Abstract
SHANK3 is located on chromosome 22q13.3 and encodes a scaffold protein that is found in excitatory synapses opposite the pre-synaptic active zone. SHANK3 is a binding partner of neuroligins, some of whose genes contain mutations in a small subset of individuals with autism. In individuals with autism spectrum disorders (ASDs), several studies have found SHANK3 to be disrupted by deletions ranging from hundreds of kilobases to megabases, suggesting that 1% of individuals with ASDs may have these chromosomal aberrations. To further analyse the involvement of SHANK3 in ASD, we screened the International Molecular Genetic Study of Autism Consortium (IMGSAC) multiplex family sample, 330 families, for SNP association and copy number variants (CNVs) in SHANK3. A collection of 76 IMGSAC Italian probands from singleton families was also examined by multiplex ligation-dependent probe amplification for CNVs. No CNVs or SNP associations were found within the sample set, although sequencing of the gene was not performed. Our data suggest that SHANK3 deletions may be limited to lower functioning individuals with autism.
Keywords: Autistic disorder, SHANK3, 22q13.3, gene copy number, association analysis, synapse
Introduction
Autism is a severe neurodevelopmental disorder and one of the most heritable neuropsychiatric syndromes, with a male to female ratio of 4:1.1 The diagnosis of autism is based on impairments in reciprocal social interaction and communication, and restricted and stereotyped patterns of interests and activities, with abnormal development apparent within the first 3 years of life.2 Autistic spectrum disorders (ASDs) include: autistic disorder, childhood disintegrative disorder, pervasive developmental disorder-not otherwise specified, Asperger syndrome and Rett syndrome. The prevalence of childhood autism is estimated at 38.9 per 10 000 with the total prevalence for all ASDs at 116.1 per 10 000.3
There is substantial evidence from twin and family studies4, 5, 6, 7 to support the involvement of genetic factors in ASDs. Statistical modelling of the pattern of inheritance in the idiopathic form of the disorder implicates 3–15 genes, suggesting an oligogenic mode of inheritance.8, 9
The contribution of chromosomal structural variation to differences in genetic makeup within the human population has recently been highlighted.10 Advances in technology have enabled the screening of large cohorts of individuals for copy number variants (CNVs) genome-wide. A number of such studies have been published,11, 12, 13, 14, 15, 16 which have found that CNVs appear to have a larger causal role within autism than previously thought.
Several CNVs have been found in SHANK3 (SH3 and multiple ankyrin repeat domains protein 3, also known as ProSAP2). SHANK3 belongs to the family of SHANK proteins, which were first identified in the rat17 and subsequently in humans.18 These proteins are believed to function as molecular scaffolds in the post-synaptic density (PSD)19 of excitatory synapses and aid in the assembly of protein complexes within cells by containing multiple binding domains for protein–protein interaction.20 SHANK3 is located on chromosome 22q13.3, spans approximately 58 Kb of genomic sequence, and contains 24 exons, 7 of which are alternatively spliced,21 which may influence the spectrum of SHANK-interacting proteins at the PSD. Its expression pattern in the brain is confined to the cortex, hippocampus and cerebellum.20
SHANK3 was first implicated in the field of neuropsychiatric disorders when a patient with terminal 22q13.3 deletion syndrome was found to have a de novo reciprocal translocation that disrupted the gene.18 22q13.3 deletion syndrome includes the phenotypes of severe expressive language delay, severe/profound mental retardation and at times autism. Since the initial report, a number of studies22, 23, 24, 25 have described further cases of 22q13.3 deletion syndrome with disruption or deletion of SHANK3. This finding has led to the hypothesis that haploinsufficiency of SHANK3 may cause the behavioural phenotypic consequences of 22q13.3 deletion syndrome.
