Abstract
Whole genome association studies (WGAS) have surged in popularity in recent years as technological advances have made large-scale genotyping more feasible and as new exciting results offer tremendous hope and optimism. The logic of WGAS rests upon the common disease/common variant (CD/CV) hypothesis. Detection of association under the common disease/rare variant (CD/RV) scenario is much harder, and the current practices of WGAS may be under-power without large enough sample sizes. In this paper, we propose a generalized linear model with regularization (rGLM) approach for detecting disease-haplotype association using unphased single nucleotide polymorphisms data that is applicable to both CD/CV and CD/RV scenarios. We borrow a dimension-reduction method from the data mining and statistical learning literature, but use it for the purpose of weeding out haplotypes that are not associated with the disease so that the associated haplotypes, especially those that are rare, can stand out and be accounted for more precisely. By using high-dimensional data analysis techniques, which are frequently employed in microarray analyses, interacting effects among haplotypes in different blocks can be investigated without much concern about the sample size being overwhelmed by the number of haplotype combinations. Our simulation study demonstrates the gain in power for detecting associations with moderate sample sizes. For detecting association under CD/RV, regression type methods such as that implemented in hapassoc may fail to provide coefficient estimates for rare associated haplotypes, resulting in a loss of power compared to rGLM. Furthermore, our results indicate that rGLM can uncover the associated variants much more frequently than can hapassoc.
Keywords: whole genome association study, interacting effects between haplotype blocks, dimension reduction, regularization/LASSO, case-control design
INTRODUCTION
The past decade has seen a surge of interest in disease-SNP and disease-haplotype association studies because of the availability of densely situated single nucleotide polymorphisms (SNPs) throughout the genome. The haplotypes in such studies are usually SNP-based in that they are specific combinations of tightly linked SNP variants, usually in the same haplotype block. It is further noted that common SNPs may lead to rare haplotypes in the population. The standard logic of whole genome association studies (WGAS) rests upon the common disease/common variant (CD/CV) hypothesis. Detection of association for the common disease/rare variant (CD/RV) scenario, on the other hand, is much harder, and the current practices of WGAS may be under-power without larger enough sample sizes. Purcell et al. [2007] offered a population based linkage analysis approach that is promising under the CD/RV scenario. Gorlov et al. [2008] also asserted the value of rare variants, and argued for a paradigm shift in association studies. These are significant path-paving developments, but there is still very limited work to date in developing statistical methods for uncovering disease associations with rare haplotypes.
It has been argued that haplotype-based procedures are more powerful than their single-SNP-based counterparts, especially when the causal polymorphism is not genotyped or when multiple causal polymorphisms act in cis fashion in the haplotype region [Akey et al., 2003; Morris and Kaplan, 2002; Rosenberg et al., 2006; Schaid et al., 2002]. A common and economical design for the study of disease-haplotype association is case-control. However, SNP genotypes on unrelated individuals are usually unphased, and as such, haplotype-based analysis is much more difficult than SNP-based analysis. More specifically, haplotypes need to be inferred from the genotype data (some of which may be missing) in some fashion, and haplotype uncertainty should be accounted for. As such, standard biostatistical methods that are applicable to SNP-based procedure are no longer feasible; instead, special methods need to be developed.
Earlier statistical methods proposed for haplotype-disease association studies based on case-control design were focused mainly on comparing the haplotype distribution for cases versus the distribution for controls [Zhao et al., 2000; Fallin et al., 2001]. These are fairly attractive methods because of their simplicity, but there are a number of drawbacks [Epstein and Satten, 2003; Schaid et al., 2002; Zaykin et al., 2002]. Due to the global nature of the tests, individual haplotype effects cannot be directly estimated, and non-genetic (e.g., environmental) covariates cannot be accounted for either. Furthermore, there are usually large degrees of freedom associated with the large number of haplotypes, reducing the powers of such tests.
