Abstract
Proenkephalin (PE) is a prohormone containing dibasic sites that are cleaved by proteases to generate peptide neurotransmitters and hormones. Little is known about the conformational features of such protease cleavage sites within prohormone substrates. Therefore, the goal of this study was to investigate the relative accessibilities of mulltiple dibasic processing sites of PE by peptide amide hydrogen-deuterium exchange mass spectrometry (DXMS). DXMS demonstrated differences in the relative accessibilities of the KR, KK, and RR cleavage sites of PE to the aqueous environment. DXMS assesses relative rates of exchange of hydrogens of the polypeptide backbone of PE with deuterium atoms from D2O (heavy water) in solvent. After hydrogen-deuterium (H-D) exchange, quenching, and pepsin digestion of PE, LC-MS/MS mass spectrometry provides relative quantification of deuterium incorporated into peptide fragments. Analyses of peptides spanning each of the twelve dibasic PE cleavage sites illustrated differences in H-D exchange rates that reflect relative solvent accessibility. The mid-domain cleavage sites showed higher accessibility to the aqueous solvent compared to particular regions of the NH2- and COOH-domains. The NH2- and COOH-terminal domains both showed relatively high H-D exchange rates. The hydrogen exchange rate profile of PE, as well as its CD (circular dichroism) features for secondary structure, were modified in trifluoroethanol, an organic solvent that represents a more hydrophobic environment. These findings suggest that the dibasic protease cleavage sites of the PE prohormone with differences in accessibility to the aqueous environment undergo proteolytic processing to generate active neuropeptides for cell-cell communication in neuroendocrine systems.
Proenkephalin and prohormones are inactive protein precursors that must undergo proteolytic processing to generate the smaller, active peptide neurotransmitters and hormones. The mature enkephalin neuropeptide functions in brain regulation of analgesia and behavior (1-3). Proenkephalin (PE) contains multiple copies of (Met)enkephalin and related peptides (4,5), which are produced by proteolytic processing of PE at multiple dibasic processing sites (6-9) that flank the enkephalin peptide sequences within PE. The processed, mature enkephalin peptides are stored within secretory vesicles and then undergo regulated secretion induced by cellular stimuli, allowing it to function as an extracellular peptide neurotransmitter.
Proteolysis of proenkephalin in the regulated secretory pathway has been extensively studied and involves the cysteine protease cathepsin L combined with the subtilisin-like prohormone convertases (6-9). The prohormone convertases known as PC1/3 and PC2 represent the primary subtilisin-like proteases (6-9) that cleave at the COOH-terminal side of the dibasic residue prohormone processing sites; their actions are followed by carboxypeptidase E (CPE) that removes COOH-terminal basic residues to generate active enkephalin and related neuropeptides (6,10). The more recently discovered cathepsin L protease pathway for PE processing involves preferential cleavage at the NH2-terminal side of dibasic residue sites (6,11), followed by aminopeptidase B to remove NH2-terminal basic residues (12) in the formation of mature enkephalin. Studies of protease gene knockout mice (6-9, 11, 13-16) have demonstrated the significant roles of secretory vesicle cathepsin L, combined with the well-known prohormone convertases, in the production of enkephalin and numerous neuropeptides functioning as key neurotransmitters and hormones.
The dibasic processing sites of PE are recognized and cleaved by the prohormone processing proteases. However, little is known about the conformational orientation and structural features of proenkephalin and prohormones at their proteolytic processing sites. For this reason, this study sought to gain knowledge of the relative accessibility of the proteolytic processing sites of proenkephalin by hydrogen-deuterium exchange mass spectrometry (DXMS) (17-19). DXMS allows evaluation of the relative rates of exchange of hydrogens of the polypeptide backbone of PE with deuterium of D2O (heavy water), and can compare protein subdomains with respect to their relative accessibility to the aqueous solvent environment. DXMS experiments demonstrated differences in the relative accessibilities of the dibasic KR, KK, and RR cleavage sites to the aqueous environment. The mid-domain processing sites of PE showed higher accessibility to the aqueous solvent compared to other NH2- and COOH-domains of PE. However, the NH2- and COOH-terminal dibasic sites showed relative high DXMS exchange rates. In the presence of the organic solvent trifluoroethanol (TFE), PE displayed differences in DXMS properties, combined with differences in secondary structural features determined by CD (circular dichroism) (20-22). These findings suggest that in more hydrophobic conditions, such as association of PE with membranes in cells, the orientation of the dibasic processing sites to the aqueous environment may differ.
The observed differences in relative accessibility of dibasic sites within PE suggest differential conformational features among these proteolytic processing sites. These findings suggest that dibasic prohormone processing sites with differences in accessibility to the aqueous solvent undergo proteolytic processing to generate mature active enkephalin and related neuropeptides.
