Abstract
Objective
The aim of this study was to investigate the type and the nature of peptides present in the in vivo formed human acquired enamel pellicle.
Design
Pellicle material was collected from 10 volunteers and subjected to sample preparations consisting of centrifugal filtration using a 10kD molecular weight cut-off membrane and high-resolution gel filtration chromatography. The fractions containing peptides < 10kD obtained by both methods were analyzed by LC-ESI-MS/MS.
Results
78 natural pellicle peptides with molecular weights ranging from 766.9 to 3981.4 Da were identified originating from 29 different proteins.
Conclusion
The number of peptides present in acquired enamel pellicle appears to be large and this is likely to enhance the functional spectrum of this protein film. The presence of small peptides in pellicle may be functionally important since structure/function studies of many salivary proteins have shown that specific domains within these native proteins retain or even exhibit enhanced biological activities. The data present the basis for determining the precise function of these pellicle peptides and for gaining insights into the role pellicle plays in the oral cavity.
Introduction
The acquired enamel pellicle is well known to be a biologically important tooth integument since it forms the interface between the enamel surface and the first layer of oral biofilm. At a functional level it is recognized that it plays a role in the mineral homeostasis of the tooth enamel (1–3). There is ample evidence that this structure is formed by the selective adsorption of proteins, peptides and other molecules present in oral fluid (4, 5). Despite the eminent importance of the acquired enamel pellicle (AEP) in oral physiology and pathological processes such as dental caries and periodontal disease insights into the molecular structure of this protein film have been difficult to obtain. The major obstacles were related to the fact that only minute quantities of in vivo formed pellicle can be harvested from tooth surfaces and this prevented the characterization of this protein film with classical biochemical technologies (6). Two recent developments have made it feasible to overcome these challenges. The first development is related to improvement of methods to harvest AEP from the tooth surface in vivo and the second development is related to the virtual explosion of new mass spectrometric techniques which allow characterization of peptides down to the femtomole level (2, 5–9).
In our first phase of investigating the in vivo human AEP proteome, we used in-gel trypsinization followed by liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). With this approach we identified 130 different proteins in the acquired enamel pellicle based on the presence of at least 2 different peptides belonging to the same protein (5). The enzymatic fragmentation approach prior to mass spectrometric (MS) analysis is essential given the size limitation of proteins/peptides amenable to full characterization. This is a disadvantage since it limits the precise information regarding their actual size and does therefore not allow to determine the amino- and carboxyl-terminal ends of the polypeptides present in the in vivo AEP. Based on previous 2-D PAGE results with in vivo pellicle material (7) and in situ formed pellicles (10) we hypothesize that small molecular weight peptides may constitute an important fraction of the in vivo formed AEP. In the current study of pellicle characterization, we omitted any fragmentation method prior to proteomic analyses in order to gain insight into the structure of protein/peptides present in the acquired enamel pellicle.
Materials & Methods
Human subjects
AEP was obtained from ten healthy male and female volunteers (4 male and 6 female), ranging in age from 24 to 40 years. The subjects exhibited neither gingivitis, periodontal disease, active dental caries, nor any other oral condition that could affect oral fluid composition. AEP collection protocols were approved by the Institutional Review Board of Boston University Medical Center, and informed consent was obtained from each subject participating in the study.
AEP collection
The procedure used for in vivo AEP collection was carried out as described previously (5). Briefly, each donor was subjected to a dental prophylaxis treatment employing coarse pumice containing no additives (Preppies, Whip Mix, Louisville, KY). AEP was then allowed to form on the enamel surfaces over a 2 h period. During this time span, the participants were asked to refrain from any consumption of food or beverages, other than water. After 2 h, teeth from each quadrant were isolated with cotton rolls, washed with water using the dental unit’s built-in spray gun, and dried by air.
For the actual removal of AEP material from the enamel surface collection strips of 0.5 × 1.0 cm (electrode wick filter paper, Bio-Rad, Hercules, CA) was folded so that one half could be held using a dental forceps (Hu-Friedy, Chicago, IL) and the other half could be brought in contact with the tooth surface. To avoid any contamination emanating from the gingival margin, only the coronal two thirds of the labial/buccal surfaces were swabbed using one collection strip per quadrant starting with the buccal area of the central incisor and ending with the buccal surface of the first molar. A total of four collection strips from each participant obtained per collection were placed into a polypropylene microcentrifuge tube and kept frozen at −20°C until used. A second collection was carried out on a separate day using the same ten subjects. For the second collection, the harvesting procedure was slightly modified by using collection strips pre-soaked in 3 % citric acid. This second procedure was carried out to promote a potentially more robust removal of pellicle from tooth surfaces.
AEP Sample Preparation
To recover pellicle proteins from collection strips, 0.5 ml of distilled water was added to each tube and extraction of pellicle was carried out by vortexing the sample for 30 s followed by sonication (Branson Cleaning Equipment Co., Shelton, CT) for 5 min in an ice bath at 4 °C (7). This procedure was repeated five times for the sample collected from each subject and the five extraction aliquots obtained from each subject were pooled to yield a total volume of approximately of 25 ml. Following concentration by speed vac to a volume of 0.5 ml, the total protein concentration was measured by the bicinchoninic acid (BCA) assay (Pierce Chemical, Co., Rockford, IL, USA) using bovine serum albumin as a protein standard.