Another class of methods for association study is based on generalized linear modeling (GLM) and likelihood [Schaid et al., 2002], which is able to incorporate non-genetic covariates and estimate individual haplotype effects directly. Most of the GLM methods use unphased SNP data and take into account uncertainty in the estimated haplotypes and associated frequencies. As such, the Expectation-Maximization (EM) algorithm is usually used to estimate the model parameters and the haplotype frequencies iteratively. With rare haplotypes, the EM estimates can be fairly unstable with large standard errors, and may even lead to non-convergence of the algorithm. This was what we observed when we applied the software hapassoc [Burkett et al., 2006] to a dataset on multiple sclerosis (unpublished data), for which about 15% of the tests failed to converge due to rare haplotypes. Solutions in the literature for this well-known problem are mostly based on clustering of haplotypes: either combining all rare haplotypes into a single group [Schaid et al., 2002] or combining each rare haplotype with its more common ancestry haplotype using an evolutionary model [Molitor et al., 2003; Seltman et al., 2003; Durrant et al., 2004; Tzeng, 2005]. These solutions may be reasonable under the CD/CV hypothesis, but they fail to accommodate the CD/RV scenario adequately and would lead to difficulty in interpreting the results.
In this paper, we propose a new approach to detect association of disease and haplotype that is applicable to both the CD/CV and the CD/RV scenarios. As already noted [Purcell et al., 2007; Gorlov et al., 2008], CD/RV is a possible scenario but rarely addressed, and therefore methods proposed in this direction would help to fill the void and would likely lead to useful applications. Our approach is based on logistic regression for case-control data, falling into the general framework of GLM, but adopting a regularization method heavily investigated in the statistical and data mining literature for dimension reduction in the past decade [Tibshirani, 1996; van de Geer, 2008]. This application represents a novel usage of regularization techniques; our main goal is not dimension reduction, but rather estimation of (rare) haplotype effects. The idea is that, even in a block with limited haplotype diversity, if a haplotype (especially a common one) is not associated with the disease (i.e. it does not increase one’s relative risk above the phenocopy rate), then it is more desirable to shrink its regression coefficient to zero. This effectively combats the problem of estimation instability and leads to an increase in the precision of the remaining parameter estimates and an improvement in power for detecting association.
Although dimension reduction per se is not the main goal of the usage of regularization methods in our application, we indeed benefit tremendously from the aspect of dimension reduction, even more so when haplotypes from multiple blocks and their interacting effects are considered jointly. In this case, the number of haplotypes can get large very quickly. Even though the regression coefficients may still be identifiable given the usually large sample size of a case-control study, a great deal of the benefits associated with regularization methods can be materialized. Regularization methods have been employed extensively for gene expression microarray analysis as it is essentially a high-dimensional data analysis problem. In comparison, applications of such methods in statistical genetics and genetic epidemiology are much more limited to date, although they have started to emerge recently. The work of Li et al. [2007] is an example, which investigated disease-haplotype association for quantitative phenotype data, but assumed that haplotypes are observed without uncertainty, an assumption unlikely to be met in reality.
MATERIALS AND METHODS
DATA, MODEL, AND LIKELIHOOD
Suppose we have a case-control sample with a total of n subjects. Let (Yi,Gi,Ei) denote the observed data for individual i, i = 1, …, n, where Yi = 1(0) if individual i is a case (control), Gi is the observed unphased genotype data over L SNPs, and Ei denotes other, non-genetic, covariates (if available). Let S(Gi) = {Hij, j = 1, …, Ji} be the set of haplotype pairs consistent with Gi. The observed data likelihood is
| (1) |
where Ψ = (θ, f) denotes the collection of model parameters (θ) and haplotype frequencies (f).
To specify the first factor, P(Yi|Hij, Ei, θ), in (1), we need to establish a model. Let µ = E[Y|H,E, θ] = P(Y = 1|H,E, θ), then the GLM for connecting µ with the predictors is
| (2) |
for some link function g, where XH (defined based on haplotype pair H), XE (defined based on non-genetic variables in E, if any), and XH.E (defined based on interaction between H and E, if any) are the predictors of the model. Since our response variable is binary (cases and controls), we adopt the usual practice of using the logistic link function, that is g(µ) = log(µ/(1 − µ)).
To specify the second factor, P(Hij | f), we assume that the haplotypes are in Hardy-Weinberg equilibrium (HWE) so that f = (f1, …, fm) with the constraint that the frequencies of the m haplotypes sum to 1. Then for a haplotype pair H = hk/hk′,
| (3) |
where I(·) is the usual indicator function giving value of 1 or 0 depending on whether the condition within the parentheses is true or not.
