Abstract
Members of the Reticulocyte Binding Protein-like (RBL) family are merozoite-expressed proteins hypothesized to be essential for effective invasion of host erythrocytes. Proteins of the RBL family were first defined as merozoite invasion ligands in Plasmodium vivax, and subsequently in P. falciparum and other malaria parasite species. Comparative studies are providing insights regarding the complexity and evolution of this family and the existence of possible functionally alternative members. Here, we report the experimental and bioinformatic characterization of two new rbl genes in the simian malaria parasite species P. knowlesi. Experimental analyses confirm that a P. knowlesi gene fragment orthologous to P. vivax reticulocyte binding protein-1 (pvrbp1) represents a highly degenerated pseudogene in the H strain as well as two other P. knowlesi strains. Our data also confirms that a gene orthologous to pvrbp2 is not present in the P. knowlesi genome. However, two very diverse but related functional rbl genes are present and are reported here as P. knowlesi normocyte binding protein Xa and Xb (pknbpxa and pknbpxb). Analysis of these two rbl genes in Southern hybridizations and BLAST searches established their relationship to newly identified members of the RBL family in P. vivax and other species of simian malaria. Rabbit antisera specific for recombinant PkNBPXa and PkNBPXb confirmed expression of the prospective high molecular weight proteins and localized these proteins to the apical end of merozoites. Their precise location, as determined by immuno-electron microscopy (IEM), was found to be within the microneme organelles. Importantly, PkNBPXa and PkNBPXb are shown here to bind to host erythrocytes, and discussion is centered on the importance of these proteins in host cell invasion.
Keywords: Plasmodium knowlesi, malaria, merozoite invasion, reticulocyte binding proteins, erythrocytes, apicomplexa
1. Introduction
Malarial merozoites gain entry into red blood cells through specific receptor-ligand interactions and a cascade of molecular interactions that are largely still undefined (reviewed in [1]). Moreover, merozoites can be characterized by their ability to invade erythrocytes of either all stages of maturation or by an evident restriction, only reticulocytes. The reticulocyte host cell specificity of human malaria P. vivax merozoites has been well known and attributed to the action of the Reticulocyte Binding Protein (PvRBP1 and PvRBP2) complex located at the invasive apical end of the merozoite [2–4]. Homologs of the P. vivax RBPs have since been characterized in the human malaria P. falciparum [5–9], non-human primate [10, 11] and rodent malaria species [12]. This family of invasion ligand proteins is now known as the Reticulocyte Binding-Like (RBL) family. Rbl genes have a characteristic small exon encoding a signal peptide, a short intron, and a second exon encoding a large (230 – 350 kDa) predominantly hydrophilic protein with a transmembrane domain and short cytoplasmic tail (reviewed in [1]).
P. cynomolgi, which is a simian malaria parasite closely related to P. vivax, also invades predominantly reticulocytes and the orthologous genes for pvrbp1 and pvrbp2 have been reported [2, 10]. In the rodent parasite P. yoelii, the Py235 gene family was shown to be most related to pvrbp2 [4, 13]. In P. falciparum, genomic and experimental studies revealed homolog genes that have been termed normocyte binding proteins (nbp1, nbp2a, nbp2b) [5, 6], also referred to as reticulocyte-binding homologs 1, 2a and 2b (rh1, rh2a and rh2b) [5–7]. Other similar P. falciparum genes were subsequently characterized as rh3/nbp3, rh4/nbp4, and rh5/nbp5, with rh3/nbp3 confirmed to be a pseudogene [8, 9, 14, 15]. Interestingly, the chimpanzee parasite, P. reichenowi, has an identical composite of family members orthologous to the P. falciparum genes. However in P. reichenowi, nbp1 is clearly a pseudogene without an open reading frame (ORF), while nbp3 has an ORF and appears to be functional [16]. The RBL proteins known to date, though quite divergent in sequence, have shared structural and biological features that are presumed to be essential for the targeting of host cells and effective invasion. The initial identification and characterization of the P. vivax RBPs supported the hypothesis that merozoites require molecules to identify the appropriate target cells and then may signal the activation of subsequent receptor-ligand interactions involved in invasion [2–4]. It is now known that several other more distantly related rbl genes are present in P. vivax [17] and it remains to be determined whether the encoded proteins serve as alternative ligands, or if some are in fact pseudogenes, as suggested by the current gene sequences reported in the P. vivax genome database. The characterization of P. falciparum RBLs has provided additional information regarding the potential complexity of the RBL family and their role in merozoite invasion. In vitro experiments focused on generating P. falciparum rbl knock out parasites have begun to clarify the role of rbls in the cascading events of merozoite invasion. PfRh1/NBP1 is an important player in invasion through a sialic acid-dependent interaction [5, 18]; however, this gene can be disrupted with continued survival of the parasites in vitro [18]. Disruption of pfrh2a/pfnbp2a and pfrh2b/pfnbp2b genes is also possible with the retrieval of live parasites, indicating that in some P. falciparum strains merozoite entry is mediated through a novel, alternative invasion pathway [19]. Recent studies have also reported regions of P. falciparum RBLs implicated in binding to erythrocytes [20–22].
Phylogenetic analysis based on the known rbl genes and encoded proteins show that they form subgroups that reflect specific relationships and a degree of similarity to the PvRBP1 or PvRBP2 prototype molecules ([10] and unpublished data). Though the overall identity between ortholog molecules in widely divergent species such as P. vivax, P. falciparum and P. yoelii is very low (23% – 30%), within a genetic clade such as between P. vivax and the Asian simian malarias or between P. falciparum and P. reichenowi, the degree of identity between orthologs within the same RBL subgroup increases dramatically to high levels of 75% or greater.
To better understand the functional role(s) and fine interactions of the RBL proteins in host cell selection and entry, we sought to identify and undertake a thorough investigation of the rbl genes and encoded proteins expressed in a simian malaria species, P. knowlesi, known to also naturally infect humans [23–27]. P. knowlesi is closely related to P. vivax and its sister parasite P. cynomolgi [28]. It is also very closely related to the simian malaria parasite P. coatneyi, which in many respects is phenotypically similar to P. falciparum [29]. As such, and given the stability of its merozoites [30], P. knowlesi has traditionally provided an exceptional instrument for the comparative study of the erythrocyte invasion mechanisms pertinent to both P. vivax and P. falciparum.
The P. knowlesi genome has recently been completed with 8-fold sequence coverage, yet the P. knowlesi rbl gene repertoire in this species remained in question, with a number of partial sequences annotated as fragments of putative rbps on chromosome 14, and no other genes recognized as members of this family [31]. Here, using a combination of experimental investigation and bioinformatics we definitively report the presence of three rbl genes in the P. knowlesi genome, and confirm that one is a pseudogene. The two expressed RBL members, PkNBPXa and PkNBPXb, have been investigated with regards to their structures and functions and localized by IEM to the microneme organelles of mature merozoites. The functional importance of these proteins in the context of the RBL superfamily, and the importance of P. knowlesi as a model for further investigations of the RBLs are discussed.
2. Materials and Methods
2.1. Parasite propagation and acquisition
P. knowlesi (H strain) infected erythrocytes were obtained fresh from Macaca mulatta monkeys or grown in vitro from cryopreserved samples and processed as described previously [32]. P. vivax Belem and Salvador I strain parasites were acquired from blood-stage infections in Saimiri boliviensis monkeys and processed to remove leukocytes and platelets prior to purification of infected erythrocytes. P. coatneyi originating from a 1961 isolation [33], P. cynomolgi (Berok strain) and P. fragile (Nilgiri strain), were expanded in M. mulatta monkeys for DNA isolation after removal of host cellular elements. All non-human primate experimental studies, infections and associated protocols were performed with the official approval of Emory University or CDC’s Institutional Animal Care and Use Committee.
2.2. Nucleic acid isolation
Genomic DNA (gDNA) was prepared from blood-stage parasites using the QIAamp DNA Blood Extraction kit (Qiagen, Valencia CA), as described previously [2]. Total RNA was prepared from mature schizonts using TRIzol (Invitrogen, Carlsbad CA) following the manufacturer’s protocol and treated with RNAase-free DNAase (Roche, Indianapolis IN).