The 0.5 ml of AEP pooled material was divided in two halves. The first half was subjected to sample preparations consisting of centrifugal filtration using a 10kD molecular weight cut-off (MWCO) membrane (Millipore, Billerica, MA) and the second half was subjected to high-resolution gel filtration chromatography (Smart System, GE Healthcare, Piscataway, NJ). The fractions containing peptides < 10 kD obtained by either method were analyzed by LC-ESI-MS/MS. For the centrifugal filtration method the sample was centrifuged for 30 min at 15,000 × g using a refrigerated Eppendorf table top centrifuge (Eppendorf, Waltham, MA) and the filtrate containing the proteins/peptides with molecular weights below 10 kD was collected. Identical procedures were employed for AEP sample preparation of pellicle collected using 3 % citric acid. Both collection techniques yielded very similar results (data not shown). The two pools resulting from centrifugal filtration were dried and subjected to MS analysis. The second half of the 0.5 ml of AEP pooled material (0.25ml) was dried by speed vac, dissolved in 100 μL of 0.05 M phosphate, 0.15 M NaCl, pH 7.0 and subjected to fractionation by high-resolution gel filtration chromatography. For this purpose, the sample was applied to a 3.2 mm × 30 mm Sephadex column (Superdex 75 PC 3.2/30, Smart System, GE Healthcare, Piscataway, NJ) which had been equilibrated with 0.05 M phosphate, 0.15 M NaCl, pH 7.0 at a flow rate of 50 μL/min. Proteins and peptides were separated over a time period of 85 min and the eluant was monitored by absorbance at 219 nm. Seventeen consecutive 5 min fractions (250 μL) were collected and the fraction containing proteins and peptides with molecular weights below 10 kD were dried and subjected to MS analysis. Identical procedures were used for the AEP proteins and peptides collected using 3% citric acid and again no differences between the collection methods could be ascertained.
Tandem Mass Spectrometry
Mass spectrometric analyses were carried out with a LTQ-linear-ion-trap (Thermo-Finnigan, San Jose, CA) which allows in-line liquid chromatography with a capillary C18 column linked to the mass spectrometer using electrospray ionization allowing survey scans in the range of 400–2000 m/z values and concomitant tandem MS/MS analyses.
All samples whether obtained by centrifugation or gel filtration chromatography were dried by speed vac and resuspended in 100 μL of 0.1 % trifluoroacetic acid and desalted using a Spin Columnc18 (The Nest Group, Inc. Southborough, MA, USA). Adsorbed proteins/peptides were eluted with a buffer containing 80% acetronitrile/19.9% water and 0.1% TFA, dried, and resuspended in 25 μL of 97.5 % H2O/2.4% acetonitrile/0.1% formic acid and then subjected to reversed-phase LC-ESI- MS/MS. The nano-flow reversed-phase HPLC capillary column, 50 μm × 10 cm (Pico Tip™ EMITTER, New Objective, Woburn, MA) was packed in-house using Magic C18 resin of 5 μm diameter and 200 Å pore size (Michrom BioResources, Auburn, CA). The column was developed with a linear 40 min gradient ranging from 5% to 50% of solvent B (97.5% acetonitrile, 0.1% formic acid) at a flow rate of 110 nL/min. Electrospray voltage and the temperature of the ion transfer capillary were 1.8 kV and 230 °C respectively.
Data analysis
The obtained MS/MS spectra were searched against human protein databases (Swiss Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://expasy.org/sprot) using SEQUEST (Bioworks Browser 3.3.1, Thermo-Finnigan, San Jose, CA). Searches were performed by selecting the following SEQUEST parameters: 1) No specific fragmentation, 2) Delta CN ≥ 0.1, 3) Peptide probability ≤ 0.5, and 4) XCorr score ≥ 2.0 and 2.6 for Z = 2 and 3, respectively. An additional inclusion criterion for positive identification and characterization of proteins/peptides was that the same AEP constituent had to be found in at least two of the four mass spectrometry analyses.
