of complex, poorly characterized clinical syndromes fall under the umbrella of “idiopathic environmental intolerance” (IEI) (a.k.a. “multiple chemical sensitivity”). Whether the etiology is psychogenic or environmental continues to be debated (1–4). Several medical conditions appear to be related to overlap with IEI—such as sick-building syndrome and nonallergic vasomotor rhinitis (nVMR). In both of these, an odor or taste often precipitates one or more organ-system responses. Examples of initiating culprits include exposure to chemicals, such as formaldehyde being released from new furniture or carpeting, pesticides in ventilation systems, sterilizing agents (quaternary amines) in indoor cleaning solutions, mildew odors found in damp environments, freon circulating in closed-ventilation systems, perfumes/potpourris, and fresh paint and volatile organic compounds released from various products used in the home and workplace.
The current consensus for establishing a diagnosis of IEI requires that the patient's syndrome satisfies six essential criteria: (1) reproducible symptoms upon repeated exposures to chemicals/irritants; (2) chronicity; (3) later manifestation of symptoms to levels of exposure lower than those previously tolerated; (4) improvement or complete resolution of symptoms upon avoidance; (5) similar symptoms when exposed to chemically unrelated substances; and (6) multiple-organ symptoms—including, but not limited to, runny nose, itchy eyes, headache, scalp pain, scratchy throat, ear ache, mental confusion or sleepiness, heart palpitations, upset stomach, nausea, diarrhea, abdominal cramping, and aching joints (5).
To study IEI as quantitatively as possible, we searched for an experimental paradigm and decided upon nVMR, a clinical phenotype that should be quantifiable and unequivocal. nVMR satisfies all of the six essential IEI criteria except neurocognitive symptoms.
nVMR is a common condition that affects between 20% and 40% of the American population suffering from chronic rhinitis symptoms. The costs of treating chronic rhinitis (both allergic and nonallergic subtypes) and its associated comorbidities (e.g., sinusitis, asthma, otitis media) exceed $10 billion annually in the United States—ranking this condition among the most expensive outpatient health care problems that physicians encounter (6–8). nVMR patients differ from allergic rhinitis patients in that they commonly experience nasal congestion, postnasal drip, sinus pressure, and ear-plugging. Triggers for nVMR include temperature and/or barometric pressure changes, postural changes, and nasal irritants, such as those listed above (9). The present study focused only on olfactory triggers. By definition, nVMR patients do not exhibit nasal eosinophilia, and allergen skin testing to seasonal and perennial allergens is negative (9). Thus, a successful diagnosis of nVMR is dependent on the exclusion of allergic rhinitis.
The mechanism of nVMR is poorly understood (10–13). It has previously been hypothesized (14) that nVMR patients, because of their intolerance to odors and irritants, might be distinguished from patients having other forms of chronic rhinitis, due to an abnormality in their olfactory transduction pathway. Paradoxically, the limited number of studies investigating odorant discrimination found a greater magnitude of olfactory loss in nVMR patients in response to common odorants, such as formalin, camphor, asafetida (India saffron spice), and oil of peppermint—as compared with allergic rhinitis patients or control subjects without rhinitis (15). The one exception was the response to musk odor; in this study (15), males manifested a greater olfactory loss than females for all odorants except asafetida. These observations are in contrast to findings by our group (16), which recently reported that nVMR patients have an olfactory threshold response similar to allergic and mixed rhinitis patients. However, both studies (16) indicate that a hyperacute sense of smell does not account for the clinical symptoms induced by odorants in nVMR patients and support our hypothesis that these patients may have olfactory receptor (OR) gene polymorphisms predisposing them to this response.
Because we chose to focus only on olfactory triggers, we searched for SNPs in OR genes. The OR gene superfamily is perhaps the largest in the mammalian genome (17, 18). The pseudogene/gene ratio is highest in human, lower in chimpanzee, and lowest in mouse—most likely reflecting the rodent's greater dependence on olfaction for survival than humans. Presently in human, there are 390 putatively functional OR genes and 465 OR pseudogenes located in multiple clusters of varying sizes scattered throughout all autosomes except chromosome (Chr) 18 and Chr 20, and on the X but not the Y chromosome (http://www.gene.ucl.ac.uk/nomenclature/). This total of 855 genes is grouped into 18 families and 238 subfamilies. By far, the two largest OR gene clusters are located on Chr 11, with at least 81 functional genes (20.7%) on 11p (Cluster II) and 89 functional genes (22.7% of total) on 11q (Cluster I). The total of 372 genes-plus-pseudogenes on Chr 11 represents 43.0% of the total OR genes in the human genome. In the hunt for candidate OR genes, we, therefore, chose Chr 11 markers within Clusters II and I as the most efficient means to screen inexpensively for the maximal number of single-nucleotide polymorphisms (SNPs) that might be associated with the nVMR phenotype. Our cohorts included first, a pedigree study, followed by a case-control study.