Experimental Procedures
Recombinant human proenkephalin (PE): expression and purification
Recombinant human PE with an N-His tag was generated by expression in E. coli, and purified by a Ni2+-affinity column. The pET19b expression vector was used to generate recombinant human PE in Rosetta 2(DE3) E. coli cells (EMD, San Diego, CA). The PE cDNA, with deletion of the signal sequence and incorporation of NdeI and BamHI restriction sites at the 5′ and 3′ ends, respectively, was generated by RT-PCR of total RNA from the human striatum (Stratagene, 540135) using Taq DNA polymerase (Qiagen, Valencia, CA) and the primers: 5′-AAAAAC-ATATGGAATGCAGCCAGGATTGCGCGAC-3′ and 5′-AAAAAGGATCCTTAAAATCTC-ATAAATCCTCCGTATCTTTTTTCC-3′ (Invitrogen, Carlsbad, CA). PCR reactions contained 1 ng cDNA as template and 0.3 μM primers with 35 thermocycles of 60 seconds at 94°C, 60 sec at 45°C, and 70 sec at 72°C. Following digestion of the amplified PE DNA with restriction enzymes NdeI/BamHI, the digested PE DNA was ligated using T4 DNA ligase (Invitrogen) to the NdeI/BamHI digested plasmid expression vector pET19b (EMD). The 732 bp PE nucleotide sequence in the PE-pET19b vector was verified by DNA sequencing (Davis Sequencing, Inc., Davis, CA) and the DNA sequence was used to deduce the primary amino acid sequence.
PE was expressed via the PE-pET19b vector by its transformation into Rosetta 2 (DE3) (EMD), a BL21 derived E. coli host. PE expression in E. coli cells was conducted in LB media with 50 μg/ml carbenicillin and 30 μg/ml chloramphenicol utilizing the T7 expression system. E. coli cells were grown at 37°C with agitation to a density of 0.6 to 0.8 optical density at 600 nm and induced with 1 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) for 3 hours at 37°C and harvested by centrifugation at 4,000 × g for 15 minutes at 4°C. The cells were resuspended in 100 mM Tris-Cl, pH 7.0 and lysed by sonication (6 × 20 sec, power level 4, microtip, Misonix, Ultrasonic Processor XL) at 4°C. The disrupted cells were centrifuged at 16,000 × g for 15 minutes at 4°C. Intact PE was localized primarily in inclusion bodies; therefore, the cell pellet was suspended in 100 mM Tris-Cl, pH 7.0, 6 M urea, and solubilized PE was obtained as as the soluble fraction after centrifugation at 20,000 × g.
Purification of N-His tagged PE was conducted by applying the solubilized PE sample to a His-bind Ni2+-affinity column (EMD) equilibrated with 100 mM Na-phosphate, 20 mM Tris, pH 8.0, 6 M urea (buffer A), and 5 mM imidazole. After washing with 40 mM imidazole in buffer A, PE was eluted with 300 mM imidazole in buffer A. Urea in the PE sample was removed in steps of lower urea concentration (5, 3, 2, 1, and 0.5 M urea) by dialysis (4°C) into a buffer consisting of 100 mM sodium phosphate, 20 mM Tris, pH 7.5, 5 mM glutathione (GSH), and 0.5 mM oxidized glutathione (GSSG) (Calbiochem, San Diego, CA). Glutathione was removed by dialysis into 50 mM Tris, pH 7.5, for storage of recombinant PE.
Purified recombinant PE was confirmed by SDS-PAGE using NuPAGE 12% Bis-Tris polyacrylamide gel (Invitrogen, Carlsbad, CA) under reducing conditions and Coomassie staining. In addition, PE was evaluated by immunoblotting with anti-(Met)-enkephalin polyclonal antibody (1:5000, Chemicon, Temecula, CA) and visualized using ECL Plus (GE Healthcare, Piscataway, NJ) detection system. The protein concentration was estimated by absorbance measurements at 280 nm using the theoretical molar extinction coefficient of 33,640 M-1cm-1 for recombinant His-tagged PE.
Optimization of pepsin digestion of proenkephalin (PE) for DXMS studies
PE digestion by pepsin is a requisite step prior to enhanced deuterium-exchange mass spectrometry (DXMS) experiments. In optimizing this process, the total number of peptides produced from pepsin digestion was evaluated under several different conditions including concentrations of denaturant and length of time for reduction of disulfide bridges. For each sample test, 50 μg of PE in 9 μl of 5 mM Tris, pH 7.2 was diluted in 27 μl of 8.3 mM Tris, 50 mM NaCl, pH 7.2 (on ice), representing the dilution of the protein into D2O based buffers in deuterium-exchange experiments. The sample was then diluted with 54 μl of a cold solution (0°C) of 0.8% formic acid, 16.6% glycerol, 1 M tris(2-carboxyethyl)phosphine (TCEP), and guanidine hydrochloride (GuHCl) at final concentrations of 0.05 M, 0.5 M, 1.0 M, 2.0 M, or 4.0 M. This quenching step represented the reduction of hydrogen-deuterium exchange with a decrease in pH to 2.2 -2.5 in addition to denaturing the protein prior to pepsin proteolysis with GuHCl and acidic conditions. The quenching process was allowed to proceed on ice for either 1 minute or 5 minutes, representing the time extremes allowed for TCEP to reduce disulfide bonds, after which, the sample was frozen by submersion into dry ice. Procedures for pepsin digestion for DXMS has been described (28, 29). Briefly, the quenched sample at 0°C was passed over a porcine pepsin immobilized column and the proteolytic peptides were collected onto a C18 column (Vydac). The separated products were mass analyzed using a Thermo Finnigan LCQ mass spectrometer and determination of pepsin-generated peptide sequences from the resulting MS:MS data sets facilitated through the use of Sequest (Finnigan, Inc.) .