Owing to its emerging role in neuropsychiatric disorders and the overlap of phenotypes between autism and 22q13.3 deletion syndrome SHANK3 was further analysed in patients with ASDs. Durand et al21 found three separate families containing disruptions or deletions of SHANK3 that were not identified in controls. Interestingly, each individual with either a deletion of SHANK3 or a truncated protein had severe language problems, whereas the individual with 22qter partial trisomy had precocious language development and fluent speech, but social communication difficulties.21 Subsequent studies have discovered de novo deletions and mutations of SHANK3 in individuals with autism, 11, 13, 26, 27 adding support to the role of the gene within the autistic phenotype.
Given the emerging evidence for SHANK3 haploinsufficiency in some individuals with autism, DNA samples from 319 multiplex families and 11 trios, from the International Molecular Genetic Study of Autism Consortium (IMGSAC) multiplex cohort, were analysed for CNVs and SNP association across this locus. DNA samples from 76 IMGSAC probands with autism from Italian singleton families were also investigated for CNVs.
Materials and methods
Subjects
Multiplex families were identified, collected and assessed by IMGSAC as described earlier.28, 29 In short, after passing an initial screen, parents were administered the Autism Diagnostic Interview-Revised (ADI-R)30 and the Vineland Adaptive Behavior Scales.31 Probands were assessed using the Autism Diagnostic Observation Schedule-Generic (ADOS-G),32 and a medical examination was carried out to exclude cases of known aetiology, for example tuberous sclerosis. IQ was assessed using the Ravens Progressive Matrices coloured33 or standard and the Picture Vocabulary Test BPVS34 or PPVT.35 Karyotyping was performed when possible on probands and molecular genetic testing for Fragile X syndrome on one affected individual in each family. Written informed consent was obtained from all parents/guardians or, where appropriate, the proband. The relevant ethical committees approved the study. A blood sample was taken from all affected individuals and first-degree relatives and a lymphoblastoid cell line created. If blood was unavailable a buccal swab was taken instead. DNA was extracted from blood, cell lines and buccal swabs by means of a DNA purification kit (Nucleon BACC2 Blood and Cell Culture DNA purification kit, Tepnel Life Sciences, Manchester, UK). A total of 319 IMGSAC multiplex families and 11 IMGSAC trios were genotyped, with a male/female ratio of 4.3:1 in probands. Seventy-six IMGSAC Italian probands from a singleton family collection, who all meet IMGSAC criteria for inclusion, were also screened for distal chromosome 22q CNVs by multiplex ligation-dependent probe amplification (MLPA).
SNP genotyping
CEU genotypes for the region on chromosome 22q13.3 covering the SHANK3 gene, including 5 kb of sequence upstream (chromosome 22: 49 398 550–49 464 810 bp, NCBI Build 35), were downloaded from the HapMap phase II (release 21). Fifteen haplotype-tagging SNPs were chosen across SHANK3 using Tagger in Haploview v4.0beta1236 (aggressive tagging, r2 > 0.8, MAF > 0.05), that tagged in total 23 SNPs (including themselves), of which 95.7% of alleles were captured with an r2>0.8. DNA was quantified using the Picogreen dsDNA Quantitation Kit (Invitrogen). A total of 40 ng of genomic DNA was genotyped using the Sequenom MassARRAY i-PLEX Gold assay (Sequenom, San Diego, USA). Two control DNA samples were included on each plate to check for inter-plate reproducibility. SNP genotype calls were verified visually using the Sequenom MassARRAY Typer software (version 3.1.4.0). Merlin error checks37 were performed to identify unlikely genotypes within the data set and Pedwipe used to remove them from the pedigree file. Genotypes were uploaded to the Integrated Genotyping System.38
Statistical analysis of SHANK3 genotypes
Hardy–Weinberg equilibrium was verified for the SNPs genotyped, in all individuals and in unrelated individuals, using PEDSTATS.39 All families were checked for Mendelian inconsistencies by PedCheck40 and two subsequently removed because of bad inheritance found in the SHANK3 SNPs and those of another gene genotyped simultaneously, thus reducing the likelihood of detection of de novo deletions. Association analysis was carried out on the IMGSAC Caucasian families. STATA (StataCorp: Stata Statistical Software Release 7.0, Stata Corporation: College Station, TX, USA) was used to perform single locus transmission disequlibrium test (TDT)41 association analysis, taking into account pedigrees, and case/pseudo–control analysis42 that uses untransmitted genotypes as pseudocontrols. Genotypes were constructed for three ‘pseudo–controls', which consist of the other possible genotypes that the offspring could have received from their heterozygous parents. Two haplotype-tagging SNPs together tested another SNP and so TDT was also performed for this haplotype within Haploview.