Materials and Methods
Subjects and DNA Isolation
Patients diagnosed with nVMR were recruited from a large community allergy practice, having 7500 active patients (72% Caucasian, 17% African-American, 10% Hispanic, and 1% Asian); about 4000 have rhinitis with an approximate breakdown of 25% allergic, 30% nonallergic, and 45% mixed rhinitis. Rhinitis was well characterized with respect to their atopic status, nasal eosinophilia, and clinical history. The nVMR phenotype is defined as having symptoms of nasal congestion and postnasal drainage triggered by one or more of 20 common irritant exposures (e.g., ammonia, antiperspirants, bleach, cold air, cooking/frying, cosmetics, crude oils, fresh newsprint, hairspray, smog, cleaning products, mildew, paint, perfume, pine, soap powder, solvent, varnish, and tobacco and wood smoke). Patients rated the severity of their symptoms in response to the 20 irritant triggers using a 10-point Likert scale (19, 20), and the total irritant index value was obtained by adding each trigger score. Concomitantly, these patients are negative to prick skin testing of common seasonal/perennial allergens and show no evidence of nasal eosinophilia. Patients were selected as having a diagnosis of nVMR if they had a negative skin-prick test (defined by allergen wheal diameter in comparison to negative saline and positive histamine controls) and an irritant index of ≥24 (20). The irritant index scale was, therefore, a quantitative gradient rather than a binary trait.
For TDT analysis (21a), the prerequisite for enrollment was having two living biologic parents who were willing to participate in the study. Bloods were obtained from 30 nVMR patients and their parents (Table 1, left). For TDT analysis, one searches for the unbalanced transfer of a specific allele to affected children, no matter what the affected status of their parents. For the case-control analysis, 103 unrelated nVMR patients and 110 unrelated allergic rhinitis patients consented to participation in this study (Table 1 right). Whole blood (3 cc) was collected in a tube containing ethylenediaminetetraacetic acid (EDTA). DNA extraction was performed using the GenomicPrep™ (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) blood DNA isolation kit. All participants signed an informed consent for genetic testing, approved by the University of Cincinnati Institutional Review Board.
Table 1.
Demographics of the pedigree study and the case-control study.
| Pedigree study | Case-control study | |||
|---|---|---|---|---|
| Patients (N = 30) | Parents (N = 58)a | nVMR cases (N = 103) | Allergic rhinitis controls (N = 110) | |
| Average age (years) | 22.8 | 43 | 48 | 44 |
| Gender (F:M) | 20:10 | 29:29 | 84:19 | 68:42 |
| Caucasian:African-American | 28:2 | 54:4 | 99:4 | 101:9 |
| Irritant index scoreb | 29 | 20.5 | — | — |
nVMR, nonallergic vasomotor rhinitis.
One subject was also the parent of a subject.
The average irritant index score is based on a scale of 1 to 100; 20 different stimuli were rated from 0 (least) to 5 (most) bothersome, as described in the text.
SNP Marker Selection and Genotyping
Clusters II and I on human Chr 11 contain a total of 372 OR genes and pseudogenes; these were selected from the Olfactory Receptor Gene Source database (ORDB) (http://senselab.med.yale.edu/senselab/ORDB/files/humanor seqanal.html), and their sequences were obtained from the Human Olfactory Receptor Data Exploratorium (HORDE) (http://bioinformatics.weizmann.ac.il/HORDE/). Of these 372 OR genes and pseudogenes, 339 were specifically mapped (Figure 1) to Human Genome Reference Sequence by Human Genome Blast (http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html). Two regions (corresponding to Clusters II and I), having the highest density of OR genes, were selected. OR Cluster II spans 1.7 Mb (from Chr 11 4.4 to 6.1 Mb); OR Cluster I spans 4.2 Mb (from 55.0 to 59.2 Mb). A total of 224 (91 + 133) OR genes and pseudogenes from the ORDB database were mapped precisely to these two regions. Forty-four SNP markers were selected from TaqMan® Validated SNP Genotyping Assays (Applied Biosystems, Foster City, California, USA) to cover these two regions (110 kb/SNP for Cluster II; 150 kb/SNP for Cluster I). The overall genotype-calling rate was 99.1%. For the purpose of quality control, we used 16 blind duplicate samples; no genotype inconsistencies were found with any of the markers. Detailed information about the 44 SNP markers is given in Appendix 1. SNPs were genotyped by TaqMan (22).
Figure 1.
