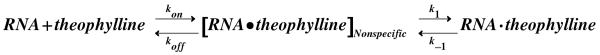Abstract
The apparent on- and off-rate constants for theophylline binding to its RNA aptamer in the absence of Mg2+ were determined here by 2D 1H-1H NMR ZZ-exchange spectroscopy. Analysis of the build-up rate of the exchange cross peaks for several base-paired imino protons in the RNA yielded an apparent kon of 600 M-1 s-1. This small apparent kon results from the free RNA existing as a dynamic equilibrium of inactive states rapidly interconverting with a low population of active species. The data here indicate that the RNA aptamer employs a conformational selection mechanism for binding theophylline in the absence of Mg2+. The kinetic data here also explain a very unusual property of this RNA-theophylline system, slow exchange on the NMR chemical shift timescale for a weak-binding complex. To our knowledge, it is unprecedented to have such a weak binding complex (Kd ≈ 3.0 mM at 15 °C) show slow exchange on the NMR chemical shift timescale, but the results clearly demonstrate that slow exchange and weak binding are readily rationalized by a small kon. Comparisons with other ligand-receptor interactions are presented.
RNAs often require divalent metal ions to fold into active conformations and efficiently carry out their biological functions.1,2 For example, divalent metal ions are required for the high affinity binding of the bronchodilator drug theophylline to its in vitro selected RNA aptamer (Figure 1A and B), which has a Kd of ∼300 nM in 10 mM Mg2+.3,4 This aptamer still binds theophylline in the absence of divalent metals ions, but with a 104 lower binding affinity.3,5 Analysis of NMR lineshapes can yield information on the kinetics of ligand binding.6 High affinity complexes (Kd < 0.5 μM) are usually in “slow exchange” and low affinity complexes (Kd > 100 μM) are usually in “fast exchange” on the NMR chemical shift timescale. Here, we observe slow exchange on the NMR chemical shift timescale for a very low affinity complex: the RNA aptamer-theophylline complex in the absence of divalent metal ions, which has a Kd of ∼ 7 mM at 25 °C. ZZ-exchange NMR experiments7,8 were used to determine the on- and off-rate constants for this complex. The results here demonstrate that a very weak binding complex can exist in slow exchange on the NMR timescale. This phenomenon arises from a slow apparent on-rate, consistent with a small population of binding-competent species and a conformational selection mechanism. This type of weak binding combined with slow exchange may be seen in other partially unstructured molecules, such as other RNA aptamers or riboswitches binding their small molecule ligands.
Figure 1.
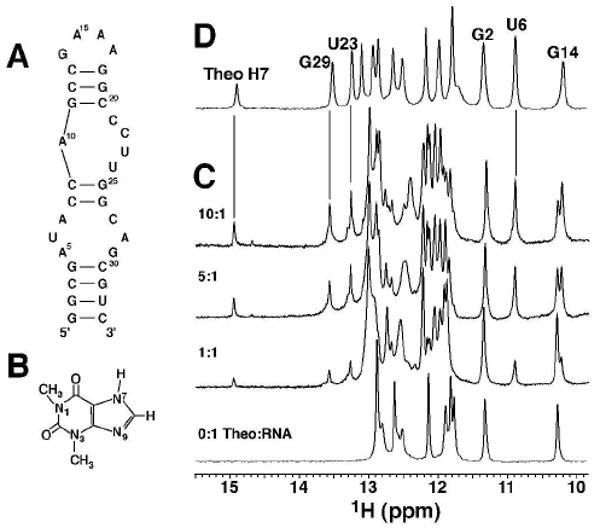
(A) Secondary structure of the theophylline-binding RNA aptamer. (B) Structure of theophylline. (C) 1D imino 1H spectra for the theophylline titration of 0.8 mM RNA in no Mg2+ for 0 to 10 molar ratios of theophylline (at 15 °C, 25 mM sodium phosphate, pH 6.8, 100 mM NaCl, 0.1 mM EDTA). (D) Spectrum of the 1:1 RNA:theophylline complex in 5 mM Mg2+.3 The vertical lines illustrate similar chemical shifts with and without Mg2+.