Results
Our first goal was to characterize naturally occurring pellicle proteins/peptides without any fragmentation prior to analysis by LC-ESI-MS/MS. Since mass spectrometry instrumentation limits the characterization of proteins/peptides to those being higher than 5000 Da, proteins/peptides exhibiting molecular sizes smaller than 10 kD were separated from the protein fraction with molecular sizes larger than 10 kD. This molecular cut-off was selected to assure that no components smaller than 5 kD were omitted from analyses.
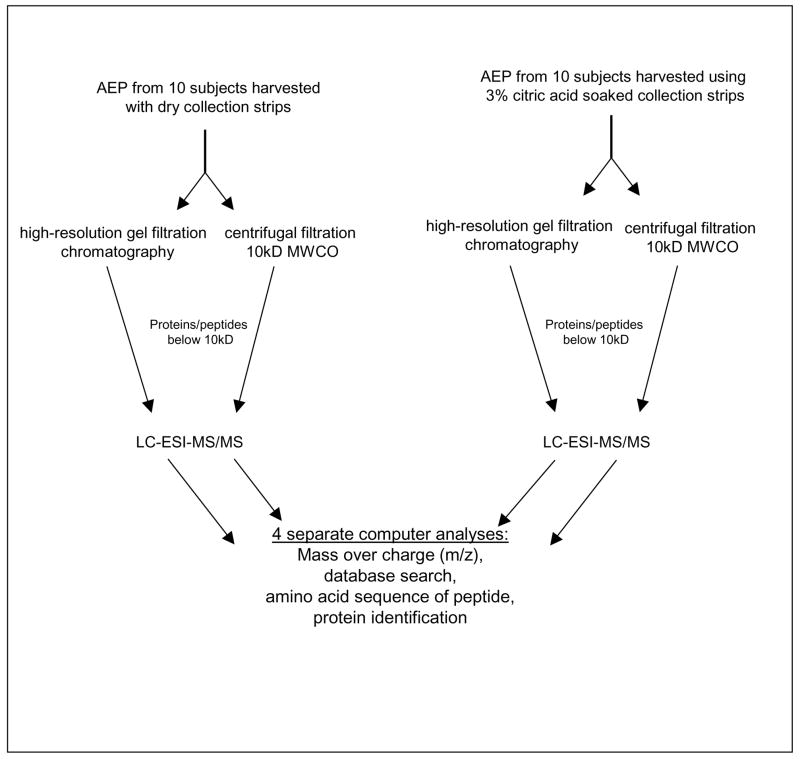
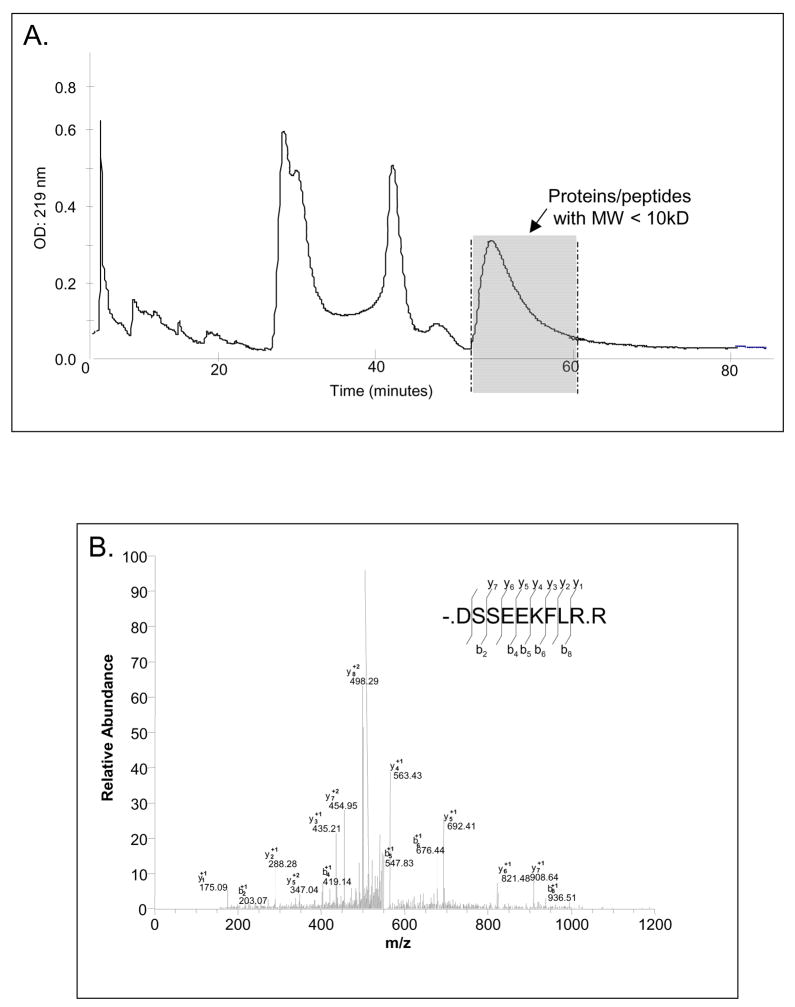
The two separation methods used were centrifugal filtration and high-resolution gel filtration chromatography and the principal steps involved for each procedure are depicted in scheme 1. The results obtained by gel filtration chromatography are shown in Figure 1. The eluant monitored at 219 nm indicates three major peaks representing 150 kD, 50 kD, and 10 kD molecular size pellicle constituents. The fractions containing proteins/peptides below 10 kD were pooled and subjected to mass spectrometric analysis. It can be seen readily that this fraction represents approximately 50% of the total AEP material. The second method to obtain small molecular weight AEP proteins/peptides was centrifugal filtration with a low protein binding membrane with a MWCO of 10 kD. The total filtrate was subjected to mass spectrometric analysis.
Scheme 1.
Summary of the AEP sample preparation for mass spectrometric analysis.
Figure 1.
(A) High resolution gel filtration chromatogram of AEP proteins and peptides. The shaded peak area of the chromatogram indicates the zone of elution of proteins/peptides with molecular weights below 10 kD. These fractions were pooled and analyzed by LC-ESI-MS/MS. (B) MS/MS spectrum and sequence analysis of a typical AEP peptide. The amino acid sequence of this peptide was -.DSSEEKLR.R which led to the identification of statherin as the parent protein (P02808). Matching b- and y- ion series are indicated in the upper right.
The LC-ESI-MS/MS analysis of AEP proteins/peptides obtained by both separation techniques resulted in the identification of 78 peptides, which originated from 29 different proteins ranging in molecular weight between 767 Da to 3981 Da (Table 1). An example of how the tandem MS results were obtained is shown with one of the AEP peptides in Figure 1B. This peptide has a sequence of nine amino acid residues that represents the N-terminal moiety of statherin. All peptides reported in Table 1 were characterized in this fashion. Table 1 is a comprehensive list of the AEP constituents found in this study. They are listed according to the number of peptides identified per protein. Column 1 lists the accession number for each protein identified, column 2 the name of the protein of origin, column 3 the structure of the natural peptide characterized, column 4 the molecular weight (MW), column 5 the isoeletric point (pI), column 6 the peptide region and column 7 the number of times that each peptide was identified by mass spectrometry. Of the 78 identified peptides, 17 were identified twice among the 4 mass spectrometric analysis, 39 were identified three times, and 22 in all four analyses.
Table 1.
List of the 78 peptides identified in the in vivo formed Human Acquired Enamel Pellicle.