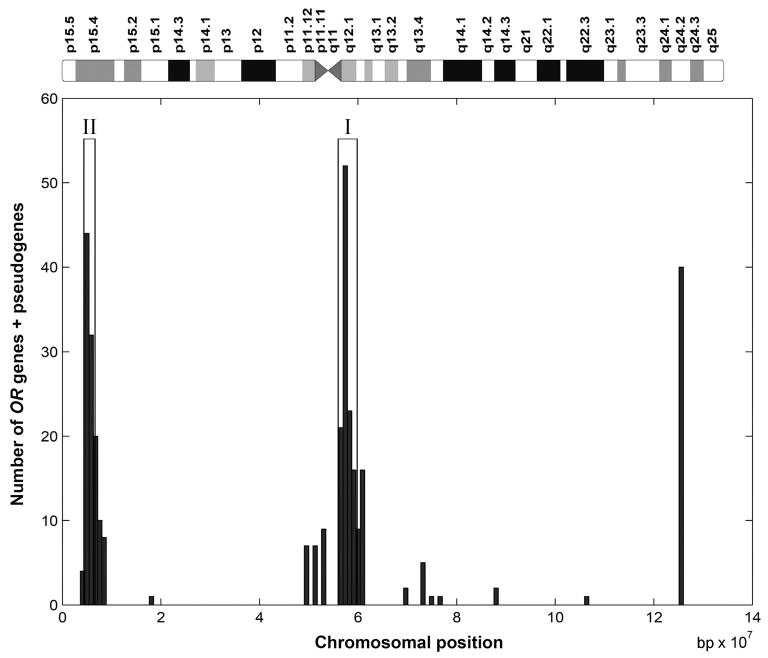
Distribution of olfactory receptor (OR) genes + pseudogenes on chromosome (Chr) 11. Total length of Chr 11 is 134 Mb. The two regions with dense OR genes correspond to OR gene Cluster II (11p, left) and Cluster I (11q, middle). The candidate regions in this study were selected as those with the highest density of OR genes, using the sliding window technique, and do not cover the entire Cluster II and I regions. Embedded within either cluster are possible candidate genes for nonallergic vasomotor rhinitis other than OR genes—such as the gene for C1 esterase inhibitor, an angiotensin II antagonist-like gene, and a purinergic receptor gene.
Statistical Analysis
Single-marker analyses of the 30 nVMR trios and the population-based association were conducted using Haploview 3.2 (22). We used the chi-squared test upon 2 × 2 allele counts (embedded within the Haploview package) in analyzing allelic associations. In order to correct for multiple-testing, the Bonferroni correction procedure was applied for the TDT analysis; empirical p-values were evaluated by the shuffling permutation test with 105 replicates for the case-control analysis (23).
Results
Demographics of the Populations Studied
For TDT analysis, the nVMR subjects were predominantly Caucasian, with two-thirds being female (Table 1, left). The patient population reported more problems than the parents with odors and irritants—based on the irritant index score. For the case-control study, the population again was predominantly Caucasian, having 81.5% nVMR female cases compared with 61.8% allergic rhinitis female controls (Table 1, right). Age and ethnicity were normally distributed between the case and control groups.
Genomic Analysis
As a preliminary screen, the pedigree study identified two SNPs (rs649358 and rs399208), 176.5 kb apart at the 3′ telomeric end of the Cluster I region; these SNPs were statistically significantly associated (p = 0.0043 and 0.0196) with the nVMR phenotype. Additional “significant” SNPs included the adjacent SNPs rs2515366 and rs1938596, both with p = 0.0143, another SNP (rs501828) having a P-value of 0.0499 in the Cluster I region, and one SNP (rs17480) in the Cluster II region (p = 0.0164).
Based on these encouraging results, we proceeded to the larger case-control population; we found no significant deviation from Hardy-Weinberg equilibrium across the 44 markers (data not shown). The SNPs that had been significant in the pedigree study were no longer significant in the case-control study; rather, four other statistically significant SNPs emerged: rs1430397 (p = 0.0094) in the Cluster II region and rs2848634 (p = 0.0490), rs1783826 (p = 0.0114), and rs2245676 (p = 0.0001) in the Cluster I region.
Table 2 summarizes the combined results for both the TDT analysis and the population-based association test. Although there were ten associations having p-values of <0.05, no marker showed significance in both studies. Moreover, no significant p-values found in the TDT analysis survived the Bonferroni correction for 44 multiple tests; this is largely due to the limited power provided by only 30 trios.
Table 2.
Combined results from TDT and population-based association tests for the SNPs in OR Cluster II (markers 1–16) and Cluster I (markers 17–44).