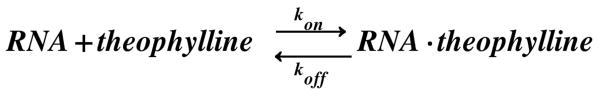
RNA-theophylline complex formation can be described by the bimolecular association reaction:
This reaction shows a strong dependence on Mg2+, as measured by equilibrium filtration binding experiments.3 NMR can be used to measure binding affinities for weak binding complexes, and Figure 1C shows the titration of RNA with theophylline in the absence of Mg2+. The aptamer is in slow exchange on the NMR chemical shift timescale, as seen for the G14 imino proton where the free and bound resonances have different chemical shifts. The relative peak volumes indicate the populations of the two states, yielding a Kd ≈ 3 mM at 15 °C (Supporting Information). To our knowledge, it is unprecedented to have such a weak-binding complex in slow exchange.
It is possible that these spectra are not reflecting the simple biomolecular binding reaction shown in Scheme 1. For example, slow exchange of the G14 imino proton resonance is also consistent with the two-step reaction shown in Scheme 2. In this scheme, the theophylline binds nonspecifically to the aptamer followed by a slow conformational change leading to the structure of the complex observed in the presence of Mg2+. The slow exchange observed for the aptamer imino resonances could then reflect a slow unimolecular conformational rearrangement of the RNA-theophylline complex (right reaction in Scheme 2). One piece of evidence against Scheme 2 is that the methyl groups of theophylline are also in slow exchange on the NMR timescale (Supporting Information).
Scheme 1.
Scheme 2.
To directly distinguish between a unimolecular (Scheme 1) and a bimolecular (Scheme 2) process, the kinetics for exchange were measured as a function of theophylline concentration using ZZ-exchange spectroscopy.7,8 This technique monitors the exchange of longitudinal magnetization between significantly populated slowly-exchanging states and can be used to determine rate constants. The imino proton-specific 2D 1H-1H ZZ-exchange experiment employs a mixing sequence that eliminates interference from 1H-1H NOEs (Supporting Information).9 2D ZZ-exchange spectra of RNA samples containing 2.6, 3.7, and 6.4 mM theophylline were acquired at 600 MHz and 15 °C. Figure 2 shows the imino proton spectra for the 2, 19, 54 and 170 ms mixing times in the 6.4 mM theophylline sample. The G4 and G25 imino protons have large chemical shift changes between the two states of the RNA. Since both residues are in canonical base pairs at the bottom and top of the lower internal loop in the RNA, their chemical environments are sensitive to theophylline binding (Figure 1). Chemical exchange between these two states leads to a build-up of cross peak intensity at shorter times with a subsequent decrease at longer times due to T1 relaxation (Supporting Information).8
Figure 2.
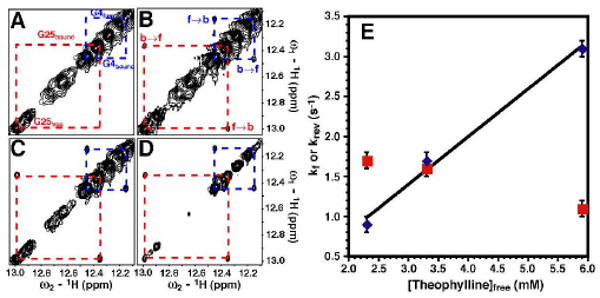
2D 1H-1H ZZ exchange spectroscopy of the theophylline-RNA sample in no Mg2+ at 10:1 theophylline:RNA at 15 °C. (A-D) Imino regions of spectra at (A) 2, (B) 19, (C) 54 and (D) 170 ms mixing times. (A) Assignments of the free and bound states for the imino protons of G25 (red) and G4 (blue) are given. (B) Exchange cross peaks originating from the free-to-bound state (f→b) and bound-to-free state (b→f) are labeled. (E) Plot of kf (blue diamonds) and krev (red squares) versus free theophylline concentration. The error bars were obtained from the 150 Monte Carlo simulations (Supporting Information).11
A two-state exchange model with a pseudo-first order rate constant was used where kf = kon·[theophylline]free. The buildup and decay for both the G4 and G25 exchange cross peaks were simultaneously fit at each theophylline concentration using a least-squares minimization routine (Supporting Information).10 The plot of kf and krev versus theophylline concentration (Fig. 2E) shows a linear dependence of kf on theophylline concentration (R2 = 0.99), where the slope gives a kon = 600 ± 57 M-1 s-1. In contrast, the krev is relatively insensitive to theophylline concentration yielding an average koff = 1.5 ± 0.3 s-1. These results rule out the unimolecular step in Scheme 2 as the source of the slow exchange process. The Kd of 2.5 mM obtained from the rates constants is in good agreement with the independently determined 3.0 mM Kd from the 1D imino spectra (Fig. 1 and Supporting Information). The apparent kon determined here is ∼310-fold less than that observed in the presence of 10 mM Mg2+ at 25 °C; whereas, the koff increased by ∼20-fold.5 Clearly, Mg2+ plays an important role in both the formation of and dissociation of the theophylline complex.