| Accession number1 |
Protein name | Peptide identified (sequence domain within parent protein) | MW (Da) |
pI | Protein region | Times identified |
|---|---|---|---|---|---|---|
| P02810 | Salivary acidic proline-rich phosphoprotein 1/2 | F.IDEERQGPPLG.G (42–52)a,b | 1211.3 | 3.8 | N-terminal/Middle | 2 |
| F.IDEERQGPPLGGQQ.S ▪ (42–55)a,b | 1524.6 | 3.8 | N-terminal/Middle | 3 | ||
| F.IDEERQGPPLGGQQSQPS.A (42–56)b | 1924.0 | 3.8 | N-terminal/Middle | 2 | ||
| G.GQQQQGPPPPQGKPQ.G (78–92)a,b | 1572.7 | 10.1 | Middle | 3 | ||
| G.PQQGPPQQGGQQQQGPPPPQG.K (53–73)a,b | 2137.3 | 6.0 | Middle | 3 | ||
| K.PQGPPPQGGRPQGPPQ.G (130–145)a,b | 1595.7 | 11.0 | C-terminal | 3 | ||
| N.QDDGPQQGPPQQGGQQQQGPPPPQGKP.Q(49–75)a,b | 2777.9 | 3.9 | N-terminal/Middle | 4 | ||
| P.PPGKPQGPPPQGGR.P(126–139)a,b | 1370.5 | 11.5 | C-terminal | 4 | ||
| Q.DDGPQQGPPQQGGQQQQGPPPPQGKPQ.G(50–76)a,b | 2777.9 | 3.9 | N-terminal/Middle | 3 | ||
| Q.GGRPQGPPQGQSPQ.-(137–150)a,b | 1391.5 | 11.0 | C-terminal | 4 | ||
| Q.GPPPPPPGKPQGPPPQ.G(121–135)a,b | 1545.8 | 10.1 | C-terminal | 3 | ||
| Q.GPPPPPPGKPQGPPPQGGRPQGPPQGQSPQ.-(121–150)b | 2918.2 | 11.5 | C-terminal | 2 | ||
| Q.GPPQQGGHQQGPPPPPPGKPQ.G (111–131)▪•a,b | 2084.3 | 10.5 | C-terminal | 4 | ||
| P02812 | Basic salivary proline-rich protein 2 | K. PQGPPPQGGNKPQGPPPPGK.P(23–43)a,b | 2810.1 | 10.6 | N-terminal | 3 |
| G.GNQPQGPPPPPGKPQ.G(278–292)a,b | 1496.7 | 10.1 | C-terminal | 3 | ||
| K.PQGPPPQGGNKPQGPPPPGK.P(167–186)a,b | 1933.2 | 10.6 | C-terminal | 3 | ||
| K.PQGPPPQGGNKPQGPPPPGKPQGPPPQGDK.S(167–196)a | 2935.2 | 10.3 | N-terminal | 2 | ||
| K.PQGPPPQGGNQPQGPPPPPGKPQ.G(84–106)a,b | 2255.5 | 10.1 | N-terminal | 3 | ||
| P.GKPQGPPPQGGNQPQGPPPPPGKPQG.P*(227–251)b | 2497.8 | 10.6 | C-terminal | 2 | ||
| Q.GPPPQGGNKPQGPPPPGKPQ.G(107–126)a,b | 1922.2 | 10.6 | N-terminal | 3 | ||
| Q.GPPPQGGNQPQGPPPPPGKPQ.G(147–168)a,b | 2030.2 | 10.1 | C-terminal | 4 | ||
| Q.GPPPQGGNQPQGPPPPPGKPQGPPPQGGNKPQ.G(147–179)a,b | 3088.4 | 10.6 | C-terminal | 3 | ||
| P02808 | Statherin | F.GYGYGPYQPVPE.Q ▪ (15–26)a,b | 1326.4 | 3.3 | Middle | 3 |
| -.DSSEEKFLR.R (1–9)a,b | 1110.2 | 4.4 | N-terminal | 2 | ||
| R.IGRFGYGYGPYQPVPEQP.L (11–28)a,b | 2025.2 | 6.8 | N-terminal/Middle | 3 | ||
| R.RIGRFGYGYGPYQPVPEQPLYPQPYQPQYQQYT.F(11–42)a,b | 3981.4 | 9.1 | N-terminal/Middle/C-terminal | 3 | ||
| Y.GPYQPVPEQPLYPQPYQPQ.Y▪ (19–37)a,b | 2226.4 | 3.3 | Middle/C-terminal | 4 | ||
| P04745 | alpha-amylase A1(salivary) | R.SGNEDEFR.N (88–95)a,b | 953.9 | 3.8 | N-terminal | 4 |
| R.ALVFVDNHDNQR.G (307–318)b | 1428.5 | 5.1 | Middle | 2 | ||
| F.IYQEVIDLGGEPIK.S (245–258)a,b | 1574.8 | 3.8 | Middle | 3 | ||
| K.AHFSISNSAEDPFIAI.H(490–505)a,b | 1719.9 | 4.1 | C-terminal | 4 | ||
| K.TGSGDIENYNDATQVR.D(158–173)a,b | 1740.8 | 3.7 | N-terminal | 3 | ||
| Q9hcy8 | S100 calcium binding protein a14 | K.NFHQYSVEGG.K(28–37)a,b | 1137.1 | 5.1 | N-terminal | 3 |
| S.FWELIGEAAK.S (84–93)a,b | 1163.3 | 4.3 | C-terminal | 3 | ||
| F.RSFWELIGEAAKSVKLE.R(82–98)a,b | 1963.2 | 7.2 | C-terminal | 2 | ||
| A.QEFSDVERAIETLI.K(13–26)a,b | 1649.8 | 3.7 | N-terminal | 4 | ||
| K.IANLGSCNDSKL.E(68–78)a,b | 1234.3 | 6.1 | Middle/C-terminal | 3 | ||
| P04083 | Annexin A1 | F.IENEEQEYVQTVK.S (14–26)a,b | 1608.7 | 3.8 | N-terminal | 4 |
| K.TPAQFDADELR.A(114–124)a,b | 1262.3 | 3.7 | Middle | 3 | ||
| W.FIENEEQEYVQTVK.S (13–26)a,b | 1755.9 | 3.8 | N-terminal | 3 | ||
| Q9UBG3 | Corunlin | R.TEGNCTALTRGE.L (21–32)a,b | 1251.3 | 4.3 | N-terminal | 4 |
| E.GNCTALTRGELKR.L(23–35)a,b | 1418.6 | 10.1 | N-terminal | 3 | ||
| R.EQGQTQTQPGS.G(373–383)a,b | 1160.1 | 12.4 | C-terminal | 4 | ||