| Markers | Allele frequencies | Association tests | ||||||
|---|---|---|---|---|---|---|---|---|
| No. | rs_id | Celera ID | Minor allele | nVMR (103) | ARa(110) | TDT p-value | nVMR vs. ARap-value | |
| 1 | rs1376681 | C___3005257_10 | C | 0.475 | 0.458 | 0.5127 | 0.7182 | |
| 2 | rs1430397 | C___8798299_10 | T | 0.328 | 0.453 | 0.5637 | 0.0094 | |
| 3 | rs1505204 | C___3187363_10 | C | 0.475 | 0.388 | 0.5775 | 0.0720 | |
| 4 | rs1389464 | C___2021191_10 | C | 0.471 | 0.432 | 0.1573 | 0.4329 | |
| 5 | rs1433897 | C___8461299_10 | A | 0.387 | 0.402 | 0.2888 | 0.7600 | |
| 6 | rs1378736 | C___8797974_10 | G | 0.460 | 0.486 | 0.6547 | 0.5947 | |
| 7 | rs2499949 | C__16029966_10 | A | 0.221 | 0.201 | 0.7630 | 0.6222 | |
| 8 | rs916111 | C___9599131_1_ | A | 0.431 | 0.458 | 0.6949 | 0.5848 | |
| 9 | rs951748 | C___1451737_10 | C | 0.475 | 0.481 | 0.8527 | 0.9053 | |
| 10 | rs934460 | C___1451536_10 | C | 0.475 | 0.500 | 0.8415 | 0.6111 | |
| 11 | rs1498553 | C___1452175_10 | C | 0.461 | 0.453 | 0.0679 | 0.8707 | |
| 12 | rs1453424 | C___7695347_10 | C | 0.279 | 0.338 | 0.2207 | 0.1965 | |
| 13 | rs1377512 | C___9604514_10 | G | 0.279 | 0.349 | 0.2207 | 0.1263 | |
| 14 | rs1463289 | C___9604619_10 | C | 0.421 | 0.386 | 0.7237 | 0.4680 | |
| 15 | rs17480 | C__12033057_10 | G | 0.446 | 0.411 | 0.0164 | 0.4715 | |
| 16 | rs1462983 | C___9604662_10 | T | 0.396 | 0.401 | 0.2393 | 0.9189 | |
| 17 | rs649358 | C___8904618_10 | A | 0.453 | 0.424 | 0.0043 | 0.5617 | |
| 18 | rs399208 | C___3109726_10 | A | 0.485 | 0.495 | 0.0196 | 0.8363 | |
| 19 | rs984371 | C___3109488_10 | C | 0.201 | 0.217 | 0.2008 | 0.6883 | |
| 20 | rs1384101 | C____268897_10 | C | 0.119 | 0.179 | 0.5271 | 0.0850 | |
| 21 | rs2460211 | C___8132897_10 | A | 0.325 | 0.386 | 0.8575 | 0.1994 | |
| 22 | rs1481928 | C___1870209_10 | C | 0.328 | 0.387 | 0.8575 | 0.2146 | |
| 23 | rs1945245 | C__11666818_10 | T | 0.267 | 0.307 | 0.4328 | 0.3774 | |
| 24 | rs637404 | C____746124_10 | C | 0.307 | 0.316 | 0.3532 | 0.8415 | |
| 25 | rs502943 | C____732012_10 | T | 0.381 | 0.429 | 1.0000 | 0.3196 | |
| 26 | rs1788998 | C___8905573_10 | C | 0.387 | 0.420 | 0.8474 | 0.4987 | |
| 27 | rs1793426 | C___8905725_10 | A | 0.307 | 0.362 | 0.8415 | 0.2372 | |
| 28 | rs501828 | C___3043337_10 | G | 0.431 | 0.395 | 0.0499 | 0.4553 | |
| 29 | rs1943482 | C___7986115_10 | G | 0.299 | 0.302 | 0.1936 | 0.9492 | |
| 30 | rs3781902 | C___8131596_10 | A | 0.500 | 0.533 | 0.3692 | 0.5005 | |
| 31 | rs2848634 | C___2950363_10 | G | 0.196 | 0.278 | 0.5127 | 0.0490 | |
| 32 | rs1783826 | C___2864618_10 | T | 0.371 | 0.495 | 1.0000 | 0.0114 | |
| 33 | rs530094 | C____924939_1_ | A | 0.275 | 0.215 | 0.2253 | 0.1565 | |
| 34 | rs1704781 | C___1983937_10 | C | 0.289 | 0.238 | 0.3173 | 0.2375 | |
| 35 | rs921134 | C___2161631_10 | G | 0.319 | 0.330 | 0.2008 | 0.8012 | |
| 36 | rs2245676 | C___1334145_10 | G | 0.480 | 0.297 | 0.8273 | 0.0001 | |
| 37 | rs1938663 | C__11388708_10 | G | 0.392 | 0.373 | 0.0719 | 0.6822 | |
| 38 | rs2515366 | C__10094781_10 | T | 0.436 | 0.429 | 0.0143 | 0.8850 | |
| 39 | rs1938596 | C___3185821_10 | G | 0.436 | 0.435 | 0.0143 | 0.9721 | |
| 40 | rs1938709 | C___8137912_10 | A | 0.230 | 0.250 | 0.8185 | 0.6399 | |
| 41 | rs1892866 | C__11330707_10 | T | 0.500 | 0.552 | 0.7055 | 0.2893 | |
| 42 | rs1938781 | C___3160372_10 | G | 0.255 | 0.298 | 0.4913 | 0.3274 | |
| 43 | rs1545527 | C___8141059_10 | C | 0.319 | 0.294 | 0.8273 | 0.5910 | |
| 44 | rs3758872 | C__11668147_10 | C | 0.000 | 0.000 | 0.0578 | n.a. | |
TDT, transmission disequilibrium test; SNPs, single-nucleotide polymorphisms; nVMR, nonallergic vasomotor rhinitis.