The data here confirm that, like many other RNAs, the free state of this aptamer does not exist in a single conformation but exists in a dynamic equilibrium of “inactive” states that rapidly interconvert with “active” species.5,11,12 Thus, Mg2+ is probably not actually changing the true kon but instead leads to an increase in the population of binding competent molecules, which then changes the apparent kon. These results indicate that the RNA employs a conformational selection mechanism for binding theophylline.
Multiple mechanisms have been proposed for ligands binding to receptors.13 For example, rapid non-specific binding followed by a slower conformation rearrangement, corresponding to Scheme 2, has previously been observed for small cationic planar aromatic molecules, such as ethidium, that intercalate into double stranded nucleic acids.14 In this system, the double stranded nucleic acid is highly ordered and intercalation requires slow conformational unstacking of neighboring base pairs. Therefore, the bimolecular process is fast and electrostatically driven, while the unimolecular conformational changes required for intercalation are slower. This contrasts with theophylline binding observed here where the uncharged theophylline is not electrostatically attracted to the RNA, and the source of the slow apparent on-rate likely arises from disorder of the RNA aptamer.
A coupled folding and binding mechanism for an intrinsically disordered protein binding to its protein partner was recently proposed based on NMR relaxation dispersion data.15 In this system, the disordered pKID peptide forms an intermediate consisting of a partially ordered complex with the KIX protein and this bimolecular reaction is in slow exchange on the NMR chemical shift timescale. The partially folded peptide-bound intermediate can then form the fully-folded complex, where this folding reaction was fast on the NMR chemical shift timescale.15 Both the peptide and RNA system involve partially unstructured molecules, but the disorder leads to different kinetics for ligand binding. For the peptide-protein system, the apparent kon for peptide is quite fast (6 × 106 M-1 s-1), where the presence of an unstructured ligand can lead to faster on-rates.16 This contrasts with the theophylline-RNA system where the absence of many imino resonances indicates that the free RNA is partially disordered (Fig. 1). The disordered RNA then leads to a slow apparent on-rate for theophylline, which arises from conformational selection for binding-competent species. This same process is likely employed by other RNAs that have conformationally heterogeneous free states, such as the aptamer domains in RNA riboswitches binding their ligands.
Supplementary Material
Acknowledgments
We thank Marella Canny for preparation of the RNA sample. This work was supported in part by NIH grant AI33098 and MPL was supported in part by a NIH training grant T32 GM65103. NMR instrumentation was purchased with partial support from NIH grant RR11969 and NSF grants 9602941 and 0230966.
Footnotes
Supporting Information paragraph NMR spectra and description of the methods used for analysis of these spectra. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Misra VK, Draper DE. Biopolymers. 1998;48:113–35. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.Woodson SA. Curr Opin Chem Biol. 2005;9:104–9. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Nat Struct Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 5.Jucker FM, Phillips RM, McCallum SA, Pardi A. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan JI, Fraenkel G. NMR of Chemically Exchanging Systems. Academic Press; New York: 1980. [Google Scholar]
- 7.Jeener J, Meier BH, Bachmann P, Ernst RR. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 8.Ernst RR, Bodenhausen G, Wokaun A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford; Oxford: 1987. [Google Scholar]
- 9.(a) Fejzo J, Westler WM, Macura S, Markley JL. J Am Chem Soc. 1990;112:2574–2577. [Google Scholar]; (b) Macura S, Westler WM, Markley JL. Methods Enzym. 1994;239:106–144. doi: 10.1016/s0076-6879(94)39005-3. [DOI] [PubMed] [Google Scholar]
- 10.Farrow NA, Zhang OW, Formankay JD, Kay LE. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 11.Leulliot N, Varani G. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 12.Williamson JR. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 13.Tsai CJ, Kumar S, Ma BY, Nussinov R. Protein Science. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porschke D. Biophys J. 1998;75:528–537. doi: 10.1016/S0006-3495(98)77542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugase K, Dyson HJ, Wright PE. Nature. 2007;447:1021–5. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 16.Pontius BW. Trends Biochem Sci. 1993;18:181–6. doi: 10.1016/0968-0004(93)90111-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.