Hydrogen-deuterium exchange mass spectrometry (DXMS) studies of proenkephalin (PE)
PE samples were prepared with three states of hydrogen-deuterium exchange in each deuterium exchange experiment, consisting of nondeuterated (ND), deuterated, and fully-deuterated (FD). The nondeuterated sample was processed exactly as described in the digestion optimization described in the previous paragraph. The FD sample represents the “maximum” hydrogen-deuterium exchange for a certain time period, which in these experiments was a period of 14 hours where the samples were allowed to exchange at room temperature. The deuterated samples represent different incubation times prior to the quenching of the exchange process. All samples used 50 μg of PE in 9 μl of 5 mM Tris, pH 7.2. The ND sample was diluted with 27 μl of H2O-based 8.3 mM Tris, 50 mM NaCl, pH 7.2, while the deuterated and the FD samples were diluted in a D2O-based buffer of the same composition. The deuterated samples were allowed to exchange at 0°C for 10 seconds, 30, 100, 300, 1000, and 3000 seconds, after which the exchange was quenched with 54 μl of 0.8% formic acid, 16.6% glycerol, 1 mM TCEP, and 0.5 M (GuHCl) and the quenching process was allowed to proceed for one minute at 0°C until being frozen by submersion into dry ice. A more detailed description of sample processing for exchange experiments has been described (23, 24). The pepsin digestion, chromatography, and the mass spectral acquisition proceeded as described in the digestion optimization section. Data processing and reduction of hydrogen-deuterium exchange experiments utilized DXMS data-reduction software (Sierra Analytics, Modesto, CA) (17, 25, 26).
Examination of PE secondary structure by circular dichroism (CD)
Circular dichroic spectra of PE were recorded with an AVIV 202 circular dichroism spectrophotometer (AVIV Biomedical) in 10 mM phosphate buffer, pH 7.1 with trifluoroethanol (TFE) at 0%, 5%, 10%, 15%, 20%, and 33%. Measurements of PE, at concentrations of 15 μM, were taken in a demountable quartz cuvette of 0.2 mm path length (Starna Cells, 20/C-Q-0.2) from 250 nm to 190 nm in a single scan at 1 nm bandwidth, 0.5 nm increments, 25°C, and with 3 second averaging. The PE CD spectra are from two experiments at each condition except for PE at 0% TFE, which was repeated three times. CD solvent baselines were obtained for all conditions and the baselines were subtracted from the sample spectra prior to spectral analysis and deconvolution; spectra were smoothed using a 5 point Mean-Movement smoothing filter in Mathematica 6.0 software (Wolfram Research). The measured ellipticities in units of mdeg were converted to the appropriate units and wavelength range in 1 nm increments for each deconvolution software programs.
The CD spectra with converted units were analyzed and deconvoluted by the CD spectra software programs CDSSTR (21, 22) and CONTIN (26). Raw CD spectra were normalized to molar circular dichroism (liter mol-1 cm-1) and mean residue ellipticity [θ] units (degrees cm2 dmol-1) for CDSSTR and CONTIN, respectively. The DOS software programs were downloaded from http://www2.umdnj.edu/cdrwjweb/. The data base employed in the CONTIN deconvolution consisted of twenty total reference proteins, which included four denatured proteins that are models of unordered segments of proteins. The data base for CDSSTR deconvolution utilized twenty-six reference proteins. The CD spectra of PE were compared under buffer conditions with and without TFE.
Results
Recombinant proenkephalin (PE): optimization of pepsin digestion of PE for DXMS studies
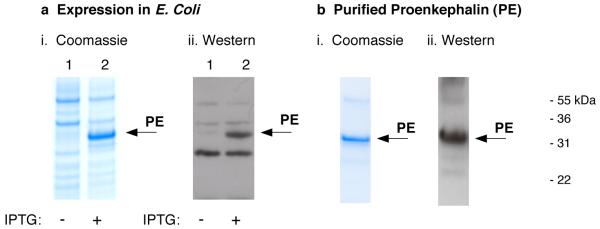
Human proenkephalin was expressed in E. coli to produce sufficient quantities for DXMS and CD studies. High level expression of N-His-tagged PE resulted in approximately 80 mg PE protein per liter of cell expression (fig. 1). IPTG induction of PE was demonstrated by production of the ∼33 kDa apparent molecular weight PE band on SDS-PAGE, that was recognized by anti-(Met)enkephalin in western blots (Fig. 1a). This recombinant PE possesses a calculated molecular weight of 31,073 daltons; thus, the relative electrophoretic mobility of PE on SDS-PAGE represents an approximation of its apparent molecular weight. After solubilization of PE from cell extracts, purification was achieved by chromatography on a Ni2+-affinity column. Purified PE was observed as a single 33 kDa band on SDS-PAGE visualized by Coomassie staining, and was recognized by anti-(Met)enkephalin in western blots (fig. 1b). This purified PE was utilized for evaluation by DXMS and CD.