Fluorescence in situ hybridisation
The extent of SNP homozygosity across SHANK3 was examined as a potential indicator of hemizygosity. Overall, 53 individuals (23 affected individuals, 25 parents and 5 unaffected siblings) were selected for analysis by FISH (fluorescent in situ hybridisation), as they were homozygous for all 14 haplotype-tagging SNPs successfully genotyped. One individual who was homozygous at only two SNPs was used as a control. Chromosome preparations for FISH analysis were obtained from the EBV-transformed lymphoblastoid cell lines following standard techniques. Two fosmid probes kindly provided by Dr. Stephen Scherer, G248P86064C8 and G248P86149G7, were labelled with Digoxigenin-11-dUTP (Roche) by nick-translation (Vysis kit) and separately co-hybridised with an FITC-labelled ‘paint' for chromosome 22 (Cambio), following standard procedures. The fosmid probe signals were detected with Rhodamin anti-Digoxigenin (Roche), and the chromosomes were counterstained with DAPI in Vectashield (Vector Laboratories). The slides were examined using an Olympus BX-51 epifluorescence microscope coupled to a Sensys charge-coupled device camera (Photometrics). At least 20 informative cells were analysed for each hybridisation experiment.
MLPA
Mulitplex ligation-dependent probe amplification reactions using probe mix SALSA P188 MLPA KIT 22q13 Lot 0407, 0906 were performed on 100 ng genomic DNA from 94 IMGSAC multiplex affected individuals, 76 Italian singleton affected individuals and one positive control individual harbouring a known SHANK3 deletion24 of 100 kb (DNA kindly provided by Dr. Maria Clara Bonaglia). The P188 kit and protocol43 were supplied by MRC-Holland (Amsterdam, The Netherlands). PCR products were separated on a 96-capillary Applied Biosystems 3700 DNA Analyzer. Peak size was verified using Genotyper 2.0 (Applied Biosystems, Foster City, USA). Coffalyser software v844 was used to analyse the MLPA data for CNVs. Bin sizes were adjusted accordingly for the peak sizes observed. Data were normalised by division of each probe's peak area by the average peak area of the seven control probes in the probe mix obtained from the sample set. The normalised data were then divided by the median peak area of all samples to obtain an indication of copy number variation for each probe. A figure of 0.7 or below and 1.3 and above were set as thresholds for losses and gains, respectively. Two consecutive probe deletions or duplications were necessary for further investigation.
The affected IMGSAC individuals were chosen for analysis through MLPA by identifying runs of consecutive homozygosity in the 14 successfully genotyped haplotype-tagging SNPs. All affected individuals were ranked according to the number of consecutive homozygous calls and the 94 individuals with the largest number selected. This sample included 18 individuals also tested by FISH and 76 others who contained between 8 and 14 homozygous calls. Only one affected individual per multiplex family was chosen; preferentially IMGSAC Case Type One (clinical diagnosis of autism, meets ADI-R and ADOS or ADOS-G criteria for autism, history of language delay, and performance IQ of >35).29 The affected individuals in total comprised 51 Case Type One, 30 Case Type Two, 2 Case Type Three and 11 Case Type Four (See Supplementary Table 1 for Case Type description). Among the affected Italian individuals, 80% were either Case Type One or Two. Overall, the probands selected were of a high functioning nature and met strict criteria for autism.