| P01040 | Cystatin a | E.KTNETYGKLE.A (30–39)a,b | 1182.3 | 7.0 | Middle | 4 |
| E.TYGKLEAVQY.K (34–43)a,b | 1171.3 | 6.8 | N-terminal/Middle | 2 | ||
| K.SLPGQNEDLVLTG.Y (72–84)a,b | 1342.4 | 3.0 | C-terminal | 3 | ||
| P04280 | Basic salivary proline-rich protein 1 | A.GNPQGPSPQGGNKPQ.G(18–32)▪a,b | 1462.5 | 10.1 | N-terminal | 3 |
| Q.GPPQQGGNRPQGPPPPGKPQ.G(226–245)▪b | 1991.2 | 11.5 | Middle/C-terminal | 3 | ||
| A.GNPQGPSPQGGNKPQGPPPPPGKPQ.G(18–48)▪b | 2415.6 | 10.6 | N-terminal | 4 | ||
| P08779 | Cytokeratin-16 | R.EVFTSSSSSSSRQ.T (442–454)a,b | 1388.4 | 7.0 | C-terminal | 3 |
| P15515 | Histatin-1 | F.YGDYGSNYLYDN.-▪(27–38)a,b | 1443.4 | 2.9 | Middle/C-terminal | 3 |
| Y.GDYGSNYLYDN.-▪• (28–38)a,b | 1280.2 | 2.9 | C-terminal | 4 | ||
| Q96QV6 | Histone H2A type1-A | I.AQGGVLPNIQAV.L (104–115)b | 1166.3 | 6.0 | C-terminal | 2 |
| V.TIAQGGVLPNIQAV.L (102–115)a,b | 1380.6 | 6.0 | C-terminal | 4 | ||
| Q9HC84 | Mucin-5B | L.SSPSPAPGCDNAIP.L (4940–4953)a,b | 1312.4 | 3.1 | C-terminal | 2 |
| R.AQAQPGVP.L (2362–2369)a | 766.9 | 6.0 | C-terminal | 2 | ||
| P62736 | Actin, aortic smooth muscle | Y.VGDEAQSKRGILTL.K (56–69)a,b | 1487.7 | 7.0 | N-terminal | 3 |
| T.AASSSSLEKSYELPDGQVI.T (232–250)a | 1982.1 | 3.8 | N-terminal | 2 | ||
| P06733 | Alpha-enolase | A.NGWGVMVSH.R (363–371)a,b | 986.1 | 7.8 | C-terminal | 4 |
| R.SERLAKYNQLLRIEEE.L(401–416)a,b | 1991.2 | 4.7 | C-terminal | 3 | ||
| P06702 | Protein S100-A9 | Y.SVKLGHPDTLNQGEFKEL.V (23–40)a,b | 2013.2 | 5.3 | N-terminal/Middle | 3 |
| V.KLGHPDTLNQGEF.K (25–37)a,b | 1456.6 | 5.2 | N-terminal/Middle | 3 | ||
| Q01546 | Cytokeratin-2P | R.GVFGGVSGSGSGGYK.G (524–538)a,b | 1316.4 | 9.7 | C-terminal | 4 |
| K.SGGGGSTSIRFSQTTSSSQHSSTK.- (615–638)a,b | 2373.4 | 11.5 | C-terminal | 4 | ||
| P02768 | Serum albumin | K.PLVEEPQNLIK.Q (403–413)a,b | 1279.5 | 4.3 | Middle/C-terminal | 3 |
| K.AVMDDFAAFVEK.C (570–581)b | 1342.5 | 3.7 | C-terminal | 2 | ||
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | A.SEGPLKGILGY.T (266–276)a,b | 1133.3 | 6.9 | C-terminal | 4 |
| V.SSDFNSDTHSSTF.D(283–295)a,b | 1431.3 | 3.9 | C-terminal | 3 | ||
| P02647 | Apolipoprotein A-I | L.REQLGPVTQEF.W (85–95)a,b | 1304.4 | 4.3 | N-terminal/Middle | 3 |
| P02788 | Lactotransferrin | R.ESTVFEDLSDEAER.D (230–243)a,b | 1626.6 | 3.4 | N-terminal | 3 |
| P20671 | Histone H2A type 1-D | I.RNDEELNKLLGKVTIA.Q(89–104)b | 1814.1 | 7.1 | Middle/C-terminal | 2 |
| P01833 | Polymeric- immunoglobulin receptor | R.ASVDSGSSEEQGGSSRAL.V(623–640)a,b | 1724.7 | 3.8 | C-terminal | 4 |
| P31151 | S100 calcium- binding protein A7 | Y.HKQSHGAAPCSGGS.Q (87–100)a,b | 1323.4 | 9.0 | C-terminal | 3 |
| P47929 | galectin-7 | V.GGDVQLDSVRIF.- (125–136)b | 1305.5 | 3.9 | C-terminal | 2 |
| P30044 | peroxiredoxin-5 | M.APIKVGDAIPAVEV.F (54–67)a | 1378.6 | 4.1 | N-terminal/Middle | 2 |
| P04080 | cystatin B | Y. QTNKAKHDELTYF.- (86–98)a,b | 1594.7 | 7.7 | C-terminal | 3 |
| P01036 | Cystatin-S | Y.EVPWEDRMSLVN.S (124–135)a,b | 1474.7 | 3.8 | C-terminal | 3 |
| P05164 | Myeloperoxidase | R.DFVNCSTLPALNLASW.R (726–741)a,b | 1751.1 | 3.1 | C-terminal | 4 |
Accession numbers in bold represent AEP proteins identified earlier by our group using in-gel trypsinization followed by LC-ESI-MS/MS (Siqueira et al., 2007).
Peptides identified by Vitorino et al., 2008 in in situ formed AEP.
Peptides identified by Helmerhorst et al., 2008 in human whole saliva.
Peptides in samples collected with dry collection strips.
Peptides which in samples collected with 3% citric acid-soaked collection strips.