2.3. Polymerase Chain Reaction (PCR) amplification and cloning of rbl genes in P. knowlesi
PCR amplification using the Expand High Fidelity System (Roche, Indianapolis IN) was performed with gene-specific primers on gDNA from P. knowlesi following the manufacturer’s instruction. Fragments were cleaned using the Qiaquick purification system (Qiagen, Valencia CA), cloned into the pCR2.1 vector (Invitrogen, Carlsbad CA) and sequenced using the ABI Prism BigDye Terminator v3.0 cycle sequencing kit (Applied Biosystems, Foster City CA). DNA fragments for pvrbp and pknbp radiolabeled probes were generated by PCR amplification using primers based on the published pvrbp1 (M88097) and pvrbp2 (AF184623) gene sequences, data retrieved from database BLAST searches and new sequencing data. Pknbp1 pseudogene (ψpknbp1) amplicons were generated using three P. knowlesi strains as templates and primers were designed according to the sequence of a 1.5 kb DNA fragment identified from the H strain genome (F 5′ GGT CGA AAC ATA ATA CGG TG 3′ and R 5′ GGA ATT CGA TGG AGT TGA TT 3′). The complete pknbpxa and pknbpxb gene structures were constructed and finalized through a combination of genome database BLAST searches of P. knowlesi contigs and shotgun sequences and new sequence data generated from amplified DNA fragments corresponding to the pknbp genes.
2.4. Confirmation of pknbpxa and pknbpxb intron/exon junctions
The 5′ ends and the exon/intron junctions of the pknbpxa and pknbpxb genes were confirmed using a 5′RACE system and reverse transcriptase PCR, RT-PCR (Invitrogen, Carlsbad CA) following the manufacturer’s recommendations. Two gene-specific primers were designed to produce cDNA, Nxa1R 5′ ATT CTG TCT ATC GTA GGA GC 3′ and Nxb2R 5′ TTG CTT CAC GGA TTT GCT 3′, followed by nested amplifications using the 5′ RACE Abridged Anchor Primer provided in the kit and gene-specific reverse primers, Nxa3R 5′ CCA ATA ATA ATT AAC AGA AG 3′ and Nxb4R 5′ TGG AGA TAG CCT CAA AT 3′.
2.5. Southern and northern blot analysis and library screening
1μg to 2μg aliquots of gDNA, digested overnight with designated restriction enzymes, were separated on 0.8% agarose gel by standard electrophoresis, essentially as described [2]. Fixation of DNA to the membranes was performed by either UV cross linking (Stratagene, La Jolla CA) or baking in vacuo at 80°C for 2 h. DNA fragments were labeled for hybridization reactions using the Prime-It II DNA labeling system (Stratagene, La Jolla CA) according to the manufacturer’s instructions. Hybridization was performed as described previously [2]. Northern blots were prepared as described previously [34]. Blots were hybridized with radiolabeled probes in 7% SDS/0.5 M NaH2PO4, pH 7.2/2% dextran sulphate at 65°C overnight. The membrane was washed three times in 6XSSC/0.1% SDS, 2XSSC/0.1%SDS and 0.2XSSC/0.1%SDS for 15 min each time at 60°C. The signals were visualized by exposure to Kodak BioMax MS film (Kodak). P. knowlesi EcoRI-digested gDNA λ Zap II libraries were constructed and screened as described previously [2] and screened with a radiolabeled probe representing the 2.5 kb from the 5′ region of the pvrbp1 gene. Three positive clones containing a 1.5 kb EcoRI fragment were identified and sequenced, and subsequently compared and shown to be identical to sequence generated from the P. knowlesi genome sequencing project.
2.6. Pulse Field Gel Electrophoresis
P. knowlesi chromosomes were size fractionated by pulse field gel electrophoresis (PFGE) and sequential Southern blot analyses were performed using probes representing the central region of the pknbpxa and pknbpxb genes. Briefly, P. knowlesi chromosomes blocks were separated by PFGE in the CHEF-DR III system (BioRad, Hercules CA). Electrophoresis was performed in 0.8% chromosomal grade agarose using 1X TAE at 14°C. Chromosomes of Hansenula wingei were used as molecular mass standards. After electrophoresis the gel was denatured and transferred to a nylon support as described previously [2]. The pknbpxa and pknbpxb probes labeled by random priming were hybridized to the membranes following standard protocols and washed under high stringency conditions (2X SSC/0.1%SDS at 60°C).
2.7. Production of fusion proteins and rabbit antisera
Two recombinant His-tagged fusion proteins, rPkNBPXa and rPkNBPXb, were produced in the Gateway (Invitrogen, Carlsbad CA) and pET (Novagen, Madison WI) systems respectively. rPkNBPXa was produced as a 1.2 kb fragment by PCR amplification using primers rNBPXaF (5′ GGG GAC AAG TTT GTA CAA AAA AGC AGG CTC CAC GTT GTT GAA AAC TGA A 3′) and rNBPXaR (5′ GGG GACCAC TTT GTA CAA GAA AGC TGG GTT GTT CAA TTT TCC TTG CAA ATC 3′). A positive recombinant clone was expressed by addition of 0.3 M NaCl for 3 h at 37°C. The inclusion bodies were purified as described previously [35], separated by SDS-PAGE and electro-eluted for 16 h. rPkNBPXb was produced as a 1.14 kb fragment by PCR amplification using primers rNBPXbF (5′ctcgagAGCTTACGCAACATATTAAAC3′) and rNBPXbR (5′ggatccGTCATC ATCATCATTATCGTG3′). A plasmid with the expected sequence was expressed using 1mM IPTG in BL21-DE3 (Novagen, Madison WI) for 3 h at 37°C. The recombinant protein was bound to Ni-NTA agarose (Qiagen, Valencia CA) and purified under native conditions using 250 mM imidazol (Sigma, St. Louis MO). The purity of the eluted proteins was assessed by SDS-PAGE. The recombinant proteins were dialyzed against PBS before inoculation in New Zealand White Rabbits (Covance, Denver PA) for production of polyclonal antisera (Rab-anti-rPkNBPXa and Rab-anti-rPkNBPXb).
2.8. SDS-PAGE and western blotting
Infected erythrocytes containing mature, segmented schizonts where harvested and extracted with reducing sample buffer. The protein extracts were analyzed by SDS-PAGE on 5% polyacrylamide gels and transferred to protean nitrocellulose (Schleicher & Schuell, Keene NH) as described previously [2]. Membranes were probed with rabbit polyclonal antisera diluted 1:500, which had been depleted of antibodies against E. coli using standard adsorption procedures. Alkaline phosphatase-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, MD) was used for detection (1:5000) and bands were visualized by adding NBT/BCIP substrate (Promega, Madison WI).
2.9. Immunofluorescence assays and immuno-electron microscopy
For indirect immunofluorescence assay (IFA), air-dried, thin blood smears prepared with mature schizont-stage parasites were incubated with rab-anti-rPkNBPXa, and rab-anti-rPkNBPXb polyclonal antisera diluted 1:500. Goat anti-rabbit IgG antibodies conjugated to FITC were added and the slides were visualized using a fluorescence equipped microscope. For immuno-electronmicroscopy, merozoites were added to rhesus RBC’s, in the presence of 5 μg ml−1 cytochalasin B (Sigma, St. Louis MO), and were fixed in 0.1% (v/v) double-distilled glutaraldehyde and 2% (w/v) paraformaldehyde prepared in culture medium (RPMI, pH 7.2), for 20 min on ice, and then washed four times in ice-cold RPMI and dehydrated through a progressively low-temperature ethanol series before being infiltrated with LR White resin (EMSCOPE, London, United Kingdom). Resin polymerization was induced by ultraviolet light at room temperature for 48 h. Sections were immune-stained with the polyclonal sera anti-rPkNBPXa and rPkNBPXb, diluted 1:50 or 1:100 in 1% BSA/PBS, followed by Protein A conjugated to 10 nm gold particles, diluted 1:70 in 1% BSA/PBS (a kind gift by Dr Pauline Bennett, King’s College London). Parallel samples were treated with pre-immunization serum antibodies for control purposes. Sections were stained for 4 min with 2% (w/v) aqueous uranyl acetate. Sections were viewed and digital images were taken using a Hitachi 7600 electron microscope.
2.10. Erythrocyte Binding Assays
Erythrocytes infected with schizont stage parasites were purified by centrifugation on Percoll gradients, as previously described [36]. The purified schizont-infected erythrocytes were placed into culture in-vitro and allowed to grow overnight as previously described [1, 5] until erythrocytes containing mature schizonts formed merozoites had completely ruptured. Cultures were centrifuged at 4000 rpm and the supernatants were removed and stored in liquid nitrogen. 500 mL of culture supernatants were rotated with 1 × 109 erythrocytes at room temperature for 4 h. The cells were washed twice by layering over and subsequent centrifugation through Dow Corning 550 silicone fluid. Bound proteins were eluted in 50 ul of 5× RPMI at room temperature and harvested by centrifugation. The resulting proteins were analyzed by SDS-PAGE on Pre-Cast 5% polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad, Hercules CA). Membranes were probed with rabbit polyclonal antisera diluted 1:500 for 2 h. Alkaline phosphatase-conjugated goat anti-rabbit IgG diluted 1:5000 (Promega, Madison WI) was used for detection of bound antibodies. Bands were visualized by addition of NBT/BCIP substrate (Promega, Madison WI) to the nitrocellulose strips.