Results
SNP genotyping
Fourteen haplotype tagging SNPs were successfully genotyped on the Sequenom system, all of which conformed to Hardy–Weinberg equilibrium (P>0.001). As only 14 of the 15 selected haplotype-tagging SNPs were successfully genotyped, there was a reduction in tagging efficiency, with 91.3% of alleles being captured with an r2>0.8 rather than 95.7%. Sample and SNP genotyping success rates were 99 and 93%, respectively, and inter-plate reproducibility 99.5%.
Association analysis of SHANK3 SNPs
Family-based association analysis was carried out using the STATA package on a subset of the samples genotyped, comprised of Caucasian individuals, which included 297 IMGSAC multiplex families and 11 IMGSAC trios. Single SNP TDT and a case/pseudo–control approach were performed and no association was found (Table 1).
Table 1. Results of single SNP TDT and case/pseudo–control analysis in Caucasian families for the 14 haplotype-tagging SNPs genotyped across SHANK3.
| Single SNP TDT | Case/pseudo control analysis | |||||
|---|---|---|---|---|---|---|
| Tagging SNP | SNPs capured | Global χ2 (1 d.f.) | P-value | OR for allele 2 vs 1 | 95% CI | P-value |
| rs9616915 | rs9616915, rs9616913, rs9628185 | 0.04 | 0.84 | 1.02 | (0.85–1.22) | 0.84 |
| rs2341009 | rs2341009 | 1.44 | 0.23 | 1.14 | (0.92–1.41) | 0.23 |
| rs2341010 | rs1001469, rs2341010 | 0.41 | 0.52 | 1.08 | (0.86–1.35) | 0.53 |
| rs2341011 | rs736334, rs2341011, rs9616816 | 1.81 | 0.18 | 0.86 | (0.69–1.07) | 0.18 |
| rs739365 | rs739365 | 1.11 | 0.29 | 0.90 | (0.75–1.09) | 0.29 |
| rs5770992 | rs5770992 | 1.81 | 0.18 | 0.79 | (0.57–1.11) | 0.18 |
| rs6010060 | rs6010060 | 0.39 | 0.53 | 0.89 | (0.63–1.27) | 0.54 |
| rs2040487 | rs2040487 | 1.35 | 0.28 | 0.90 | (0.75–1.09) | 0.28 |
| rs6010061 | rs6009951, rs6010061 | 1.35 | 0.25 | 0.90 | (0.75–1.07) | 0.25 |
| rs2106111 | rs2106111 | 0.27 | 0.60 | 0.95 | (0.80–1.14) | 0.60 |
| rs6010063 | rs6010063, rs6010065 | 0.47 | 0.49 | 1.06 | (0.89–1.27) | 0.49 |
| rs715586 | rs715586 | 0.03 | 0.87 | 0.98 | (0.76–1.26) | 0.87 |
| rs8137951 | rs8137951 | 0.48 | 0.49 | 0.93 | (0.76–1.14) | 0.49 |
| rs756638 | rs756638 | 0.22 | 0.64 | 1.05 | (0.86–1.28) | 0.64 |
| rs739365, rs6010061 | rs5770820 | 0.48 | 0.49 | |||
CI, confidence interval; df, degrees of freedom; OR, odds ratio.
The SNPs captured by the tagging SNPs are also shown.
Copy number analysis by FISH mapping
Fluorescent in situ hybridisation was used to test for chromosome rearrangements in individuals identified by the SNP genotyping as displaying potential hemizygosity in SHANK3. Out of the 53 individuals, 44 respective lymphoblastoid cell lines were tested, consisting of 21 affected individuals (16 male and 5 female), 12 fathers, 6 mothers and 5 unaffected siblings. Eight individuals did not have a cell line available and one cell line failed to grow. A sample that was heterozygous at 12 out of 14 SNPs in SHANK3 was also probed as a negative control.