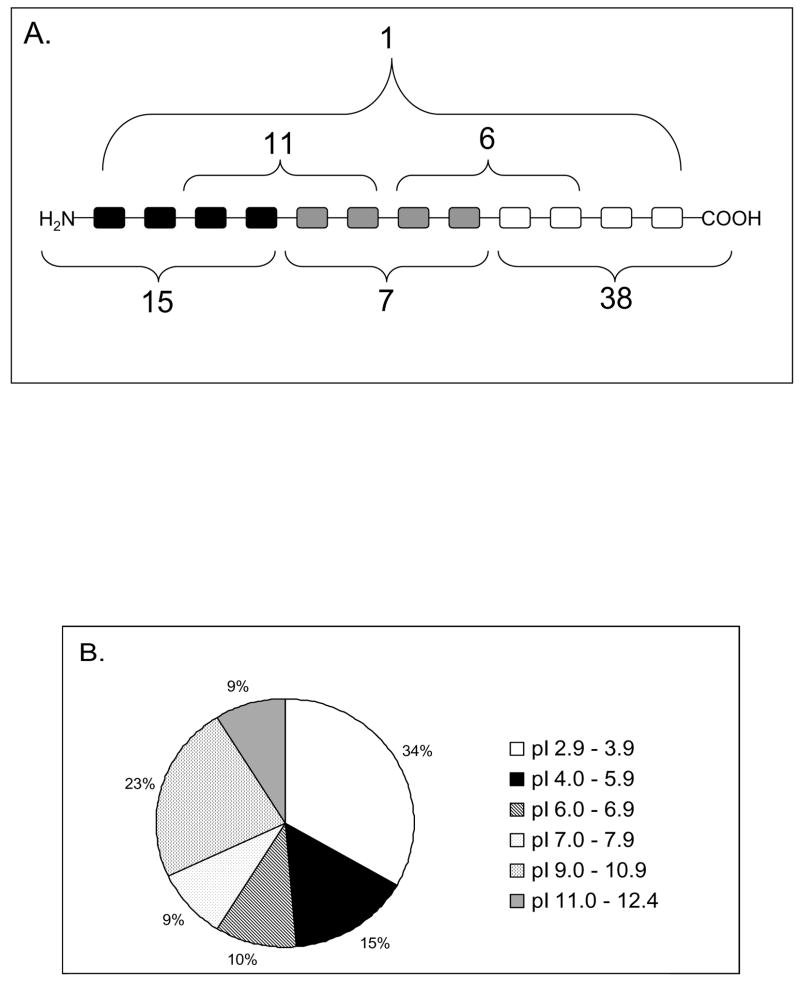
To see whether the pellicle peptides showed a preferential location within the full primary structure of the parent molecules its polypeptide structure was arbitrarily divided into 3 equal parts. Each of the pellicle peptides was matched against the primary structure of the parent protein and the results of this are shown in Figure 2A. The most frequently encountered domain of origin was the C-terminal region (48%) followed by N-terminal region (19.2%) and the least frequent origin was the middle region (9%). Peptides derived from overlap regions comprising N-terminal and middle domains accounted for 14% and those overlapping the middle and C-terminal region accounted for 7.6% (Figure 2A).
Figure 2.
(A) Localization of each AEP peptide in the primary structure of the parent protein. (B) Distribution of the AEP peptides according to their isoeletric points (pI).
The pellicle peptides identified were also grouped according to their isoeletric points (Figure 2B). Clearly 50% of all these peptides had isoeletric points bellow 5.9 and therefore exhibit a negative charge at a pH range between 6.8 to 7.2, the pH conditions which prevail in the oral cavity (11). Only one-third of the small molecular weight components exhibit basic characteristics ranging in pI between 9.8 to 12.4.
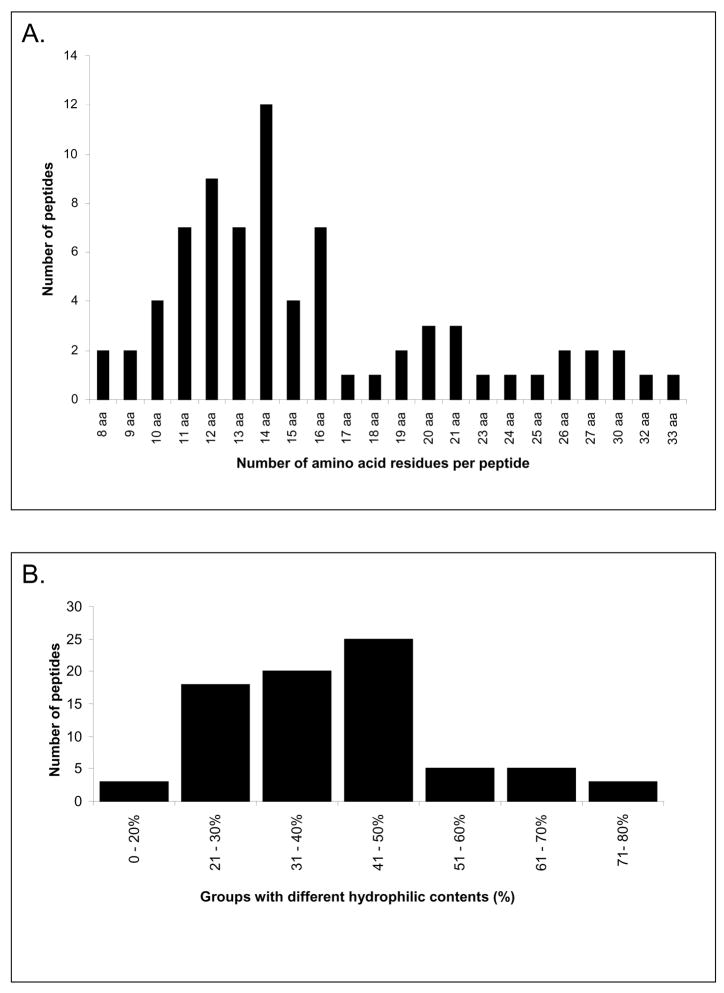
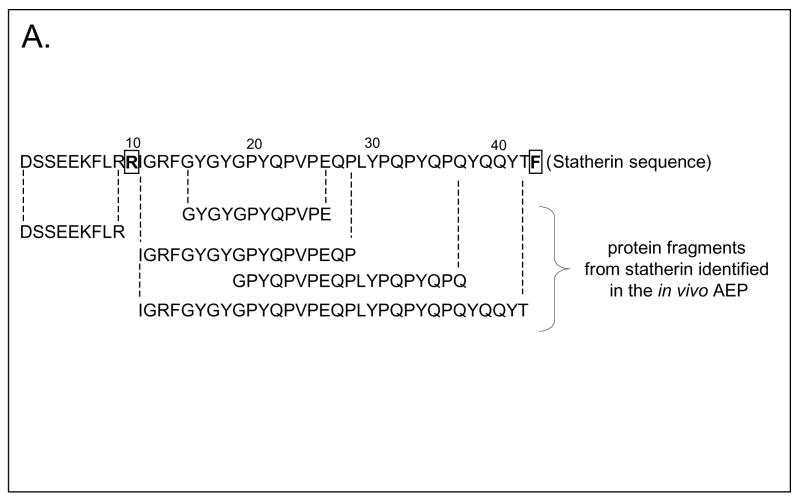
The frequency of occurrence of the AEP peptides according to their number of amino acid residues is shown in Figure 3A. Overall, peptides containing 8 to 33 amino acid residues were identified with a predominant occurrence of peptides comprising 10 to 16 amino acid residues and a calculated molecular size range of 1137 to 1814 Da. With respect to hydrophilicity, the majority of the AEP peptides characterized contained 20 to 50% of hydrophilic residues indicating that the bulk of the peptides identified contained a higher proportion of hydrophobic to hydrophilic residues (Figure 3B). In general, the pellicle peptidome as determined in this study provided no evidence for the presence of intact salivary proteins in the molecular size range investigated. The data obtained with respect to statherin fragments are shown in Figure 4. These peptides represent almost a full coverage of statherin since only arginine (residue 10) and phenylalanine (residue 43) were not accounted for.
Figure 3.
(A) Size distribution of AEP peptides according to their number of amino acid residues per peptide. (B) Grouping of pellicle peptides according to their percentage of hydrophilic amino acid residues.
Figure 4.
Amino acid sequence of statherin (P02808). The peptide fragments identified by LC-ESI-MS/MS are aligned below the protein sequence. Note the almost full coverage of statherin obtained with the exception of Arg10 and Phe43 (boxed).
Discussion
Among the 78 peptides characterized as pellicle constituents 61 were identified in at least 3 of the 4 analyses. This is an amazingly consistent result considering that these data were derived from 4 separate clinical collections and in addition it is well known that multiple repeat experiments of the same sample with tandem MS show a relatively high variability (12–14). The data obtained instill therefore considerable confidence that the components identified are true pellicle constituents with defined primary structures.
Since the size limitations for a direct characterization by MS/MS is an obstacle not easily overcome, the restriction to the analysis of the less than 5000 Da molecules provided a more realistic insight into the composition of at least small pellicle components than endeavors into the full spectrum of pellicle proteins dependent on tryptic fragmentation prior to MS analysis. Despite the limitations in size of the pellicle protein/peptides studied these small pellicle constituents did represent a very significant portion of the pellicle proteome.