SPECIFICATION OF XH
Before we continue with the model development, it is helpful to give some examples on the predictors in (2) to facilitate understanding of the model. The formula given in (2) is completely general, which can accommodate different settings, depending on the hypotheses of interest about the haplotypes, and depending on whether non-genetic covariate information E is available and how it interacts with the haplotypes. In this paper, because our focus is on the detection of disease-rare haplotype association, we assume there is no non-genetic covariate (i.e., E = ø) to streamline our description and investigation.
Suppose the goal is to perform an omnibus test to investigate whether the haplotypes defined by the SNPs are associated with the disease, and further suppose that the effects of haplotypes are additive in the logit scale. Let us assume that there are m possible haplotypes, h1, h2, …, hm. Without loss of generality, hm is the most common haplotype and will be used as the reference haplotype. Then XH = (x1, x2, …, xm−1), where xk is the number of copies of haplotype hk in haplotype pair H. For example, let G = (0/1, 1/1, 0/1) be the observed unphased genotype of an individual at three SNPs (with the alleles of each SNP coded as 0 and 1). Then since the genotypes at the first and the third SNPs are heterozygous, there are two possible pairs of (unordered) haplotypes consistent with G, that is, S(G) = {010/111, 011/110}, where the allelic combinations before and after the “/” are the two haplotypes that make up the pair. Suppose all 8 haplotypes, {000, 001, 010, 011, 100, 101, 110, 111} are available, and 111 is the most common one and treated as the reference haplotype. Then for H1 = 010/111,XH1 = (0, 0, 1, 0, 0, 0, 0). On the other hand, for H2 = 011/110,XH2 = (0, 0, 0, 1, 0, 0, 1).
Although our focus in the current paper is the omnibus test, the model as specified in (2) can be used for testing other hypotheses, such as the effect of a particular haplotype. For example, Suppose one is interested in assessing the effect of haplotype h* relative to all the other haplotypes and H = hk/hk′. Then all the haplotypes other than h* will be lumped into the reference category, and we let XH = I(hk = hk′ = h*) if a recessive model is hypothesized, or XH = I(hk = h* or hk′ = h*) if a dominant model is entertained. Additive, co-dominant, and other models can also be set up.
REGULARIZED REGRESSION
An EM algorithm for finding the maximum likelihood estimates (MLEs) of (1) using the GLM of (2) has been proposed and implemented by Burkett et al. [2004, 2006] in the software package hapassoc. When the software was applied to a multiple sclerosis data set (unpublished data) to investigate haplotype and disease associations using the omnibus test, about 15% of the runs did not converge due to the existence of rare haplotypes. Instead of grouping rare haplotypes together or with their similar common haplotypes as suggested in the literature, we propose the following regularized procedure so that we can uncover associations even when rare haplotypes are involved in leading to the association, thus giving a more complete picture. Due to the fact that most of the haplotypes are not associated with increased disease risk, obtaining their regression coefficient estimates is unimportant and could in fact adversely affect the precision of the estimates of the associated haplotypes. Thus, it is highly desirable to shrink the coefficient estimates of the non-associated haplotypes to 0 to increase the precision of the estimates of the remaining coefficients, especially if the sample size is limited. Doing so would also alleviate the problem of non-convergence, and would lead to an increase in power for detecting association.
One way of doing so is to adopt a regularized regression method, such as LASSO [Tibshirani, 1996; van de Geer, 2008] through maximizing a penalized likelihood. Note that regularized methods are usually proposed for high dimensional data problems. Here, however, we are proposing a novel usage of this type of methods because the number of haplotypes, especially if the underlying SNPs are within a haplotype block, is usually quite limited and much smaller than the number of subjects. The penalized log-likelihood function using the LASSO penalty is
| (4) |
To maximize the above penalized likelihood, we employ the EM algorithm after suitably devising the “complete data” as follows.
Let Zi be the (missing) haplotype of individual i. Then (Gi, Zi) is considered as the complete data and its corresponding complete data likelihood (assuming E = ø) is
where I(Zi = Hij) is an indicator function taking the value of 1 (0) if the condition inside the parentheses is true (false). For an omnibus test, the parameter vector θ = {α, β = (β1, …, βm−1)}. The penalized complete data log-likelihood based on LASSO is then given as
| (5) |
where λ is a tuning parameter denoting the amount of penalty. We will discuss the selection of λ in a later subsection.