Figure 1. Expression and purification of recombinant human proenkephalin (PE).
a. Expression of human PE. Expression of PE in E. coli was induced in the presence of IPTG for 2 hours (panel i, lane 2), shown by the 33 kDa PE band on SDS-PAGE stained with Coomassie blue. Cells grown in the absence of IPTG did not show PE (lane 1). The induced PE band was recognized by anti-(Met)enkephalin in western blots (panel ii, lane 2). Controls without IPTG showed no PE induction (panel ii, lane 1).
b. Affnity purification of PE. The N-His tagged PE was purified by a Ni2+-affinity column. Purified PE was illustrated by SDS-PAGE gels stained with Coomassie blue (panel I, 1 μg purified PE per gel lane). The purified PE was recognized by anti-(Met)enkephalin by a western blot (panel ii). Purification yielded 30-50 mg of recombinant PE per liter of E. coli.
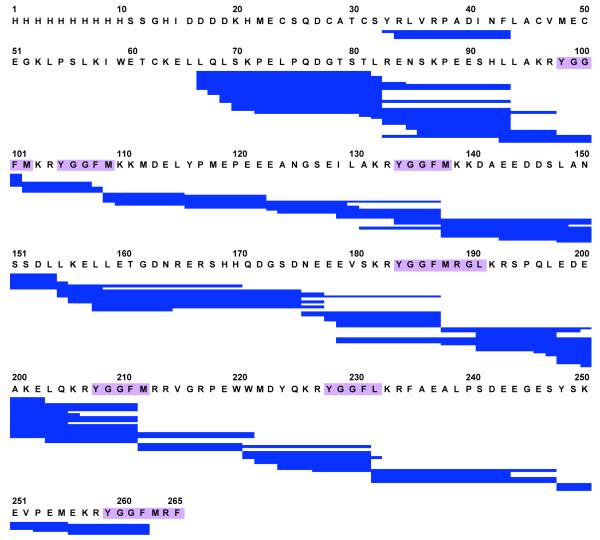
The extent of PE domains that can be studied by DXMS is determined by the peptide coverage of the protein that results from the pepsin digestion process. This important step utilizes pepsin that can function under acidic pH conditions used to quench hydrogen-deuterium exchange. Optimization of pepsin digestion of PE was assessed under several conditions including 0.5 M GuHCl and 1 M TCEP (reducing agent) with a pepsin column, with the peptide products then undergoing LC-MS/MS analyses. PE-derived peptides produced from digestion optimization enabled 87% coverage with over 100 overlapping peptides (fig. 2). Importantly, coverage included all the dibasic processing sites of PE. Therefore, the pepsin-generated peptides could allow evaluation of H-D exchange at regions of dibasic processing sites of PE.
Figure 2. Pepsin digestion map encompasses dibasic cleavage sites of proenkephalin (PE).
Optimization of conditions (0.5 M GuHCl, 1 M TCEP, for 1 min) for pepsin digestion of PE resulted in peptides encompassing all the dibasic cleavage sites flanking enkephalin peptides (purple) within PE. These peptides (123 peptides) provided 87% coverage of the PE sequence, and included all of the dibasic sites which allowed their analyses by DXMS.
Deuterium-exchange mass spectrometry (DXMS) reveals different accessibilities of dibasic sites of PE to the aqueous environment
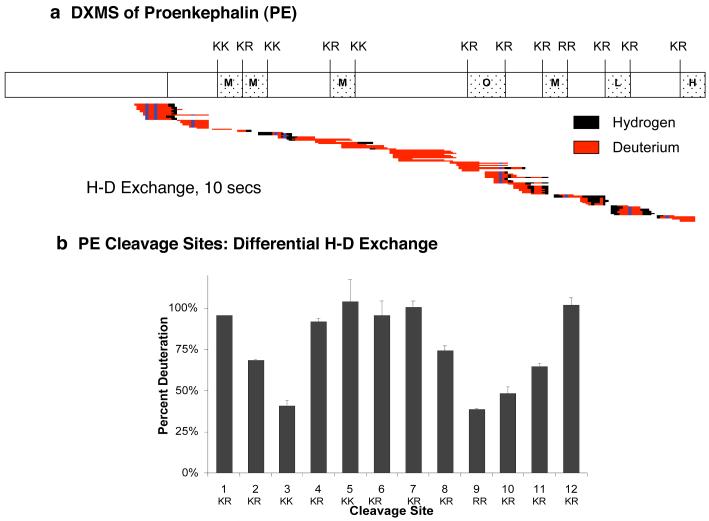
DXMS was utilized to compare the relative accessibilities of different domains of PE and its dibasic processing sites to the aqueous environment. The exchange between deuterium isotopes (from heavy water, D20) and PE amide polypeptide backbone hydrogens was rapid with extensive hydrogen-deuterium (H-D) exchange after only 10 seconds for the exchange period (fig. 3). H-D exchange is illustrated by peptide regions containing deuterium shown in red, and regions containing hydrogen (not deuterium) are shown in black (fig. 3a). During increased times for H-D exchange, the high level of deuteration was maintained (data not shown). Interestingly, some differences in H-D exchange were observed throughout the PE protein sequence.