The characterization of the pellicle fraction studied here revealed some new insights into their salient properties. E.g. most pellicle precursor proteins have previously been shown to carry a net negative charge and show functional domains to be localized in the N-terminus. These characteristics have been recognized widely but were based mostly on in vitro pellicle studies (15–17). The novel and different findings with in vivo formed pellicle reported here are in part contradicting the classical concepts. The first discrepancy relates to the fact that 48% of the peptides identified originated from the C-terminal domains of pellicle precursor proteins. Nevertheless 50% of these peptides carry a net negative charge at neutral pH. This fraction may exhibit higher affinity to hydroxyapatite based on charge interactions than the more neutral or even basic components derived from C-terminal protein moieties. The second surprise relates to the large fraction of hydrophobic peptides which were encountered indicating that ionic interactions with hydroxyapatite are not the only driving force for pellicle formation. Support for hydrophobic forces being possibly critical for the adsorption process derives from thermodynamic work with single proteins and mineral interactions (18). In such cases the exclusion of ordered water concomitant with an increase in entropy represent possible driving forces for hydroxyapatite adsorption of protein and peptides. It is well recognized that the AEP peptides characterized here constitute a mixture of components directly adsorbing to hydroxyapatite and components interacting with other pellicle constitutuents. The characteristics of hydrophobicity and net charge are equally important for peptide-peptide interactions leading to poly-disperse structures typical for the protein film formed on the tooth surface.
In our previous pellicle study (5) we identified 130 proteins based on at 2 different peptides derived from the same protein using tryptic fragmentation prior to MS analysis. Of these 130 proteins, 21 proteins were also identified in the present study where we focused only on the unmodified small molecular weight fraction. This supports the notion that at least 21 of the previously identified pellicle components are unlikely to represent intact primary structures of precursor proteins. Proteolytic fragmentation before or after the adsorption process is vital for truncation whether from the N-terminal, C-terminal or both polypeptide ends. Such oral fluid related proteolysis has been shown to be extensive and important in the fragmentation of salivary proteins (19, 20). The selectivity of the well defined protein fragments in pellicle characterized by direct LC-ESI-MS/MS shows that pellicle formation underlies strongly controlled biological processes. Earlier studies on in situ formed pellicles have indicated similar selectivities among the peptides incorporated into the protein film formed on the enamel surfaces. (10,21). The present study represents the first detailed characterization of small molecular components present in the in vivo formed human acquired enamel pellicle. These data provide a basis for expanding into more detailed investigations into the structure-function relationships of pellicle peptides. Encouragement for such studies derives from the fact that many salivary proteins exhibit specific functional domains which not only retain but in some cases augment the activities of the native protein (20,22,23).