Two fosmid clones, G248P86064C8 and G248P86149G7, were used as FISH probes across a region of ∼64 kb covering the SHANK3 gene to search for deletions in the selected IMGSAC samples. Of the 45 lymphoblastoid cell lines probed, including the control, no deletions were identified.
MLPA analysis
Multiplex ligation-dependent probe amplification was used as a second method to look for smaller deletions across SHANK3 not detectable by FISH. The 37 probes within the P188 probe set cover a larger distance than the fosmids, approximately 19 Mb, enabling characterisation of potential deletions. MLPA is also a higher throughput technique than FISH; therefore, a greater number of individuals could be analysed simultaneously.
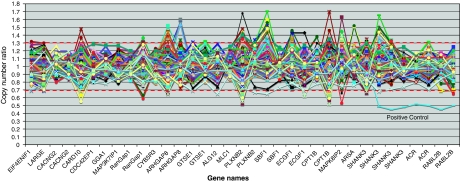
Ninety-four affected individuals from IMGSAC multiplex families, seventy-six affected individuals from IMGSAC Italian singleton families and one positive control were analysed using MLPA (Figure 1). Two individuals were removed as they had SDs for the copy number ratios above 0.25, which was used as the cutoff for this sample. All probes performed accurately apart from two within the gene MLC1, MLC1 Probe 6335-L5910 and MLC1 Probe 6338-L5913, which have been removed from Figure 1. No individual had two or more consecutive probes showing deletions or duplications within SHANK3, apart from the positive control, which showed all seven probes deleted from exon 9 in SHANK3 to the telomere. Three affected individuals exhibited two probes duplicated consecutively and one affected individual had two probes deleted; however, these events were all more than 150 kb proximal to SHANK3.
Figure 1.
Copy number analysis using MLPA on the region chr22q12.2-q13.33 in 168 autistic probands from IMGSAC multiplex families and Italian singleton families. The thresholds for gains and losses are marked as 1.3 and 0.7, respectively. The positive control sample clearly shows the loss of SHANK3, ACR and RABL2B. Graph not to scale. For a complete list of the probes and their positions see MRC-Holland probe mix P188 lot 0407, 0906.
Discussion
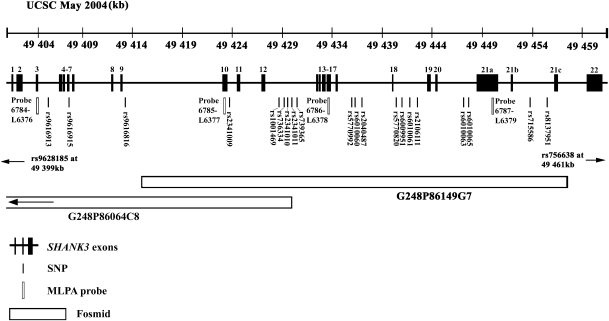
The IMGSAC multiplex family collection was analysed for association and CNVs within SHANK3, using SNP genotyping, FISH and MLPA analysis. The SNP, fosmid and MLPA probe positions are shown in Figure 2, comprehensively covering the gene SHANK3. A smaller group of affected individuals from an Italian cohort of IMGSAC singleton families was also analysed by MLPA for CNVs in SHANK3.
Figure 2.
Positions of SNPs, fosmids and MLPA probes within SHANK3 (UCSC May 2004, NCBI Build 35). Exons annotated as those in the study by Durand et al.21
The association analysis results from the SNP genotyping in the IMGSAC families presented here were all nonsignificant. Nevertheless, no previous reports of association with SHANK3 have been published, and one would not expect to observe association of a rare deletion, if only present in a small number of individuals. Sequence analysis was not conducted, and so rare mutations potentially could have been missed within SHANK3, such as those found in previous studies.21, 26, 27 The sample size of the IMGSAC cohort of families is relatively small in comparison with larger genome-wide association and CNV detection studies that are currently being carried out,12, 16 and so this must also be taken in to consideration.