Figure 3. Hydrogen-deuterium exchange mass spectrometry (DXMS) reveals differential aqueous solvent accessibilities of proenkephalin (PE) intervening domains compared to dibasic processing sites.
a. DXMS of the PE protein precursor. DXMS of PE was conducted by incubating recombinant PE in D2O for 10 seconds, followed by quenching, pepsin digestion, and LC-MS/MS. Results display pepsin-generated peptide fragments containing deuterium that was exchanged for hydrogen (H-D exchange) shown in red, and peptide regions without H-D exchange shown in black (dark blue indicates non-exchangeable prolines). The DXMS peptide map illustrates a large extent of H-D exchange throughout PE. Enkephalin-related peptide sequences within PE are indicated as (Met)enkephalin (‘M’), (Leu)enkephalin (‘L’), Octapeptide (‘O’, M-Arg-Gly-Leu), and Heptapeptide (‘H’, M-Arg-Phe).
b. Differential H-D exchange at the different dibasic processing sites of PE. Differences in the relative extent of H-D exchange was observed among the twelve dibasic PE cleavage sites. High levels of deuteration were observed among the dibasic sites located in the mid-region of PE (sites #4-8), as well as the NH2- and COOH-terminal sites (#1 and 12). Lower levels of deuteration were observed for dibasic residues #2, 3, and 9-11.
Notably, comparison of H-D exchange at the 12 dibasic processing sites, represented by peptides that span these sites, showed differences in the relative accessibilities of the dibasic sites to the aqueous environment (fig. 3b). The dibasic sites #4-8 (dibasic sites numbered from NH2- to COOH-domains of PE) in the mid-region of PE showed greater H-D exchange compared to the much lower level of deuteration at the dibasic sites #2,3 and #9-11 located at NH2- and COOH-domains of PE. However, the NH2-terminal and COOH-terminal dibasic sites #1 and #12 show a high level of deuteration similar to dibasic sites in the mid-region of PE. Thus, the 12 dibasic processing sites of PE display a pattern of differential accessibilities to the aqueous solvent, suggesting differential conformational orientations of these cleavage sites.
Altered dynamics of proenkephalin domains and processing sites to the aqueous solvent environment in the presence of trifluoroethanol (TFE)
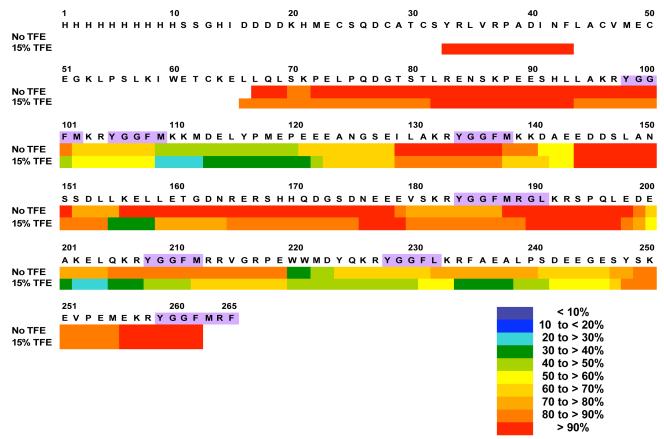
The effects of increasing the hydrophobicity of the solvent environment of PE was investigated in DXMS experiments in the presence of TFE (15%). Color representations of relative H-D exchange illustrates decreased amounts of H-D exchange in the presence of TFE for most regions of PE, and no change in a few regions of PE (fig. 4). TFE apparently restricted overall H-D exchange of PE.
Figure 4. Restricted H-D exchange of PE peptide domains in the presence of TFE (trifluoroethanol).
H-D exchange was conducted in the presence of TFE (15%, trifluoroethanol), an organic solvent, to represent a more hydrophobic environment compared to without TFE. The H-D exchange period in D2O was 10 secs (same time as in fig. 3, without TFE). Different ranges of the percent deuteration is illustrated in a color-coded manner (see key in figure). Comparisons of percent deuteration of PE peptide domains in the absence and presence of TFE revealed changes in H-D exchange, represented by reduced deuteration of the majority of PE domains in the presence of TFE.
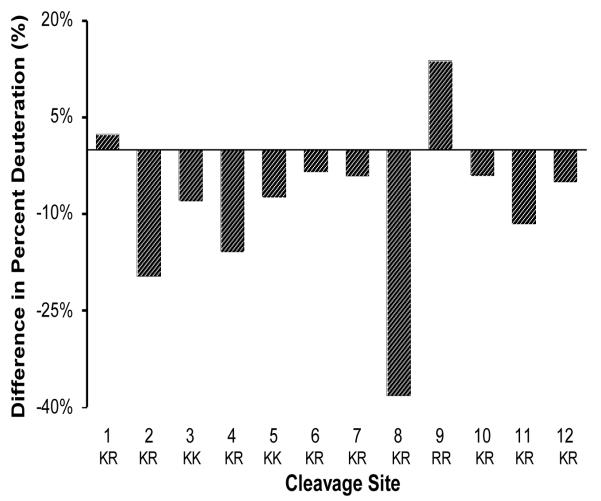
Comparison of the relative amounts of H-D exchange at dibasic processing sites of PE in the presence of TFE (15%) showed primarily decreased H-D exchange compared to controls without TFE (fig. 5). Dibasic site #8 showed the largest decrease in H-D exchange, and dibasic sites #2-7 and #10-12 showed more modest decreases in H-D exchange. The dibasic sites #1 and #9, however, showed modest increases in H-D exchange. These results demonstrate that TFE, an organic solvent, modified the relative accessibilities of the dibasic processing sites of PE to the aqueous solvent environment.