2.11. Bioinformatics and sequence analysis
TBlastN searches of preliminary and finalized P. knowlesi sequence database (8x coverage) at Sanger Center (http://www.sanger.ac.uk), the P. vivax sequencing project at TIGR Center (http://www.tigr.org) and PlasmoDB (www.PlasmoDB.org) database were performed by using translated sequence from pvrbp1 and pvrbp2 and the newly identified pkrbl genes. Signal peptide cleavage sites and transmembrane domains were predicted with SignalP V2.0 (www.cbs.dtu.dk/services/SignalPV2.1) and TMpred (www.ch.embnet.org/software/TMPRED_form.html) software, respectively. Multiple alignments of nucleic acid and protein sequences were generated using ClustalW 1.8.2 with all default parameters. Tree topology was generated using the Neighbor-Joining algorithm in the MEGA 2.2 software on distances calculated with a Poisson correction [37].
3. Results
3.1. The ortholog of pvrbp1 is a highly degenerated pseudogene in P. knowlesi, but a functional full-length gene in the sister taxon P. coatneyi
To identify and characterize a pvrbp1 ortholog in P. knowlesi, Southern hybridization assays were initially performed under high stringency conditions, and no hybridization signals were observed, suggesting that a highly related intact orthologous gene was not present in this species [2]. However, when a 2.5 kb pvrbp1 probe representing the 5′ end of the gene was subsequently evaluated using lowered stringency conditions, clear distinct bands were detected in each gDNA sample digested with restriction enzymes (Fig. 1A). In contrast, pvrbp1 probes representing other downstream regions of the gene, also evaluated at lower stringencies, did not result in hybridization signals (data not shown).
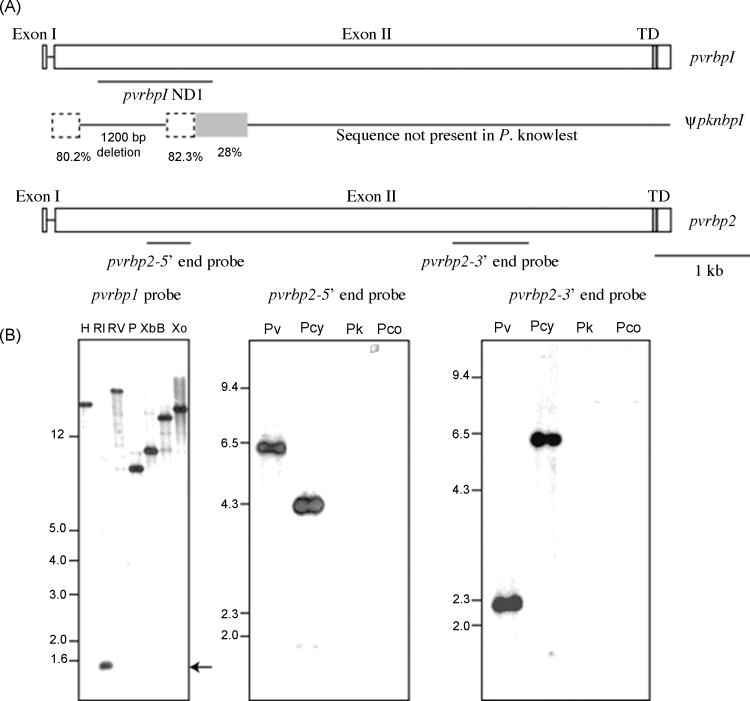
Figure 1.
The P. knowlesi gene orthologous to pvrbp1 in P. vivax is a pseudogene. (A) Schematic representation of pvrbp1 and pvrbp2 genes and the remnant pknbp1 locus with locations of DNA probes used in Southern blot analysis. The comparison of the 1.5 kb fragment of the nbp1 locus in P. knowlesi with P. vivax rbp1 revealed two remnant regions (dashed lines) in the P. knowlesi gene ψpknbp1 comprising part of the intron and 5′ end of coding region. (B) Southern blot analysis (bottom left) of P. knowlesi H strain gDNA digested with HindIII (H), EcoRI (RI), EcoRV (RV), PstI (P), XbaI (Xb), BamHI (B) and XhoI (Xo) hybridized with a 5′-end pvrbp1 probe identifying a 1.5 kb fragment after EcoRI restriction analysis (arrow). P. vivax (Pv), P. cynomolgi (Pcy), P. knowlesi (Pk), and P. coatneyi (Pc) gDNA digested with HindIII restriction enzyme was analyzed by Southern blot analysis using a 5′-end (pvrbp2–5′end) or a 3′-end (pvrbp2–3′end) probe from the pvrbp2 gene, only P. cynomolgi showed cross-hybridization under moderately stringent conditions.
To evaluate this putative related but divergent ortholog further, the 1.5 kb EcoRI fragment detected by Southern hybridization was cloned and sequenced. Instead of an expected single ORF, multiple ORFs were detected. In addition, 1200 bases of sequence present in pvrbp1 and the orthologous genes in P. cynomolgi, P. coatneyi and P. fragile [10], though expected, were not present in the degenerate pknbp1 sequence. In conclusion, a total of 626 bp of the 1.5 kb cloned sequence showed identities of 80.2% and 82.3% when aligned with the pvrbp1 sequence. The remaining 900 bp of downstream sequence showed no apparent relationship (<28% identity) to pvrbp1 (Fig. 1B). DNA samples from the Hackeri and Philippine strains of P. knowlesi also hybridized with the 1.5 kb DNA probe (data not shown). Although some single nucleotide polymorphism was observed, as would be expected between strains of parasites, amplified gene fragments corresponding to the 1.5 kb EcoRI clones were essentially identical (99% identity) and also showed no continuous ORF.
Contig sequences recently made available in the P. knowlesi (H strain) genome database (http://www.sanger.ac.uk/Projects/P_knowlesi/) [31] are consistent with our experimental analyses, supporting the presence of a degenerated pseudogene. Our searches of the genome database identified an additional 200 bp upstream corresponding to pvrbp1 sequence, but no related sequences were present downstream. Thus, we conclude that the ortholog of pvrbp1 in P. knowlesi (H strain, Hackeri and Philippine strains) is a highly degenerate pseudogene, with only short segments of the 5′ region maintained in the genome for each strain tested.
P. coatneyi is a sister taxon of P. knowlesi [29], yet it displays unique biological and phenotypic characteristics distinct from P. knowlesi (reviewed in [38]). In contrast to P. knowlesi data, the Southern hybridizations suggested that a complete ortholog of pvrbp1 existed in the P. coatneyi genome (data not shown). To characterize the P. coatneyi rbp1 (pcrbp1) gene, overlapping gene regions were amplified by PCR using primers designed from the pvrbp1 sequence, cloned and sequenced (Genbank Accession number DQ973816). This sequence proved to be highly homologous to the pvrbp1 gene, showing that pcrbp1 is an intact, likely functionally expressed gene. The pcrbp1 homolog exhibits a nucleotide identity of 84% with the pvrbp1 gene, and the deduced protein sequence shows 75 % identity to PvRBP1. P. fragile, also a genetically close parasite in the simian malaria clade, has an intact and presumably functional nbp1 gene (Genbank Accession number DQ973815).
3.2. A pvrbp2 ortholog is not present in the P. knowlesi or P. coatneyi genomes
To determine if a rbp2 ortholog was in P. knowlesi (and P. coatneyi), enzyme restricted P. knowlesi H strain and P. coatneyi DNA were analyzed by Southern blotting hybridization using 5′ through 3′-end amplicons of the pvrbp2 gene as probes. No distinct bands could be discerned using conditions of lowered stringency, which previously permitted easy detection of the ψpknbp1 gene or the pvrbp1 and pvrbp2 orthologs in P. cynomolgi [10] (Fig. 1B). These data indicate that no highly similar counterpart or ortholog genes for pvrbp2 exist in P. knowlesi or P. coatneyi, and are at least for P. knowlesi, in complete agreement with BLAST searches of the P. knowlesi genome database [31].