AR, allergic rhinitis; these patients do not have nVMR.
For the population-based association test, the correlation of rs2245676 with the phenotype was still significant after correcting for multiple testing, the empirical p-value being 0.0066 by shuffling cases and controls with 105 replicates; however, this SNP showed a p-value of 0.83 in the pedigree study. This marker is located within the Cluster I pseudogene, OR9I2P.
Discussion
The present study was carried out prior to the release of any HapMap consortium data. We reasoned that genetic differences in olfaction might be associated with the nVMR phenotype triggered by various potent odors. Moreover, we posited that the nVMR trait might be associated with SNPs in one or more of the OR genes. Just recently, in fact, olfactory detection threshold phenotypes of four odorants were screened against 43 OR genes, genomewide, and a strong association was found between the odorant isovaleric acid and SNPs in OR11H7P; this predicted receptor-ligand functional relationship was then validated using the Xenopus oocyte expression system, in which the intact allele of OR11H7P exhibited a positive response to isovaleric acid (24).
Because OR genes are dispersed over 20 of 22 autosomes plus the X chromosome, it was too expensive for an initial screening study to select SNPs for all of the known 390 functional OR genes and 465 OR pseudogenes. We, therefore, chose to focus on the two most densely populated OR gene-pseudogene clusters, examining SNP frequencies at a density of 110-150 kb per SNP, in hopes of finding some region that might be highly statistically significant in a genotype-phenotype association study.
We hypothesized that TDT analysis of 30 VMR patients and their unaffected biologic parents would be sufficient as a first-level screen: Could a very highly significant OR gene be identified within either of these clusters? Encouragingly, six statistically significant markers were identified by TDT analysis; in fact, two adjacent SNPs (rs649358 and rs399208), 176.5 kb apart at the 3′ telomeric end of the Cluster I region, showed an association with the nVMR phenotype, having p-values of 0.0043 and 0.0196, respectively. At this point, some laboratories would publish these data. In fact, hundreds of such publications have appeared—especially in journals having “pharmacology,” “toxicology,” “drug,” “environmental,” “pharmacogenetics,” or “pharmacogenomics” in their titles. Such studies should be regarded as inconclusive, underpowered, and in need of further replication before the findings can be regarded as “informative” (25).
None of the six sites in the pedigree study survived the Bonferroni multiple-tests correction analysis. Most likely, this is due to the low SNP marker density in this pilot study. From the data obtained by the HapMap consortium, the general rule for genomewide association studies now is to screen the genome at a density of one tag SNP per 5 kb, and for Africans the density should be one tag SNP per 2.5 kb (25).
In the present study, we then compared 103 nVMR cases with 110 allergic rhinitis controls and found that the original SNPs uncovered via TDT analysis had disappeared, whereas four new p < 0.05 SNPs emerged. Combination of the two sets of data (Table 2) gave no consistently statistically significant variant site—with the exception of one pseudogene; although the p-value for rs2245676 was 0.0001 in the case-control study, the p-value was 0.83 in the pedigree study. The allelic frequency of this SNP is significantly different between Caucasians and Africans; we, therefore, cannot rule out the possibility of a false-positive finding due to population stratification. Thus, no definitive conclusions can be drawn due to the limited power of both the pedigree study and the case-control study. A list of power calculations is presented in Table 3, indicating that both the pedigree and population studies were underpowered.
Table 3.
Power calculations for this study, if we assume the test marker is in perfect linkage disequilibrium with the true causative locus.a
| A: where α = 0.05 (without correction for multiple-testing) | ||
|---|---|---|
| H2 | TDT | Case-control |
| 0.02 | 0.121 | 0.464 |
| 0.05 | 0.231 | 0.849 |
| 0.10 | 0.403 | 0.989 |
| 0.20 | 0.669 | 1.000 |
| B: where α = 0.001 (with correction for multiple-testing) | ||
| H2 | TDT | Case-control |
| 0.02 | 0.006 | 0.078 |
| 0.05 | 0.019 | 0.383 |
| 0.10 | 0.058 | 0.836 |
| 0.20 | 0.186 | 0.997 |
TDT, transmission disequilibrium test.
The optimal situation is calculated with a K = 0.20 (incidence of nonallergic vasomotor rhinitis in the United States is estimated to be between 20% and 25%) and the minor allele frequency = 0.1. H2 denotes heritability (the proportion of phenotypic variation ascribed to the genetic variability at the putative locus under investigation). The reason for including an H2 of 0.10 and 0.20 is to attain “the highest achievable” statistical power for our small-sample TDT analysis—although such high heritability levels seldom occur in common diseases. Additivity is assumed in these power calculations. These calculations underscore the fact that the power of the TDT analysis is far lower than that of the case-control study.