The SNP genotyping was used as a crude indication of potential hemizygosity across SHANK3 within the IMGSAC multiplex families. Unfortunately, the Sequenom system was unable to provide quantitative information about signal intensity from the genotype calls, and so the distinction between a true homozygous call and a hemizygous call could not be distinguished by simple analysis of an output file. Therefore, those individuals homozygous for all haplotype-tagging SNPs across the region were probed using FISH.
The two probes used for FISH are each approximately 40 kb in length, and so if the deletion occurred within the probe sequence, but still allowed the probe to hybridise, a fluorescent signal would still be seen and a deletion missed. To overcome this problem, MLPA was used to potentially identify smaller aberrations. This technique relies on hybridisation of probes on either side of a target sequence and subsequent ligation that permits amplification. The amount of PCR product, dependent on ligation, indicates copy number. No CNVs within SHANK3 were identified by this technique. This finding could have been due to the method of selection of the affected individuals tested; 94 individuals with the most consecutive homozyogous SNPs. Individuals with just a few homozygous calls at either end of SHANK3 were not included in the analysis, but could be representative of a deletion extending outwards from the extremities of the gene. This method of choosing affected individuals preferentially selected for deletions and so duplications may have been missed, whereas the Italian singleton sample was not preselected, hence equally likely to harbour deletions or duplications. Four affected individuals did demonstrate events of two consecutive probes duplicated or deleted, positioned at greater than 150 kb proximal to SHANK3, these regions were deemed not to be directly affecting the expression of the SHANK3 gene when interrogated by the use of an expression database (mRNA_bySNP_browser_v1.0.1.exe45).
The majority of IMGSAC families analysed were multiplex in origin. Recent reports have suggested that the prevalence of de novo CNVs in singleton families is between 7 and 10%, whereas in multiplex families it is 2–3%.11, 13 As the majority of SHANK3 CNVs identified are de novo in origin, this would decrease the probability of detecting an event within our multiplex family sample set, particularly as no Mendelian errors were found during the SNP genotyping. Although the Italian individuals that were tested by MLPA originated from singleton families, no CNVs were identified within these affected individuals.
Another reason for the lack of CNVs identified within this study could be because of the inclusion criteria used to select IMGSAC multiplex and singleton families. The majority of affected individuals are higher functioning and meet a more narrowly defined diagnosis of autism than those that have been reported with SHANK3 CNVs in previous papers.21, 26 Seventy six percent of the IMGSAC affected individuals assessed by MLPA had performance IQ data available with a mean of 78 (SD=30), whereas severe mental retardation was described for some autistic individuals with SHANK3 aberrations by Durand et al.21
SHANK3 is a binding partner of the neuroligins,46 which in turn have ligands in the form of the neurexins. Both of these protein families contain genes within which rare mutations have been identified in autism; NLGN347, 48 and NLGN4,47, 48, 49 CNTNAP250 and NRXN1.12, 51 Nevertheless, coding mutations in NLGN3 and NLGN4 have not been found to be associated with autism in IMGSAC families,52 and no CNVs have been identified in SHANK3 using the methodology described here, although sequence analysis may have detected rare variants. Despite this negative finding, a number of genes within the synaptic network could still be harbouring mutations, which could lead to a similar phenotypic end point. Further analysis of these genes for CNVs within the IMGSAC cohort is warranted.