3.3. Characterization of two novel P. knowlesi rbl genes, denoted pknbpxa and pknbpxb
To search for possible distantly related rbl genes in P. knowlesi, which may serve as alternatives to pvrbp2, we used a combination of methods including library screening based on pvrbp2 probes, PCR amplification based on pvrbp2 gene sequences and database mining. A probe of about 1.5 kb representing the 3′ end of the pvrbp2 gene hybridized weakly to P. knowlesi gDNA in Southern blots using low stringency conditions (data not shown). This sequence was used to BLAST the P. knowlesi sequencing project database using both nucleotide and translated sequences. ORFs on two separate contigs showed significant, but not particularly strong scores by BLAST based searching. The sequences were used as templates for further probe design and PCR based amplification of gDNA. Screening P. knowlesi EcoRI digested-gDNA λ Zap II libraries identified two clones, pknbpxa and pknbpxb. The sequences of the two cloned DNA fragments were utilized to search the P. knowlesi sequence database to extend the primary sequences and fill in the gaps.
The pknbpxa clone contains an EcoRI-fragment of 6641 nt with an ORF of 5742 nt but missing the expected exon I, intron, and 5′ end of exon II. PCR from lambda phage and BLAST search of the P. knowlesi database provided enough further sequence to design gene-specific reverse primers for RT-PCR experiments and determination of the exon/intron junctions. The 9578 nt contig encodes exon I from nt 130 to nt 184, a short intron of 186 nt, and an exon II from nt 371 to nt 8679. Thus, the total coding sequence of pknpbxa is 8364 nt. The predicted PkNBPXa protein is 2788 amino acids, the putative signal peptide cleavage site located between S27 and E28 and the transmembrane region predicted to be 21 amino acids long (amino acid 2724 to amino acid 2745) (Fig. 2A).
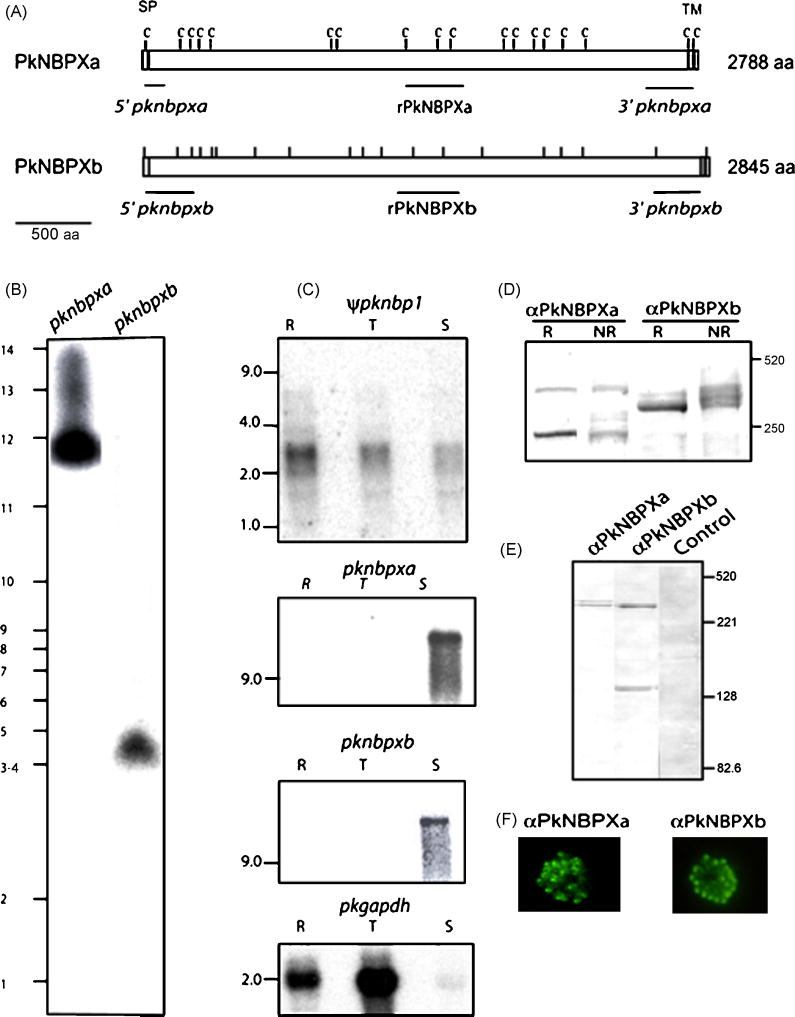
Figure 2.
(A) Schematic of the P. knowlesi PkNBPXa and PkNBPXb RBL proteins with cysteine residues (tick marks), positions of the signal peptides (SP), a large extracellular domain and a transmembrane domain (TM) shown. Probes used for chromosomal Southern and Northern blot hybridizations are indicated in italics and the fragments expressed as recombinant proteins are indicated in bold letters. (B) The pknbpxa and pknbpxb genes were localized in chromosomes 3–4 and 12, respectively, by PFGE using the pknbpxa and pknbpxb probes. (C) Total RNA from ring stages (R), late trophozoites and early schizonts (T) and late schizonts (S) was hybridized with radiolabeled probes representing the 5′ end of the ψpknbp1 gene, and the central regions of pknbpxa and pknbpxb genes using high stringency conditions. Apparent ψpknbp1 partial transcripts (~3 kb) were detected in ring, trophozoite and schizont stages, compared to pknbpxa and pknbpxb transcripts of >9kb, which were detected only in the matured schizonts. The Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as positive control (pkgapdh). PkNBPXa and PkNBPXb were detected by western blot in P. knowlesi-schizont-infected erythrocytes solubilized with SDS-PAGE sample buffer with (R, reduced) or without (NR, non-reduced) 2-mercaptoethanol (D), and in supernatant collected after schizont rupture (E). The NBPXa1 antisera recognized specific bands of ~300 kDa and 250 kDa, and the NBPXb1 antisera recognized bands of ~300k Da and 140 kDa (D and E). PkNBPXa and PkNBPXb native proteins were identified by IFA using rabbit antisera PkNBPXa1 or PkNBPXb1, respectively. The robust single dot pattern typical for the PvRBPs at the apical pole was observed in segmented schizont-stage parasites; pre-immune sera were used as negative control (F).
The pknbpxb clone contains 9479 nt with a single ORF of 8681 nt, but no clear typical small exon I or intron consensus splice site sequence. We therefore generated 5′ RACE clones to verify the 5′UTR sequence, signal peptide-encoding exon I and the exon/intron junctions. Thus, PkNBPXb is encoded by exon I (nt 49 to nt 103) and by exon II (nt 347 to nt 8826) with an intron of 243 nt, which has a 3′ AAG splice site instead of the usual TAG or CAG 3′ splice motifs. Thus, the total coding sequence of pknpbxb is 8535 nt. The PkNBPXb protein has 2845 amino acid residues with a predicted signal peptide cleavage site between C21 and K22 and a hydrophobic transmembrane domain between amino acid 2784 and amino acid 2801 (Fig. 2A).
Overall, the gross structural characteristics of PkNBPXa and PkNBPXb proteins are remarkably similar to those of PvRBP1 and PvRBP2 and the other members of the RBL family. Similarly, these proteins are likely membrane-anchored as determined by a hydrophobic region at the carboxy-terminal ends (Fig. 2A). Chromosomal mapping localized pknbpxa in chromosome 12 and pknbpxb in chromosomes 3–4 (Fig. 2B). Currently, the pknbpxa gene is annotated in the P. knowlesi database as two unlinked fragments (PKH146970 and PKH146980). A contig containing the pknbpxb gene assembly is not currently retrieved through BLAST searches, although it is noted that shotgun sequences for this gene are present at in the Sanger database and, surprisingly, cover the complete gene.
3.4. The pknbpxa and pknbpxb genes are transcribed and expressed in mature schizonts
Transcription of both pknbpxa and pknbpxb was verified by RT-PCR using schizont stage RNA (data not shown). Northern blot analyses were then performed and demonstrated the schizont-specific nature of the transcripts. Total RNA was extracted over the 24-hour life cycle period from synchronous parasites representing ring stages (R), late trophozoite and early schizont stages with two nuclei (T), and mature schizonts with eight or more nuclei (S). Large transcripts (> 9 kb) were detected for both genes, suggesting the presence of long untranslated regions, and they were present only in the mature schizont-stage RNA samples (Fig. 2C).