However, we believe that our hypothesis of a relationship between OR genetic polymorphisms and nVMR remains feasible. Either a future genomewide association study of all OR gene-pseudogene regions throughout the genome—at the current recommended density (25, 26) of 2.5–5 kb per tag SNP—or a microarray analyses of the entire “OR genome” in well-characterized nVMR patients is required to uncover an important OR polymorphism associated with the nVMR trait. Selection of these subjects might be based on their olfactory threshold responses and diagnosed using standardized psychophysical tests currently available for the assessment of olfactory function. Very recently, automated self-administered instruments, using the staircase approach for measuring olfactory responses (based on the validated 40-odor University of Pennsylvania Smell Identification Test (UPSIT), the Cross-Cultural Smell Identification Test™, and the Smell Threshold Test™), have facilitated our ability to investigate olfaction in humans (16). The current understanding of olfactory function in humans has controlled for factors such as age, gender, ethnicity, exposure to toxic agents, and various other disease states, such as Parkinson disease and Alzheimer disease (27–30).
Conclusion
In summary, further investigation of the role of OR gene polymorphisms in the pathogenesis of nVMR may provide valuable insight into a better understanding of this disorder. Moreover, any highly statistically significant genotype-phenotype association with an OR gene and nVMR may also provide important inroads into the more complex, yet poorly characterized, clinical syndromes that fall under the umbrella of IEI or multiple chemical sensitivity.
Acknowledgments
The authors thank their colleagues for valuable discussions and critical reading of this manuscript. This work was supported, in part, by Pilot Project funds from NIH Grant P30 ES06096 (J.A.B., L.J., and D.W.N.).
Appendix I.—Detailed information of the 44 SNP markers
| No. | dbSNP rs_id | Celera ID | Chr | NCBI Pos. | Genetic Pos. | NCBI Gene | Celera Pos. | Celera Gene |
|---|---|---|---|---|---|---|---|---|
| 1 | rs1376681 | C__3005257_10 | 11p | 4395403 | 8.64 | 4555293 | ||
| 2 | rs1430397 | C__8798299_10 | 11p | 4534144 | 8.335 | 4694035 | ||
| 3 | rs1505204 | C__3187363_10 | 11p | 4578975 | 7.965 | LOC390037 | 4738872 | RNF137 |
| 4 | rs1389464 | C__2021191_10 | 11p | 4693821 | 7.1 | LOC390040 | 4853710 | |
| 5 | rs1433897 | C__8461299_10 | 11p | 4906213 | 9.115 | 5065714 | ||
| 6 | rs1378736 | C__8797974_10 | 11p | 5030396 | 9.845 | LOC119679 | 5181285 | |
| 7 | rs2499949 | C__16029966_10 | 11p | 5115825 | 10.345 | 5266666 | ||
| 8 | rs916111 | C__9599131_1_ | 11p | 5233652 | 11.04 | HBG1 | 5388010 | HBG1 |
| 9 | rs951748 | C__1451737_10 | 11p | 5381476 | 11.525 | 5535635 | ||
| 10 | rs934460 | C__1451536_10 | 11p | 5499491 | 11.105 | MGC20470 | 5653762 | MGC20470 |
| 11 | rs1498553 | C__1452175_10 | 11p | 5673337 | 11.735 | 5828125 | TRIM22 | |
| 12 | rs1453424 | C__7695347_10 | 11p | 5781842 | 12.215 | 5936563 | ||
| 13 | rs1377512 | C__9604514_10 | 11p | 5783926 | 12.215 | LOC390076 | 5938647 | |
| 14 | rs1463289 | C__9604619_10 | 11p | 5876789 | 12.625 | 6032055 | ||
| 15 | rs17480 | C__12033057_10 | 11p | 5990420 | 12.775 | LOC120793 | 6145694 | |
| 16 | rs1462983 | C__9604662_10 | 11p | 6094146 | 12.435 | LOC196335 | 6249340 | |
| 17 | rs649358 | C__8904618_10 | 11q | 55024839 | 59.955 | LOC219425 | 52592871 | |
| 18 | rs399208 | C__3109726_10 | 11q | 55202882 | 59.665 | 52769375 | ||
| 19 | rs984371 | C__3109488_10 | 11q | 55353058 | 59.415 | LOC219437 | 52917849 | |
| 20 | rs1384101 | C___268897_10 | 11q | 55569236 | 59.055 | 53134041 | ||
| 21 | rs2460211 | C__8132897_10 | 11q | 55670193 | 58.885 | 53234988 | ||
| 22 | rs1481928 | C__1870209_10 | 11q | 55797294 | 58.675 | 53362064 | ||
| 23 | rs1945245 | C__11666818_10 | 11q | 55924487 | 58.465 | LOC390159 | 53496810 | |
| 24 | rs637404 | C___746124_10 | 11q | 56246728 | 58.4 | 53830121 | ||