Acknowledgments
We thank all of the families and the clinical teams who have contributed to this IMGSAC study. We also thank: Chris Allan, Lyn-Louise Buckingham and the Genomics Core facility at the Wellcome Trust Centre for Human Genetics, Oxford, for technical support; Jordana Tzenova Bell, Denise Brocklebank and Andrew Morris for their help and advice on statistical methodologies; Maria Clara Bonaglia for providing the control DNA sample for the MLPA analysis; Stephen Scherer for the provision of the fosmid clones; Barbara Parrini and Roberta Igliozzi for their contribution to the collection of the Italian IMGSAC families; Jordy Coffa for his advice on MLPA analysis; and Janine Lamb for her helpful comments and guidance. This work was funded by the Nancy Lurie Marks Family Foundation, the Simons Foundation, Autism Speaks UK and the Wellcome Trust.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Volkmar FR LC, Bailey A, Schultz RT, et al. Autism and pervasive developmental disorders. J Child Psychol Psychiatry. 2004;45:135–170. doi: 10.1046/j.0021-9630.2003.00317.x. [DOI] [PubMed] [Google Scholar]
- Lord C, Cook EH, Leventhal BL, et al. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A, et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368:210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism. a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Rutter M, Silberg J, O′Connor T, et al. Genetics and child psychiatry: II Empirical research findings. J Child Psychol Psychiatry. 1999;40:19–55. [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, et al. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet. 1995;57:717–726. [PMC free article] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Nickerson DA, Altshuler D, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Brune CW, Sudi J, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Boeckers TM, Kreutz MR, Winter C, et al. Proline-rich synapse-associated protein-1/cortactin binding protein 1 (ProSAP1/CortBP1) is a PDZ-domain protein highly enriched in the postsynaptic density. J Neurosci. 1999;19:6506–6518. doi: 10.1523/JNEUROSCI.19-15-06506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, et al. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113 Part 11:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning MA, Cassidy SB, Clericuzio C, et al. Terminal 22q deletion syndrome: a newly recognized cause of speech and language disability in the autism spectrum. Pediatrics. 2004;114:451–457. doi: 10.1542/peds.114.2.451. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, et al. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderlid BM, Schoumans J, Anneren G, et al. FISH-mapping of a 100-kb terminal 22q13 deletion. Hum Genet. 2002;110:439–443. doi: 10.1007/s00439-002-0713-7. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2008. [DOI] [PubMed]
- IMGSAC A full genome screen for autism with evidence for linkage to a region on chromosome 7q International Molecular Genetic Study of Autism Consortium. Hum Mol Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- IMGSAC A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Sparrow SBD, Cicchetti D. Vineland Adaptive Behaviour Scales. Circle Pines, MN: American Guidance Service; 1984. [Google Scholar]
- Lord C, Risi S, Lambrecht L, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- Raven J, Raven JC, Court JH. Manual for Raven's Progressive Matrices and Vocabulary Scales. Section 2: The Coloured Progressive Matrices. San Antonio, TX: Harcourt Assessment; 1998. [Google Scholar]
- Dunn LIM, Dunn LM, Whetton C, Burley J.The British Picture Vocabulary Scale, 2nd edn (BPVS-II). NFER-Nelson; 1997 [Google Scholar]
- Dunn LM, Hottel JV. Peabody picture vocabulary test performance of trainable mentally retarded children. Am J Ment Defic. 1961;65:448–452. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, et al. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Fiddy S, Cattermole D, Xie D, et al. An integrated system for genetic analysis. BMC Bioinformatics. 2006;7:210. doi: 10.1186/1471-2105-7-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa J, van de Wiel MA, Diosdado B, et al. MLPAnalyzer: Data analysis tool for reliable automated normalization of MLPA fragment data. Cell Oncol. 2008;30:323–335. doi: 10.3233/CLO-2008-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, et al. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 2004;47:724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Oliveira G, Coutinho A, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkaloglu B, O'Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Blasi F, Bacchelli E, Pesaresi G, et al. Absence of coding mutations in the X-linked genes neuroligin 3 and neuroligin 4 in individuals with autism from the IMGSAC collection. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:220–221. doi: 10.1002/ajmg.b.30287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.