Native and reduced protein was detected using rabbit anti-rNBPXa and rabbit anti-rNBPXb antisera on mature schizont extracts, and culture supernatants harvested 14 h after erythrocyte rupture by western immunoblotting. Both PkNBPXa and PkNBPXb are predicted to be high molecular weight proteins of 324 kDa and 334 kDa, respectively. The two polyclonal antisera recognized proteins of ~300 kDa as predicted by the software algorithms without noting any migration differences between the reduced and non-reduced samples. However, rabbit anti-rPkNBPXa also recognized an additional band of 220 kDa which could be a processed fragment of PkNBPXa or attributed to spurious reactivity with the erythrocyte protein, spectrin (although there is no evidence of this from immuno-fluorescence or immuno-electronmicroscopy, see below). Rabbit anti-rNBPXb recognizes an additional band of ~140 kDa in these immunoblots (Fig. 2D). Soluble PkNBPXa and PkNBPXb of the expected size (~300 kDa) were also detected in culture supernatants by western blot by using rabbit anti-rNBPXa and rabbit anti-rNBPXb antisera (Fig. 2E).
3.5. PkNBPXa and PkNBPXb localize to the apical microneme organelles of merozoites
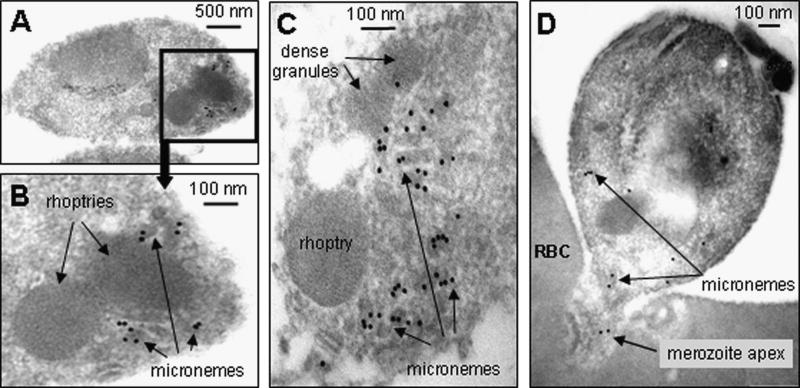
Rabbit antibodies specific for rPkNBPXa and rPkNBPXb located the native proteins at the apical end of merozoites in segmented schizont stage parasites by IFA (Fig. 2F). A single robust dot of fluorescent signal sometimes seemingly capping the merozoite, which is typical for the PvRBPs at the apical pole of P. vivax merozoites [2], was also observed in P. knowlesi mature segmented schizonts with these two antisera. No reactivity was detected with pre-immune rabbit sera and with the P. knowlesi RBL antisera on earlier stages of parasite development. Immuno-electron microscopy of ex-vivo parasite preparations showed that anti-PkNBPXb was predominantly located in the micronemes (Fig. 3A–D). Anti-rPkNBPXa antibodies also reacted with micronemes, but less strongly (data not shown). Significantly, in the invading merozoite (Fig. 3D), very little labeling remains within the merozoite, mostly restricted to micronemes that have apparently failed to locate apically. This suggests that PkNBPXb had already been secreted, as would be expected if the protein is important in early red cell adhesion.
Figure 3.
Immuno-electron microscopic localization of the PkRBL protein, PkNBPXb. In A – C, PkNBPXb is shown to be strongly localized to the apical micronemes of free P. knowlesi merozoites, using rabbit anti- NBPXb anti-serum (1:50) as primary antibody, and Protein A -10 nm gold for detection.. In an invading merozoite (D), very little labelling remains (rabbit anti-NBPXb anti-serum 1:100 dilution used), and is mostly detected on micronemes that have apparently failed to locate apically, suggesting that the RBL had already been mostly secreted and lost, as expected if the protein is important in red cell adhesion. Note the absence of general labelling around the merozoite perimeters in all examples. RBC – red blood cell.
3.6. PkNBPXa and PkNBPXb bind to host erythrocytes
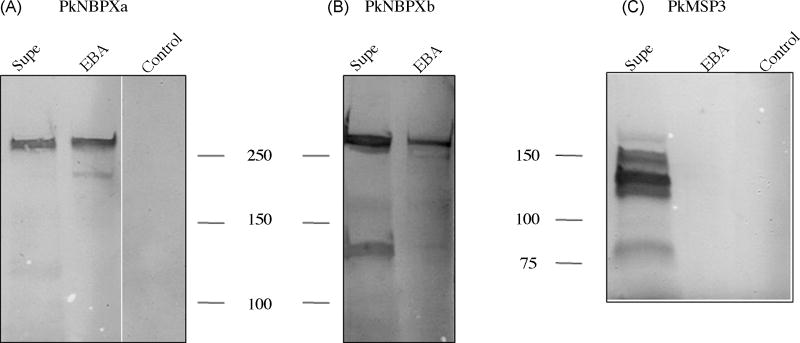
The capacity of the two expressed RBL proteins to bind to rhesus macaque erythrocytes was determined by traditional erythrocyte binding assays. Intense bands were detected in immunoblots probed with rabbit anti-rNBPXa and rabbit anti-rNBPXb antisera (Fig. 4A and 4B). No bands were detected with erythrocytes that were incubated without supernatant and subsequently washed and eluted under the same conditions as the samples incubated with culture supernatants. There were no differences detected in the migration of PkNBPXa and PkNBPXb proteins between reduced and non-reduced samples (data not shown). To ensure that the binding detected was not due to carryover contamination, immunoblots were also probed with antiserum against the 140kDa PkMSP3 [39]. PkMSP3 is known to be present in culture supernatants but does not bind to the surface of erythrocytes. As expected, antibody reactivity with PkMSP3 was detected when testing culture supernatants (~140kD) but not the erythrocyte binding assay eluates (Fig. 4C and unpublished data). This indicates the binding of expressed P. knowlesi RBLs to rhesus macaque erythrocytes is specific.
Figure 4.
PkNBPXa and PkNBPXb bind to host erythrocytes. Culture supernatants (Supe) containing soluble proteins released from free merozoites were electrophoresed by SDS-PAGE and transferred to nitrocellulose membranes as were protein samples eluted from rhesus macaque erythrocytes after incubation with supernatants in binding assays (EBA). Western blots using NBPXa1 or NBPXb1 antisera demonstrate that PkNBPXa and PkNBPXb bind to rhesus erythrocytes. PkMSP3 140 was abundantly detected in the culture supernatants by PkMSP3 140 rabbit antisera but was not detected in the eluted protein samples. Rhesus monkey erythrocytes were incubated within culture medium (Control) and eluted under the same conditions as erythrocytes incubated with culture supernatants. No protein bands were detected by any of the antisera (data not shown for PkNBPXb).
3.7. Identification of a rbl gene in P. vivax that is an ortholog to pknbpxa
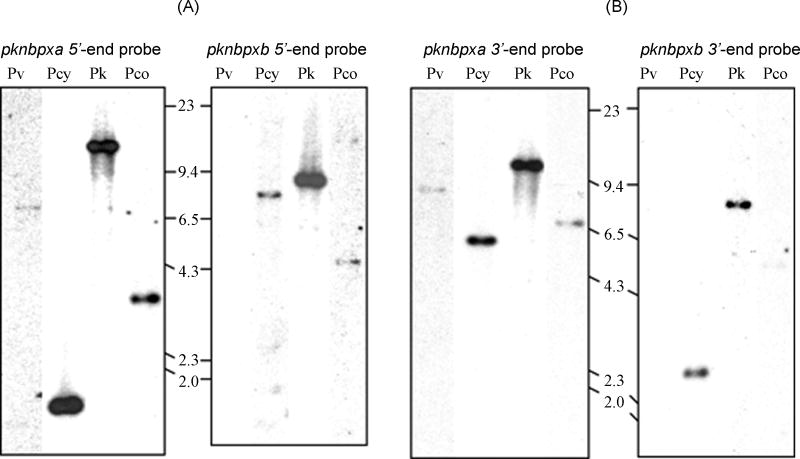
P. vivax gDNA was examined by Southern hybridization with fragments of the 5′ and 3′ portions of the P. knowlesi rbl genes as probes. Weak bands of approximately 7.0 kb were noted on P. vivax digested gDNA when probed with a pknbpxa 5′ DNA fragment (Fig. 5A) or approximately 9.0 kb, when probed with a pknbpxa 3′ fragment (Fig. 5B). These data suggested the presence of at least one more rbl gene more closely related to pknbpxa in this species. No hybridization was observed when pknbpxb probes were used at low stringency on the P. vivax DNA. Database search of the P. vivax genome sequencing project (www.plasmodb.org/P.vivax) using pknbpxa sequences provided a contig of 11726 bp containing several large ORFs totaling about 8366 bp. Though the gene predicted for the protein has a potential consensus start methionine (TATAATG), the expected intron/exon 5′ and 3′ consensus splicing sequences were not readily observed, and two stop codons at positions 5503 and 7071 interrupt a potential 8366 bp ORF [17]. Although pknbpxb was not detected in P. vivax, homologous sequences to both pknbpxa and pknbpxb were detected by Southern hybridization in the simian malaria parasites, P. cynomolgi and P. coatneyi (Fig. 5A and 5B).