| 25 | rs502943 | C___732012_10 | 11q | 56354976 | 58.4 | 53938382 | ||
| 26 | rs1788998 | C__8905573_10 | 11q | 56363335 | 58.4 | OR5G3P | 53946741 | |
| 27 | rs1793426 | C__8905725_10 | 11q | 56459110 | 58.4 | LOC390180 | 54042474 | |
| 28 | rs501828 | C__3043337_10 | 11q | 56572812 | 58.4 | 54156411 | ||
| 29 | rs1943482 | C__7986115_10 | 11q | 56770969 | 58.4 | 54354555 | AGTRL1 | |
| 30 | rs3781902 | C__8131596_10 | 11q | 56895407 | 58.4 | P2RX3 | 54479076 | P2RX3 |
| 31 | rs2848634 | C__2950363_10 | 11q | 57027688 | 58.4 | SLC43A1 | 54608883 | RTN4RL2 |
| 32 | rs1783826 | C__2864618_10 | 11q | 57178324 | 58.4 | 54759451 | FLJ30213 | |
| 33 | rs530094 | C___924939_1_ | 11q | 57306397 | 58.4 | CTNND1 | 54888301 | CTNND1 |
| 34 | rs1704781 | C__1983937_10 | 11q | 57422668 | 58.4 | 55004335 | ||
| 35 | rs921134 | C__2161631_10 | 11q | 57570756 | 58.4 | 55152459 | ||
| 36 | rs2245676 | C__1334145_10 | 11q | 57688053 | 58.4 | OR9I2P | 55269667 | |
| 37 | rs1938663 | C__11388708_10 | 11q | 57860843 | 59.495 | LOC219964 | 55443912 | |
| 38 | rs2515366 | C__10094781_10 | 11q | 58049730 | 58.705 | LOC219968 | 55633478 | |
| 39 | rs1938596 | C__3185821_10 | 11q | 58157046 | 58.4 | ZFP91-CNTF | 55740786 | |
| 40 | rs1938709 | C__8137912_10 | 11q | 58311191 | 58.4 | 55894916 | ||
| 41 | rs1892866 | C__11330707_10 | 11q | 58457509 | 58.4 | 56041242 | MGC15937 | |
| 42 | rs1938781 | C__3160372_10 | 11q | 58690470 | 58.4 | FLJ22794 | 56274211 | FLJ22794 |
| 43 | rs1545527 | C__8141059_10 | 11q | 59029755 | 58.4 | LOC401696 | 56615677 | — |
| 44 | rs3758872 | C__11668147_10 | 11q | 59196535 | 58.4 | FLJ36874 | 56783923 | FLJ36874 |
Footnotes
Publisher's Disclaimer: Full terms and conditions of use: http://www.informaworld.com/terms-and-conditions-of-access.pdf
This article may be used for research, teaching and private study purposes. Any substantial or systematic reproduction, re-distribution, re-selling, loan or sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
References
- 1.Staudenmayer H, Binkley KE, Leznoff A, Phillips S. Idiopathic environmental intolerance: part 1: a causation analysis applying Bradford Hill's criteria to the toxicogenic theory. Toxicol Rev. 2003;22:235–246. doi: 10.2165/00139709-200322040-00005. [DOI] [PubMed] [Google Scholar]
- 2.Staudenmayer H, Binkley KE, Leznoff A, Phillips S. Idiopathic environmental intolerance: part 2: a causation analysis applying Bradford Hill's criteria to the psychogenic theory. Toxicol Rev. 2003;22:247–261. doi: 10.2165/00139709-200322040-00006. [DOI] [PubMed] [Google Scholar]
- 3.Binder LM, Campbell KA. Medically unexplained symptoms and neuropsychological assessment. J Clin Exp Neuropsychol. 2004;26:369–392. doi: 10.1080/13803390490510095. [DOI] [PubMed] [Google Scholar]
- 4.Moen BE. Chemical sensitivity and the work place environment: research needs. Psychoneuroendocrinology. 2005;30:1039–1042. doi: 10.1016/j.psyneuen.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Committee on MCS. Multiple chemical sensitivity: a 1999 consensus. Arch Environ Health. 1999;54:147–149. doi: 10.1080/00039899909602251. [DOI] [PubMed] [Google Scholar]
- 6.Weiss KB, Sullivan SD. The health economics of asthma and rhinitis. I Assessing the economic impact. J Allergy Clin Immunol. 2001;107:3–8. doi: 10.1067/mai.2001.112262. [DOI] [PubMed] [Google Scholar]
- 7.Brandt D, Bernstein JA. Questionnaire evaluation and risk factor identification for nonallergic vasomotor rhinitis. Ann Allergy Asthma Immunol. 2006;96:526–532. doi: 10.1016/S1081-1206(10)63546-6. [DOI] [PubMed] [Google Scholar]
- 8.Reed SD, Lee TA, McCrory DC. The economic burden of allergic rhinitis: a critical evaluation of the literature. Pharmacoeconomics. 2004;22:345–361. doi: 10.2165/00019053-200422060-00002. [DOI] [PubMed] [Google Scholar]