Figure 5. Relative accessibilities of dibasic cleavage sites to the aqueous solvent environment are altered in the presence of TFE, as revealed by H-D exchange.
The average level of deuteration of each dibasic cleavage site in the presence of TFE (15%) was subtracted from that in the absence of TFE. These differences are illustrated in the bar graph plots. Reduced deuteration in the presence of TFE was observed for most cleavage sites, especially for sites #2 and #8. Modest increases in percent deuteration were observed for dibasic sites #1 and #9. These data illustrate differential effects of TFE to modify relative accessibilities of PE dibasic cleavage sties to the aqueous environment.
Secondary structure features of PE examined by circular dichroism
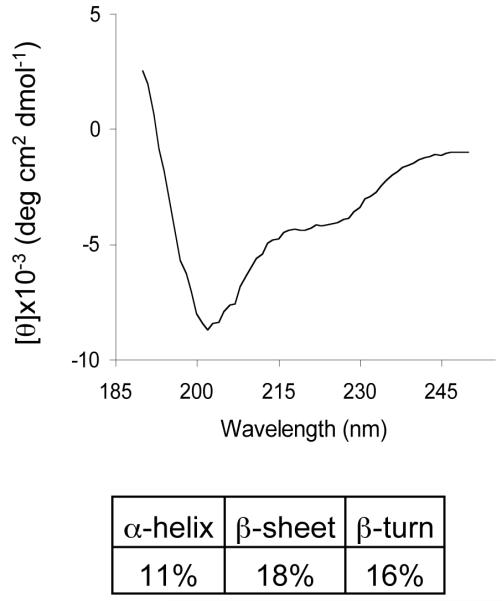
The extent of secondary structures of PE was examined under conditions utilized for DXMS studies, in the absence and presence of TFE. CD spectra (fig. 6) data suggested extensive secondary structural features of PE consisting of alpha-helix (11%), β-sheet (18%), and β-turns (16%).
Figure 6. Circular dichroism (CD) illustrates secondary structural features of proenkephalin (PE).
Analysis of the CD spectrum of PE revealed secondary structures of PE. The CD spectrum of recombinant PE was obtained, and was deconvoluted using CDSSTR (21) to reveal secondary structures consisting of 11% alpha-helix, 18% beta-sheet, 16% beta-turn, and 55% disordered structures.
CD analyses of PE in the presence of TFE, an organic solvent, was also conducted to observe PE secondary structure under a more hydrophobic condition. Increasing concentrations of TFE, an organic solvent, resulted in substantial alterations in CD spectra (fig. 7a) that reflect changes in secondary structure. As the concentration of TFE increases, the percent of alpha-helix secondary structure was substantially increased from 11% to approximately 45% (fig. 7b). In contrast, the relative degrees of β-sheet and β-turn secondary structures were not increased with increasing TFE; rather, a modest decrease in β-sheet and β-turn structures was observed with TFE. These findings suggest that increases in alpha-helix structure of PE in the presence of TFE may be related to the TFE-induced restriction in overall H-D exchange of PE domains. Moreover, TFE-induced alterations in H-D exchange at dibasic processing sites of PE may relate to changes in its secondary structure. Overall, DXMS and CD structural studies of PE have revealed dynamic H-D exchange properties of PE domains to the aqueous environment.
Figure 7. Changes in secondary structure of PE in the presence of TFE as illustrated by CD analyses.
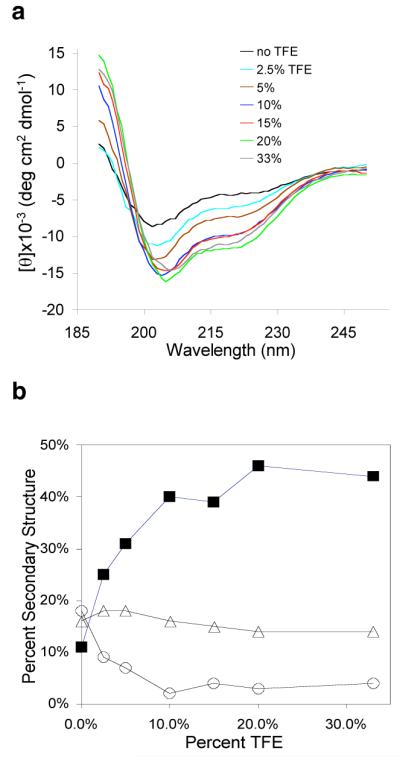
a. CD spectra of PE in the presence of TFE (trifluorethanol). CD spectra for PE were obtained under different concentrations of the trifluoroethanol (TFE), an organic solvent. TFE of 0% to 33% was found to alter the CD spectra of PE, indicating alterations in secondary structure.
b. Increase in alpha-helical structure of PE in the presence of TFE. The relative extent (percent) of changes in secondary structural features with increasing concentrations of TFE (0 to 33%) are illustrated for alpha-helix (■), beta-sheet (○), and beta-turn (△) features. TFE substantially increases the alpha-helical secondary structure of PE.