Acknowledgments
NIH/NIDCR grants DE05672, DE07652, and DE17788.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lendenmann U, Grogan J, Oppenheim FG. Saliva and dental pellicle--a review. Adv Dent Res. 2000;14:22–28. doi: 10.1177/08959374000140010301. [DOI] [PubMed] [Google Scholar]
- 2.Siqueira WL, Helmerhorst EJ, Zhang W, Salih E, Oppenheim FG. Acquired enamel pellicle and its potential role in oral diagnostics. Ann N Y Acad Sci. 2007;1098:504–509. doi: 10.1196/annals.1384.023. [DOI] [PubMed] [Google Scholar]
- 3.Hannig M, Joiner A. The structure, function and properties of the acquired pellicle. Monogr Oral Sci. 2006;19:29–64. doi: 10.1159/000090585. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hashimi I, Levine MJ. Characterization of in vivo salivary-derived enamel pellicle. Arch Oral Biol. 1989;34:289–295. doi: 10.1016/0003-9969(89)90070-8. [DOI] [PubMed] [Google Scholar]
- 5.Siqueira WL, Zhang W, Helmerhorst EJ, Gygi SP, Oppenheim FG. Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res. 2007;6:2152–2160. doi: 10.1021/pr060580k. [DOI] [PubMed] [Google Scholar]
- 6.Yao Y, Grogan J, Zehnder M, Lendenmann U, Nam B, Wu Z, et al. Compositional analysis of human acquired enamel pellicle by mass spectrometry. Arch Oral Biol. 2001;46:293–303. doi: 10.1016/s0003-9969(00)00134-5. [DOI] [PubMed] [Google Scholar]
- 7.Yao Y, Berg EA, Costello CE, Troxler RF, Oppenheim FG. Identification of protein components in human acquired enamel pellicle and whole saliva using novel proteomics approaches. J Biol Chem. 2003;278:5300–5308. doi: 10.1074/jbc.M206333200. [DOI] [PubMed] [Google Scholar]
- 8.Vitorino R, de Morais Guedes S, Ferreira R, Lobo MJ, Duarte J, Ferrer-Correia AJ, et al. Two-dimensional electrophoresis study of in vitro pellicle formation and dental caries susceptibility. Eur J Oral Sci. 2006;114:147–153. doi: 10.1111/j.1600-0722.2006.00328.x. [DOI] [PubMed] [Google Scholar]
- 9.Vitorino R, Calheiros-Lobo MJ, Williams J, Ferrer-Correia AJ, Tomer KB, Duarte JA, et al. Peptidomic analysis of human acquired enamel pellicle. Biomed Chromatogr. 2007;21:1107–1117. doi: 10.1002/bmc.830. [DOI] [PubMed] [Google Scholar]
- 10.Vitorino R, Calheiros-Lobo MJ, Duarte JA, Domingues PM, Amado FM. Peptide profile of human acquired enamel pellicle using MALDI tandem MS. J Sep Sci. 2008;31:523–37. doi: 10.1002/jssc.200700486. [DOI] [PubMed] [Google Scholar]
- 11.Dawes C. What is the critical pH and why does a tooth dissolve in acid? J Can Dent Assoc. 2003;69:722–724. [PubMed] [Google Scholar]
- 12.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 13.Hu J, Coombes KR, Morris JS, Baggerly KA. The importance of experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief Funct Genomic Proteomic. 2005;3:322–331. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- 14.Prakash A, Piening B, Whiteaker J, Zhang H, Shaffer SA, Martin D, et al. Assessing bias in experiment design for large scale mass spectrometry-based quantitative proteomics. Mol Cell Proteomics. 2007;6:1741–1748. doi: 10.1074/mcp.M600470-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Yin A, Margolis HC, Grogan J, Yao Y, Troxler RF, Oppenheim FG. Physical parameters of hydroxyapatite adsorption and effect on candidacidal activity of histatins. Arch Oral Biol. 2003;48:361–368. doi: 10.1016/s0003-9969(03)00012-8. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons RJ, Hay DI. Adsorbed salivary acidic proline-rich proteins contribute to the adhesion of Streptococcus mutans JBP to apatitic surfaces. J Dent Res. 1989;68:1303–1307. doi: 10.1177/00220345890680090201. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz SS, Hay DI, Schluckebier SK. Inhibition of calcium phosphate precipitation by human salivary statherin: structure-activity relationships. Calcif Tissue Int. 1992;50:511–517. doi: 10.1007/BF00582164. [DOI] [PubMed] [Google Scholar]
- 18.Moreno EC, Kresak M, Hay DI. Adsorption thermodynamics of acidic proline-rich human salivary proteins onto calcium apatites. J Biol Chem. 1982;257:2981–2989. [PubMed] [Google Scholar]
- 19.Helmerhorst EJ, Sun X, Salih E, Oppenheim FG. Identification of Lys-Pro-Gln as a novel cleavage site specificity of saliva-associated proteases. J Biol Chem. 2008;283:19957–19966. doi: 10.1074/jbc.M708282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helmerhorst EJ, Alagl AS, Siqueira WL, Oppenheim FG. Oral fluid proteolytic effects on histatin 5 structure and function. Arch Oral Biol. 2006;51:1061–1070. doi: 10.1016/j.archoralbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Hannig C, Spitzmüller B, Miller M, Hellwig E, Hannig M. Intrinsic enzymatic crosslinking and maturation of the in situ pellicle. Arch Oral Biol. 2008;53:416–22. doi: 10.1016/j.archoralbio.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Groot F, Sanders RW, ter Brake O, Nazmi K, Veerman EC, Bolscher JG, et al. Histatin 5-derived peptide with improved fungicidal properties enhances human immunodeficiency virus type 1 replication by promoting viral entry. J Virol. 2006;80:9236–9243. doi: 10.1128/JVI.00796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stallmann HP, Faber C, Bronckers AL, de Blieck-Hogervorst JM, Brouwer CP, Amerongen AV, et al. Histatin and lactoferrin derived peptides: antimicrobial properties and effects on mammalian cells. Peptides. 2005;26:2355–2359. doi: 10.1016/j.peptides.2005.05.014. [DOI] [PubMed] [Google Scholar]