Figure 5.
Genes homologous to pknbpxa and pknbpxb are in P. vivax and other related simian malaria species. P. vivax (Pv), P. cynomolgi (Pcy), P. knowlesi (Pk), and P. coatneyi (Pco) gDNAs were digested with the restriction enzyme HindIII, blotted and probed with the 5′ (Panel A) and 3′ (Panel B) probes from pknbpxa and pknbpxb genes as indicated in Fig. 2. Sizes are in kilobases. The hybridization temperature was 60°C followed by low stringency washes as described in the materials and methods. The 5′ and 3′ probes from pknbpxa hybridized strongly with P. vivax and P. cynomolgi, and with less intensity to P. coatneyi gDNA. The 5′ and 3′ probes from pknbpxb hybridized with P. cynomolgi and P. coatneyi gDNA but not with P. vivax gDNA.
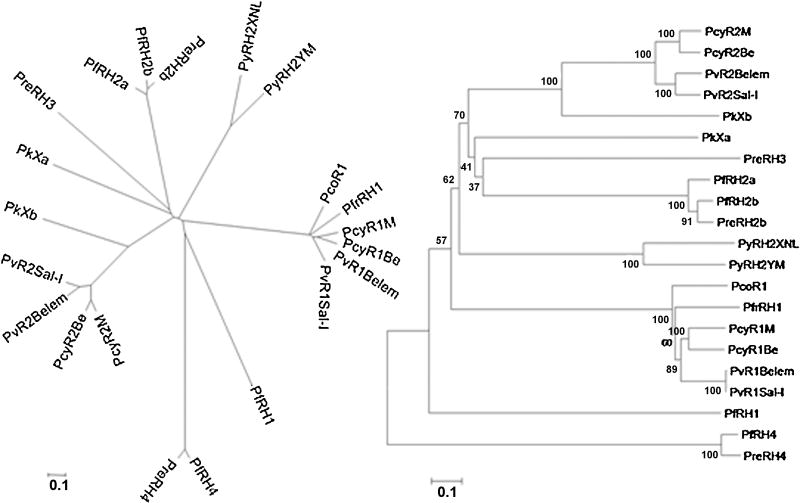
3.8. Phylogenetic relationship of pknbpxa and pknbpxb within the RBL family of proteins
To achieve a comprehensive updated phylogenetic analysis of known rbl genes, the rbl sequences from human, simian, chimpanzee, and rodent malaria parasites were aligned and analyzed. These alignments included orthologs of pvrbp1 from P. coatneyi (GenBank accession number DQ973816) and P. fragile (GenBank accession number DQ973815). The analysis of the RBL proteins demonstrates that these proteins comprise a large family of two distinct groups, namely, PvRBP1-like and PvRBP2-like homologs. Functional pvrbp1-like genes have been identified in P. falciparum, P. fragile, P. cynomolgi and P. coatneyi, but are degenerated pseudogenes in P. knowlesi and P. reichenowi. In contrast, various paralogs and orthologs of pvrbp2 have been identified in P. vivax, P. falciparum, P. reichenowi, P. cynomolgi, P. knowlesi, and P. yoelii (Fig. 6). Importantly, the genes designated here as pknbpxa and pvrbpxb share a paralogous relationship with pvrbp2, and pknbpxa may constitute a new rbl2 subgroup sharing greater affinity with pfnbp3/rh3 and pfnbp2a-b/rh2a-b. Though quite divergent, pknbpxb is clearly more related to rbp2 of P. vivax and P. cynomolgi than is pknbpxa.
Figure 6.
Phylogenetic relationships among members of the RBL invasion ligand family. Unrooted neighbor-joining tree obtained from the alignment of RBL protein sequences from P. vivax, P. cynomolgi, P. coatneyi, P. knowlesi, P. fragile, P. yoelii, P. falciparum, and P. reichenowi. The cladogram (right) was generated using the Neighbor-Joining method following a Poisson model with pairwise deletion from MEGA 4 software. Bootstrap values for 1000 replicas are indicated in the tree. Distinct groups can definitely be identified for RBL1, RBL2, RBL3 and RBL4.
4. Discussion
Here we report the experimental characterization of the rbl family in P. knowlesi and show it is comprised of one pseudogene related to pvrbp1 and two functional intact members, which we have called pknbpxa and pknbpxb. These two functional genes encode large RNA transcripts and high molecular weight proteins as predicted, and we have confirmed that they become expressed in the late schizont stage. Their products localize to the microneme organelles of mature merozoites and bind specifically to rhesus monkey erythrocytes.
Our initial identification of these genes was through hybridization of pvrbp gene fragments to restriction enzyme digests of P. knowlesi gDNA (and other species), followed by sequencing and RT-PCR to fully characterize the genes. These experimental findings have been important alongside the development of the P. knowlesi genome project [31], since this genome-wide sequencing and annotation effort alone failed to recognize the ψpknbp1 pseudogene and the pknbpxb gene reported here, and annotate them accordingly. On the other hand, two fragments of the pknbpxa gene were annotated by the genome project, but not recognized as segments of one gene. Our experimental and bioinformatic work was required to identify unannotated members of the rbl family (ψpknbp1 and pknbpxb), correctly link annotated fragments of the pknbpxa gene, and confirm all the sequences, structures, and importantly, their expression and functional properties.
The P. knowlesi ortholog of pvrbp1 is only a relic pseudogene (ψpknbp1) in the H, Hackeri, and Philippine strains of P. knowlesi. What is left of this pseudogene has a high level of identity with ~0.8 kb of the 5′ end of pvrbp1 (a gene of ~8.6 kb) with no ORF. The ψpknbp1 sequence downstream has been entirely lost in relationship with rbp1 gene structure, presumably through an ancient recombination event, or extensive drift over time through mutations. Interestingly, this pseudogene is differentially transcribed as a small fragment that is consistent with the size of the gene remnant. This truncated transcript is primarily produced during ring-stage parasite growth, as opposed to schizont stage development for bona fide full-length rbl genes. The presence of rbl pseudogenes has been recognized before in P. falciparum and P. reichenowi, and different levels of degeneracy have been observed. For example, the P. falciparum rbl3 (pfnbp3/rh3) gene transcript is present, yet not translated into protein in the 3D7 strain due to two frameshift mutations [8]. The P. reichenowi rbl1 (prnbp1/rh1) gene, however, is much more degenerated containing numerous frameshifts leading to termination codons throughout the sequence [11].
Regardless of the mechanism by which rbl pseudogenes are generated and maintained, it has now become evident that P. knowlesi parasites can survive without an rbl1 ortholog. Furthermore, this apparently extends also to a pvrbp2 ortholog as our data shows that the P. knowlesi genome lacks completely a direct counterpart of pvrbp2, although an ortholog is in P. cynomolgi. Initial hybridization experiments were surprising, since the closely related simian malaria parasite P. cynomolgi [10], and the sister taxon P. coatneyi (unpublished), clearly had orthologs of rbp1 and rbp2 in their genomes. Thus, only two rbl genes function in P. knowlesi, pknbpxa, reported in the genome database as PKH_146970 and PKH_146980, and pknbpxb as reported here but remains unannotated. Clearly, the loss of the rbp1 and rbp2 orthologs in P. knowlesi is a recent evolutionary event occurring sometime after the divergence of P. coatneyi and P. knowlesi, perhaps two to three million years ago [40, 41].
Our data indicate P. knowlesi has evolved to maintain just two functioning members of the rbl gene family (pknbpxa and pknbpxb), while losing the rbp1 and rbp2 orthologs and perhaps others present in the other members of the vivax-simian malaria clade of Plasmodium species. In contrast, most other species of Plasmodium seem to have a larger repertoire of expressed rbl genes, including P. vivax. P. yoelii has at least a dozen rbl genes that may be functional; all falling within the rbp2-like grouping of rbl genes. P. falciparum has a group of six genes of which five are deemed functional and likely serve to interact with alternative receptors in whatever role they play in erythrocyte invasion. P. vivax also apparently has more than two functional rbl genes.