Discussion
Proenkephalin possesses multiple dibasic proteolytic processing sites that require cleavage to generate the smaller, active enkephalin opioid peptide neurotransmitters. DXMS studies of proenkephalin (PE) have revealed the relative accessibilities of the dibasic processing sites of PE, and have illustrated the dynamic properties of PE of highly exchangeable hydrogens (of the polypeptide backbone) for deuteriums from the D2O aqueous environment. Comparisons among the dibasic cleavage sites of PE showed clear differences in their relative accessibility to the aqueous environment. The dibasic residue sites in the mid-region of PE showed high relative levels of H-D exchange. However, dibasic sites near the N- and C-domains of PE, corresponding to sites #2-3 and sites #9-11, respectively, showed lower extents of H-D exchange. Interestingly, the dibasic sites closest to the NH2- and COOH-termini of PE showed high levels of H-D exchange. Under conditions of increased hydrophobicity with TFE (15%, trifluoroethanol), marked decreases in relative extents of H-D exchange at the majority of dibasic sites of PE were observed. CD analyses of PE showed that it possesses secondary structure features of alpha-helix, β-sheet, and β-turns which are present in the PE protein that displays differential accessibilities of its dibasic processing sites to the aqueous environment. It was of interest to find that TFE substantially increased the extent of alpha-helical secondary structure of PE, which occurred with the more restricted H-D exchange properties of PE in the presence of TFE. The combined DXMS and CD data demonstrate the dynamic nature of the dibasic processing sites of PE with respect to differential H-D exchange with its aqueous environment, which occurs within the PE protein possessing alpha-helix, β-sheet, and β-turn secondary structures. These conformational features of PE differ in more hydrophobic environments (with TFE) that may represent PE associated with soluble and membrane components of cells.
DXMS showed that the PE protein displays rapid exchange of hydrogens for deuteriums. Extensive H-D exchange was observed with only 10 seconds as the exchange period. Longer exchange periods of 100 seconds or greater, showed nearly maximal deuteration levels of PE that were similar to the extent of H-D exchange occurring after several hours (14 hrs). In contrast, many other proteins, such as a-synuclein amyloid (25) and C-terminal Src kinase (27), undergo slower rates of H-D exchange during minutes and hours. The rapid rate of H-D exchange for PE suggests that the majority of its domains possess conformations that are highly accessible to the aqueous environment. These H-D exchange properties of PE occur with its secondary structures of alpha-helix, β-sheet, and β-turn. These secondary structural features occur with PE conformations that allow rapid H-D exchange.
Proenkephalin is synthesized at the rough endoplasmic reticulum (RER) from its mRNA, and is then routed into the RER and Golgi apparatus where it is packaged into newly formed secretory vesicles. The pH at the RER and Golgi apparatus is near neutral, at approximately pH 7.0-7.2 (28). Thus, the neutral pH conditions for DXMS and CD studies of PE in this project may reflect PE’s cellular environment at the RER or Golgi apparatus which represent the early stage of the regulated secretory pathway. Subsequent to the RER and Golgi apparatus, PE is then packaged into newly formed secretory vesicles which undergo maturation with internal pH conditions changing to lower pH conditions. It will be of interest in future studies to examine the H-D exchange and CD properties of proenkephalin under different pH conditions.
Proenkephalin in vivo is processed to several high molecular weight intermediates of 4.5 to 23 kDa (29, 30). Recombinant PE expressed and purified from E. coli has been used as substrate for in vitro prohormone processing studies (29), which show that recombinant PE is converted by processing proteases to its known in vivo intermediate products (4.6 to 22.5 kDa) via cleavage at dibasic sites. These data indicate that recombinant PE expressed in E. coli possesses a conformation that allows its appropriate proteolytic processing in vitro that resembles its in vivo processing. These findings illustrate that the dibasic sites of recombinant PE are capable of proteolytic processing to generate in vivo PE-derived intermediate products.
Among protein substrates of proteolytic enzymes, there is little knowledge of the relative accessibilities of multiple substrate cleavage sites to the aqueous environment. The results of this DXMS study of proenkephalin provide significant new knowledge of conformational H-D exchange of different protease cleavage sites within a protein substrate. The differential accessibilities of dibasic processing sites of proenkephalin suggest that a protease enzyme possesses the ability to properly interact with each dibasic site to achieve enzymatic cleavage.
In summary, DXMS studies have revealed differential accessibilities of dibasic processing sites of proenkephalin. Moreover, H-D exchange is restricted under more hydrophobic conditions, represented by inclusion of TFE (trifluoroethanol) in the DXMS buffer conditions. These H-D exchange properties of the dibasic sites reside within PE that possesses secondary structural features of alpha-helix, β-sheet, and β-turn. These findings suggest that dibasic PE prohormone processing sites with differences in accessibilty to the aqueous environment undergo proteolytic processing to generate active neuropeptides for cell-cell communication in the nervous and endocrine systems.
Abbreviations
- PE
proenkephalin
- DXMS
hydrogen-deuterium exchange mass spectrometry
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- TFE
trifluoroethanol
- CD
circular dichroism
Footnotes
This research was supported by grants from the National Institutes of Health (NIH) NS24553, DA04271, and HL58120 to V.H., and CA099835, CA118595 and AI076961 to V.L.W. W.D. Lu was supported by a postdoctoral fellowship from NIDA, NIH (F32DA02496).