- 9.Bachert C. Persistent rhinitis—allergic or nonallergic? Allergy. 2004;59(Suppl 76):11–15. doi: 10.1111/j.0108-1675.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 10.Sanico AM, Philip G, Proud D, Naclerio RM, Togias A. Comparison of nasal mucosal responsiveness to neuronal stimulation in non-allergic and allergic rhinitis: effects of capsaicin nasal challenge. Clin Exp Allergy. 1998;28:92–100. doi: 10.1046/j.1365-2222.1998.00182.x. [DOI] [PubMed] [Google Scholar]
- 11.Campbell JD, Stinson MJ, Simons FE, HayGlass KT. Systemic chemokine and chemokine receptor responses are divergent in allergic versus non-allergic humans. Int Immunol. 2002;14:1255–1262. doi: 10.1093/intimm/dxf098. [DOI] [PubMed] [Google Scholar]
- 12.Riccio AM, Tosca MA, Cosentino C, Pallestrini E, Ameli F, Canonica GW, Ciprandi G. Cytokine pattern in allergic and non-allergic chronic rhinosinusitis in asthmatic children. Clin Exp Allergy. 2002;32:422–426. doi: 10.1046/j.1365-2222.2002.01315.x. [DOI] [PubMed] [Google Scholar]
- 13.Zakrzewska A, Kobos J, Gryczynska D. Evaluation of CD25, CD152, Fas-ligand expression in the adenoids of allergic, and non-allergic children: a pilot study. Int J Pediatr Otorhinolaryngol. 2003;67(Suppl 1):S205–S208. doi: 10.1016/j.ijporl.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Higo R, Ichimura K, Ota Y, Ishizuka T, Shimazoki Y. Investigation of “anosmic zones” associated with nasal allergy. Nippon Jibiinkoka Gakkai Kaiho. 1996;99:1648–1652. doi: 10.3950/jibiinkoka.99.1648. [DOI] [PubMed] [Google Scholar]
- 15.Mann SS, Maini S, Nageswari KS, Mohan H, Handa A. Assessment of olfactory status in allergic and non-allergic rhinitis patients. Ind J Physiol Pharmacol. 2002;46:186–194. [PubMed] [Google Scholar]
- 16.Resvani M, Brandt D, Bernstein JA. Investigation of olfactory threshold responses in chronic rhinitis subtypes. Ann Allergy Asthma Immunol. 2007;99:571–572. doi: 10.1016/S1081-1206(10)60389-4. [DOI] [PubMed] [Google Scholar]
- 17.Gaillard I, Rouquier S, Giorgi D. Olfactory receptors. Cell Mol Life Sci. 2004;61:456–469. doi: 10.1007/s00018-003-3273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henion TR, Schwarting GA. Patterning the developing and regenerating olfactory system. J Cell Physiol. 2007;210:290–297. doi: 10.1002/jcp.20888. [DOI] [PubMed] [Google Scholar]
- 19.Turner CJ, Ellis S, Giles J, Altiere R, Sintek C, Ulrich H, Valdez C, Zadvorny E. An introductory pharmacy practice experience emphasizing student-administered vaccinations. Am J Pharm Educ. 2007;71:1–6. doi: 10.5688/aj710103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein JA, Brandt DM, Martin V. Differentiation of chronic rhinitis subtypes using an irritant index scale. Proceedings of Annual Meeting. Ann Allergy Asthma Immunol. 2007 Abstract 38:12. [Google Scholar]
- 21.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 21a.Guo W, Fung WK. Combining the case-control methodology with the small size transmission/disequilibrium test for multiallelic markers. Eur J Hum Genet. 2005;13:1007–1012. doi: 10.1038/sj.ejhg.5201453. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 24.Menashe I, Abaffy T, Hasin Y, Goshen S, Yahalom V, Luetje CW, Lancet D. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:2462–2468. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev. 2008 doi: 10.1080/03602530801952864. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat Rev Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 27.Amoore JE, Ollman BG. Practical test kits for quantitatively evaluating the sense of smell. Rhinology. 1983;21:49–54. [PubMed] [Google Scholar]
- 28.Doty RL, Shaman P, Dann M. Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol Behav. 1984;32:489–502. doi: 10.1016/0031-9384(84)90269-5. [DOI] [PubMed] [Google Scholar]
- 29.Doty RL, Marcus A, Lee WW. Development of the 12-item Cross-Cultural Smell Identification Test (CC-SIT) Laryngoscope. 1996;106:353–356. doi: 10.1097/00005537-199603000-00021. [DOI] [PubMed] [Google Scholar]
- 30.Doty RL, Mishra A. Olfaction and its alteration by nasal obstruction, rhinitis, and rhinosinusitis. Laryngoscope. 2001;111:409–423. doi: 10.1097/00005537-200103000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]