We have already noted here that the P. vivax (Sal I) genome has an nbpxa ortholog gene, although it seems to be a non-functional pseudogene. Hypothetical translation of the long ORF, across the stop codons and without frameshifts gives an amino acid identity of 63% between PkNBPXa and the P. vivax homologue. It remains to be demonstrated if the pvrbpxa gene is transcribed, as is the nbp3/rh3 gene in P. falciparum [8]. The two frameshift mutations and lack of evident intron splice sites indicate this is likely a pseudogene and probably does not produce a functional protein. This gene has been designated as pvrbp3 [16]. At least four other gene sequences in the P. vivax (Sal I) genome [17] are annotated as rbp genes, besides those discussed above (pvrbp1, pvrbp2, and pvrbp3), but none are orthologs of pknbpxb. Importantly, of these four additional genes designated as pvrbp1b, pvrbp2a, pvrbp2b and pvrbp2d, two, pvrbp1b and pvrbp2d are likely non-functional genes with mutations that disrupt the reading frame. Thus, P. vivax may have a larger set of functional rbl genes than previously realized, but less than recently reported [16], whereas P. knowlesi has reduced its rbl allotment to only two ligands. Nevertheless, P. knowlesi has compensated by retaining its invasion ligand diversity through three dbl genes, while P. vivax only has one.
The pknbpxa and pknbpxb genes have the signature structure of rbl genes but they do not exhibit a high degree of homology between them, as is also the case for the rbp1 and rbp2 genes in P. vivax or P. cynomolgi [2, 4, 10]. The absence of the rbp1 and rbp2 family members in P. knowlesi may simply be a reflection of the adaptation of P. knowlesi to invade all red blood cells and not be restricted to reticulocytes like P. vivax and P. cynomolgi. However, P. coatneyi does not preferentially invade reticulocytes and it has a complete rbl1 gene structure that is functional from the perspective that there is a complete ORF. The presence of distant members in P. knowlesi may also simply be indicative of a dynamic evolutionary process and the parasite’s strategies to maintain a minimum number of functional members of this gene family. Experiments aiming to disrupt pknbpxa and pknbpxb genes support the hypothesis that the encoded proteins are essential for P. knowlesi survival, as no parasites were retrieved after selection for disrupted parasite phenotypes in in vivo and in vitro experiments, although disruptions of other genes performed at the same time were successful in each case (unpublished data).
Phylogenetically, these two rbl genes have helped to define new ortholog genes in P. vivax, P. cynomolgi, P. coatneyi and P. fragile, which complement the pvrbp2 subgroup of the RBL family (Fig. 6). The data presented here and contained in genome databases suggest that the rbl gene family in Plasmodium is dynamic, in that gene duplications, recombination events and mutations have been common occurrences; yet all species investigated to date seem to minimally maintain at least two bona fide family members.
A micronemal location is evident for PkNBPXb by IEM analysis, and a similar pattern is observed for PkNBPXa, though the antibody reactivity was most clear for PkNBPXb. The micronemal location of these proteins contrasts with the rhoptry localization reported for PfNBP2a/b [19] and Py235 [42]. IFA patterns in merozoites for RBP1 and 2, and for RH1 and RH4 that colocalize with micronemal invasion ligands such as DBP, EBA-175 and AMA1 suggest a microneme location for these rbl gene products. Further studies are necessary to thoroughly evaluate the locations of different RBLs in and between different species of Plasmodium to reliably locate them by IEM. If RBLs are stored in different locations as seems to be the case (i.e. in the micronemes versus rhoptries), or potentially in different microneme organelles, this may reflect different functional roles in invasion attributable to specific family members. More so than rhoptries, the micronemes appear to store invasion ligands that interact with specific RBC receptors to initiate actions during the early stages of recognition and entry. Precise localization data could be helpful for formulating and testing hypotheses relating to the function of the RBLs in the cascade of events that dictate erythrocyte invasion.
RBL proteins have been hypothesized to be critical components in host cell identification and invasion, and accumulating evidence with counterpart molecules present in each malaria species examined supports this premise. As their names denote, reticulocyte or normocyte binding proteins are predicted to function by binding to (as yet undefined) receptors on erythrocytes. The specific adhesion of RBL proteins to host erythrocytes in in vitro erythrocyte binding assays has been demonstrated for P. vivax (with reticulocyte specificity) [2], P. falciparum [5, 20–22], and P. yoelii [12], and here we demonstrate that the expressed RBLs of P. knowlesi bind to erythrocytes from the rhesus macaque model.
P. knowlesi is the traditional model for investigating Plasmodium merozoite invasion of erythrocytes [1, 43, 44]. Ex-vivo P. knowlesi merozoites have a biological half-life of about 20 minutes and remain viable in the extracellular milieu much longer than other species [43]. This trait allows for the unique experimental capture and analysis of merozoites during different stages of attachment and entry of erythrocyte host cells [1, 43, 44] (and see Fig. 3D). Of interest, our IEM data shown here, depicting an invading parasite suggests that the RBLs are released from microneme storage at a time before the merozoite begins to enter the host red blood cell (Fig. 3D). It is also worth noting that labeling was absent from the general surface of free merozoites, consistent with the notion that the RBLs are likely to function in apically-related events rather than initial red blood cell adhesion. Considering the likelihood that binding of the RBLs may be critical for invasion of erythrocytes, further investigation to define the binding properties and characteristics of these molecules is warranted.
With the goal of using the P. knowlesi model to improve our understanding of the invasion of red blood cells, we set out to define the rbl genes and proteins present in this species. Moreover, the importance of the RBL family as potential vaccine candidates is emphasized by the parasite’s maintenance of functional members, ability to bind erythrocytes, proposed role in signaling release of other merozoite proteins early during invasion [1–4], and evidence suggesting their critical role in parasite survival. P. knowlesi has become a public health concern since a series of recent reports have confirmed the zoonotic transmission in human populations throughout South East Asia, and severe disease and four deaths have so far been attributed to these infections [25–27]. It is possible that expressed RBLs characterized in this report are the ligands used to gain entry into human erythrocytes, as well as the erythrocytes of simian hosts, be they the experimental rhesus monkey host (Macaca mulatta) or the natural host (M. fascicularis or M. nemestrina) in South East Asia. Further studies are in progress to define the host cell binding specificities of the P. knowlesi RBLs, identify the P. knowlesi RBL red blood cell binding domain(s), and maximize the use of this model parasite to pave the way for the possible inclusion of RBL molecules in future malaria vaccines.
Acknowledgments
This work was supported by National Institutes of Health grant # R01AI247 and the Yerkes National Primate Research Center Base grant #RR-00165 awarded by the National Center for Research Resources of the National Institutes of Health. The authors thank Claudia Corredor-Medina for technical assistance and Vladimir Corredor for helpful discussions. The authors are also indebted to Graham Mitchell (Guy’s, London) with respect to provision of laboratory facilities for ARD, and to John Hopkins and the Centre for Ultrastructural Imaging (King’s College London) for electron microscopic assistance.