References
- 1.Beaumont A, Hughes J. Biology of opioid peptides. Annu. Rev. Pharmacol. Toxicol. 1979;19:245–67. doi: 10.1146/annurev.pa.19.040179.001333. [DOI] [PubMed] [Google Scholar]
- 2.Akil H, Watson SJ, Young E, Lewis ME, Khachaturia H, Walker JM. Endogenous opioids: biology and function. Annu. Rev. Neurosci. 1984;7:223–55. doi: 10.1146/annurev.ne.07.030184.001255. [DOI] [PubMed] [Google Scholar]
- 3.Steiner H, Gerfen CR. Role of dynorphin and enkephalin in the regulation of striatal output pathways and behavior. Exp. Brain Res. 1998;123:60–76. doi: 10.1007/s002210050545. [DOI] [PubMed] [Google Scholar]
- 4.Gubler U, Seeburg P, Hoffman BJ, Gage LP, Udenfriend S. Molecular cloning establishes proenkephalin as a precursor of enkephalin-containing peptides. Nature. 1982;295:206–208. doi: 10.1038/295206a0. [DOI] [PubMed] [Google Scholar]
- 5.Yoshikawa K, Williams C, Sabol SL. Rat brain preproenkephalin mRNA, cDNA cloning, primary structure, and distribution in the central nervous system. J Biol Chem. 1984;259:14301–14308. [PubMed] [Google Scholar]
- 6.Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Tox. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R, Prat A. The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol. 2008;40:1111–25. doi: 10.1016/j.biocel.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J. Biol. Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 9.Steiner DF. The proprotein convertases. Curr. Opin. Chem. Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 10.Fricker LD. Carboxypeptidase E. Annu. Rev. Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- 11.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. USA. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang SR, O’Neill A, Bark S, Foulon T, Hook V. Secretory vesicle aminopeptidase B related to neuropeptide processing: molecular identification and subcellular localization to enkephalin- and NPY-containing chromaffin granules. J. Neurochem. 2007;100:1340–1350. doi: 10.1111/j.1471-4159.2006.04325.x. [DOI] [PubMed] [Google Scholar]
- 13.Furuta M, Yano H, Zhou A, Rouille Y, Holst JJ, et al. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc. Natl. Acad. Sci. USA. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X, Orci L, Carroll R, Norrbom C, Ravazzola M, Steiner DF. Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc. Natl. Acad. Sci. USA. 2002;99:10299–10304. doi: 10.1073/pnas.162352799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen RG, Peng B, Pellegrino MJ, Miller ED, Grandy DK, et al. Altered processing of pro-orphanin FQ/nociceptin and pro-opiomelanocortin-derived peptides in the brains of mice expressing defective prohormone convertase 2. J. Neurosci. 2001;21:5864–5870. doi: 10.1523/JNEUROSCI.21-16-05864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scamuffa N, Calvo F, Chretien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 17.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods VL. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Nat. Acad. Sci. USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch. Biochem. Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Maity H, Lim WK, Rumbley JN, Englander SW. Protein hydrogen exchange mechanism: local fluctuations. Protein Sci. 2006;12:153–160. doi: 10.1110/ps.0225803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brahms S, Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular-dichroism. J. Molec. Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 21.Compton LA, Johnson WC. Analysis of protein circular-dichroism spectra for secondary structure using a simple matrix multiplication. Ana.l Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 22.Hennessey JP, Johnson WC. Information-content in the circular-dichroism of proteins. Biochemistry. 1981;20:1085–1094. doi: 10.1021/bi00508a007. [DOI] [PubMed] [Google Scholar]
- 23.Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL. Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Science. 2005;14:2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derunes C, Burgess R, Iraheta E, Kellerer R, Becherer K, Gessner CR, Li S, Hewitt K, Vuori K, Pasquale EB, Woods VL, Ely KR. Molecular determinants for interaction of SHEP1 with Cas localize to a highly solvent-protected region in the complex. FEBS Lett. 2006;580:175–178. doi: 10.1016/j.febslet.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 25.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL. Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantazato D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL. Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci USA. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong L, Lieser SA, Miyashita O, Miller M, Tasken K, Onuchic JN, Adams JA, Woods VL, Jennings PA. Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J. Mol. Biol. 2005;351:131–143. doi: 10.1016/j.jmb.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 28.Alberts, Johnson, Lewis, Raff, Roberts, Walter . Molecular Biology of the Cell. Fourth edition Garland Science Publisher; New York: 2002. pp. 729–730. [Google Scholar]
- 29.Schiller MR, Mende-Mueller L, Moran K, Meng M, Miller KW, Hook VYH. “Prohormone thiol protease” (PTP) processing of recombinant proenkephalin. Biochemistry. 1995;34:7988–7995. doi: 10.1021/bi00025a004. [DOI] [PubMed] [Google Scholar]
- 30.Hwang SR, Garza C, Mosier C, Toneff T, Wunderlich E, Goldsmith P, Hook V. Cathepsin L expression is directed to secretory vesicles for enkephalin neuropeptide biosynthesis and secretion. J Biol Chem. 2007;282:9556–9563. doi: 10.1074/jbc.M605510200. [DOI] [PubMed] [Google Scholar]