Footnotes
Note: Nucleotide sequence data reported in this paper are available in the EMBL, GenBank™ and DDBJ databases under the accession numbers AY151130, EU867791, EU867792, DQ973816, and DQ973815.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Galinski MR, Dluzewski AR, Barnwell JW. A mechanistic approach to merozoite invasion of red blood cells: merozoite biogenesis, rupture, and invasion of erythrocytes. In: Sherman IW, editor. Molecular approaches to malaria. ASM Press; 2005. pp. 113–168. [Google Scholar]
- 2.Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69(7):1213–26. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 3.Galinski MR, Barnwell JW. Plasmodium vivax: Merozoites, invasion of reticulocytes and considerations for malaria vaccine development. Parasitol Today. 1996;12(1):20–9. doi: 10.1016/0169-4758(96)80641-7. [DOI] [PubMed] [Google Scholar]
- 4.Galinski MR, Xu M, Barnwell JW. Plasmodium vivax reticulocyte binding protein-2 (PvRBP-2) shares structural features with PvRBP-1 and the Plasmodium yoelii 235 kDa rhoptry protein family. Mol Biochem Parasitol. 2000;108(2):257–62. doi: 10.1016/s0166-6851(00)00219-x. [DOI] [PubMed] [Google Scholar]
- 5.Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J Exp Med. 2001;194(11):1571–81. doi: 10.1084/jem.194.11.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc Natl Acad Sci U S A. 2000;97(17):9648–53. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Triglia T, Thompson J, Caruana SR, Delorenzi M, Speed T, Cowman AF. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect Immun. 2001;69(2):1084–92. doi: 10.1128/IAI.69.2.1084-1092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor HM, Triglia T, Thompson J, Sajid M, Fowler R, Wickham ME, Cowman AF, Holder AA. Plasmodium falciparum homologue of the genes for Plasmodium vivax and Plasmodium yoelii adhesive proteins, which is transcribed but not translated. Infect Immun. 2001;69(6):3635–45. doi: 10.1128/IAI.69.6.3635-3645.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneko O, Mu J, Tsuboi T, Su X, Torii M. Gene structure and expression of a Plasmodium falciparum 220-kDa protein homologous to the Plasmodium vivax reticulocyte binding proteins. Mol Biochem Parasitol. 2002;121(2):275–8. doi: 10.1016/s0166-6851(02)00042-7. [DOI] [PubMed] [Google Scholar]
- 10.Okenu DM, Meyer EV, Puckett TC, Rosas-Acosta G, Barnwell JW, Galinski MR. The reticulocyte binding proteins of Plasmodium cynomolgi: a model system for studies of P. vivax. Mol Biochem Parasitol. 2005;143(1):116–20. doi: 10.1016/j.molbiopara.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Rayner JC, Huber CS, Galinski MR, Barnwell JW. Rapid evolution of an erythrocyte invasion gene family: the Plasmodium reichenowi Reticulocyte Binding Like (RBL) genes. Mol Biochem Parasitol. 2004;133(2):287–96. doi: 10.1016/j.molbiopara.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Ogun SA, Holder AA. A high molecular mass Plasmodium yoelii rhoptry protein binds to erythrocytes. Mol Biochem Parasitol. 1996;76(1–2):321–4. doi: 10.1016/0166-6851(95)02540-5. [DOI] [PubMed] [Google Scholar]
- 13.Keen JK, Sinha KA, Brown KN, Holder AA. A gene coding for a high-molecular mass rhoptry protein of Plasmodium yoelii. Mol Biochem Parasitol. 1994;65(1):171–7. doi: 10.1016/0166-6851(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 14.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124(4):755–66. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez M, Lustigman S, Montero E, Oksov Y, Lobo CA. PfRH5: a novel reticulocyte-binding family homolog of plasmodium falciparum that binds to the erythrocyte, and an investigation of its receptor. PLoS ONE. 2008;3(10):e3300. doi: 10.1371/journal.pone.0003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayner JC, Huber CS, Barnwell JW. Conservation and divergence in erythrocyte invasion ligands: Plasmodium reichenowi EBL genes. Mol Biochem Parasitol. 2004;138(2):243–7. doi: 10.1016/j.molbiopara.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang’a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455(7214):757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Triglia T, Duraisingh MT, Good RT, Cowman AF. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol Microbiol. 2005;55(1):162–74. doi: 10.1111/j.1365-2958.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- 19.Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. Embo J. 2003;22(5):1047–57. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, Yeo KP, Aw SS, Kuss C, Iyer JK, Genesan S, Rajamanonmani R, Lescar J, Bozdech Z, Preiser PR. Antibodies targeting the PfRH1 binding domain inhibit invasion of Plasmodium falciparum merozoites. PLoS Pathog. 2008;4(7):e1000104. doi: 10.1371/journal.ppat.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaur D, Singh S, Jiang L, Diouf A, Miller LH. Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion. Proc Natl Acad Sci U S A. 2007;104(45):17789–94. doi: 10.1073/pnas.0708772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, Nawaz F, Mu J, Jiang L, Miller LH, Wellems TE. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe. 2008;4(1):40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin W, Contacos PG, Coatney GR, Kimball HR. A Naturally Acquited Quotidian-Type Malaria in Man Transferable to Monkeys. Science. 1965;149:865. doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- 24.Fong YL, Cadigan FC, Coatney GR. A presumptive case of naturally occurring Plasmodium knowlesi malaria in man in Malaysia. Trans R Soc Trop Med Hyg. 1971;65(6):839–40. doi: 10.1016/0035-9203(71)90103-9. [DOI] [PubMed] [Google Scholar]
- 25.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017–24. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 26.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46(2):165–71. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox-Singh J, Singh B. Knowlesi malaria: newly emergent and of public health importance? Trends Parasitol. 2008;24(9):406–10. doi: 10.1016/j.pt.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 1993;10(4):914–23. doi: 10.1093/oxfordjournals.molbev.a040038. [DOI] [PubMed] [Google Scholar]
- 29.Vargas-Serrato E, Corredor V, Galinski MR. Phylogenetic analysis of CSP and MSP-9 gene sequences demonstrates the close relationship of Plasmodium coatneyi to Plasmodium knowlesi. Infect Genet Evol. 2003;3(1):67–73. doi: 10.1016/s1567-1348(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 30.Johnson JG, Epstein N, Shiroishi T, Miller LH. Factors affecting the ability of isolated Plasmodium knowlesi merozoites to attach to and invade erythrocytes. Parasitology. 1980;80(3):539–50. doi: 10.1017/s0031182000000998. [DOI] [PubMed] [Google Scholar]
- 31.Pain A, Bohme U, Berry AE, Mungall K, Finn RD, Jackson AP, Mourier T, Mistry J, Pasini EM, Aslett MA, Balasubrammaniam S, Borgwardt K, Brooks K, Carret C, Carver TJ, Cherevach I, Chillingworth T, Clark TG, Galinski MR, Hall N, Harper D, Harris D, Hauser H, Ivens A, Janssen CS, Keane T, Larke N, Lapp S, Marti M, Moule S, Meyer IM, Ormond D, Peters N, Sanders M, Sanders S, Sargeant TJ, Simmonds M, Smith F, Squares R, Thurston S, Tivey AR, Walker D, White B, Zuiderwijk E, Churcher C, Quail MA, Cowman AF, Turner CM, Rajandream MA, Kocken CH, Thomas AW, Newbold CI, Barrell BG, Berriman M. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455(7214):799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnwell JW, Galinski MR, DeSimone SG, Perler F, Ingravallo P. Plasmodium vivax, P. cynomolgi, and P. knowlesi: identification of homologue proteins associated with the surface of merozoites. Exp Parasitol. 1999;91(3):238–49. doi: 10.1006/expr.1998.4372. [DOI] [PubMed] [Google Scholar]
- 33.Eyles DE, Dunn FL, Warren M, Guinn E. Plasmodium Coatneyi from the Philippines. J Parasitol. 1963;49:1038. [PubMed] [Google Scholar]
- 34.Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105(2):311–5. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 35.Altman JD, Henner D, Nilsson B, Anderson S, Kuntz ID. Intracellular expression of BPTI fusion proteins and single column cleavage/affinity purification by chymotrypsin. Protein Eng. 1991;4(5):593–600. doi: 10.1093/protein/4.5.593. [DOI] [PubMed] [Google Scholar]
- 36.Barnwell JW, Howard RJ, Miller LH. Altered expression of Plasmodium knowlesi variant antigen on the erythrocyte membrane in splenectomized rhesus monkeys. J Immunol. 1982;128(1):224–6. [PubMed] [Google Scholar]
- 37.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17(12):1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 38.Galinski MR, Corredor V. Variant antigen expression in malaria infections: posttranscriptional gene silencing, virulence and severe pathology. Mol Biochem Parasitol. 2004;134(1):17–25. doi: 10.1016/j.molbiopara.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Johnson JG, Epstein N, Shiroishi T, Miller LH. Identification of surface proteins on viable Plasmodium knowlesi merozoites. J Protozool. 1981;28(2):160–4. doi: 10.1111/j.1550-7408.1981.tb02825.x. [DOI] [PubMed] [Google Scholar]
- 40.Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA. A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci U S A. 2005;102(6):1980–5. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, Su XZ. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22(8):1686–93. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- 42.Oka M, Aikawa M, Freeman RR, Holder AA, Fine E. Ultrastructural localization of protective antigens of Plasmodium yoelii merozoites by the use of monoclonal antibodies and ultrathin cryomicrotomy. Am J Trop Med Hyg. 1984;33(3):342–6. doi: 10.4269/ajtmh.1984.33.342. [DOI] [PubMed] [Google Scholar]
- 43.Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. 1978;77(1):72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell GH, Bannister LH. Malaria parasite invasion: interactions with the red cell membrane. Crit Rev Oncol Hematol. 1988;8(4):225–310. doi: 10.1016/s1040-8428(88)80011-8. [DOI] [PubMed] [Google Scholar]