ESTIMATION OF PARAMETERS
The EM algorithm is used to estimate the parameters that maximize the penalized log-likelihood objective function in (4). From an initial guess of the parameter value Ψ(0), the EM algorithm iterates between the following E and M steps until convergence.
E-Step. For i = 1, …, n, compute the contribution (weight) of haplotype pair Hij given the current estimate of the parameter Ψ(t) = (θ(t), f(t)) and the observed data Gi. Let
then the ’s are normalized to obtain the weights
M-Step. Find the next iterate of estimate, Ψ(t+1), of Ψ by maximizing
| (6) |
Specifically, maximization with respect to θ and f can be accomplished separately. We note that the first component of (6), a weighted penalized log-likelihood for regression, involves θ only and can be maximized using the glmpath R package [Park and Hastie, 2007]. On the other hand, the second component of (6) is a weighted multinomial log-likelihood that involves the f parameters only, which can be easily maximized using the R software.
TUNING PARAMETER DETERMINATION
Determining the amount of penalty is extremely important in balancing the elimination of unimportant covariates while retaining the important ones. Methods based on cross validation or generalized cross validation exist, but they are usually very computationally intensive [Stone, 1974]. Instead, we make use of a recent result in Zou et al. [2007], which shows that the number of non-zero coefficients in a LASSO regression is an unbiased estimate of the degrees of freedom. Specifically, for each λ value considered, let be the MLEs and pλ be the corresponding number of non-zero coefficient estimates in . Since a larger penalty (large λ) will result in a smaller model (smaller pλ) and hence smaller likelihood, one can use the AIC criterion [Akaike, 1973] to choose an appropriate λ to avoid over-penalization as follows:
TESTING FOR ASSOCIATION
To test the null hypothesis (H0: β = 0; there is no association between the haplotypes and the disease) versus the alternative hypothesis (H1: β ≠ 0; there is at least one non-zero β coefficients to signify association), we can make use of the likelihood ratio test statistic
| (7) |
where is the MLE of (1) when β is set to zero. Although it is well known that T follows a chi-square distribution asymptotically, how well the asymptotic can approximate the actual null distribution is unknown under the current setting. Therefore, we choose to evaluate the strength of association signals based on a permutation procedure.
For a single analysis sample (i.e., for a real data analysis) we propose to adopt the standard permutation procedure for our task. Specifically, we permute the disease labels (cases or controls) among the n individuals (keeping their observed SNP genotypes unchanged) B times to obtain Tb, b = 1, …,B. These can be regarded as a sample of size B from the distribution of T (as defined in (7)) under the null hypothesis. Then
provides an estimate of the p-value for testing the hypotheses.
For a simulation study with R replicates, the above procedure will be rather computationally expensive. Therefore, we propose the following procedure to reduce the computational burden by pooling permutation samples from all replicates to form a joint sample from the null distribution. More specifically, for the rth replicate with observed test statistic Tr, we permute the labels B times as above to obtain Tr,b, b = 1, …,B. Then the collection {Tr,b; r = 1, …,R, b = 1, …,B} can be regarded as a random sample of size BR from the common null distribution, i.e., the distribution under the null hypothesis H0. Consequently, for r = 1, …,R,
| (8) |
provides an estimate of the p-value for the rth replicate. Since the permutation samples are pooled across all replicates to form a sample from the null, B can be set to be much smaller than the situation when only one sample is analyzed. For example, if R = 500 and the desire number of permutations for estimating the p-values is also 500, then we set B = 1 for each replicate. In other words, we do not need to perform 500 permutations for each replicate to obtain the p-value. Instead, we only need to perform one permutation per replicate to achieve the same precision. This pooling idea is similar to that used in Becker and Knapp [2004].
SIMULATION STUDY
We performed a simulation study to evaluate the statistical properties and the performances of the proposed method. For ease of reference, our generalized linear model with regularization approach will be termed rGLM, and the results from rGLM will be compared and contrasted with those from hapassoc [Burkett et al., 2006] which uses a logistic regression without regularization. Although rGLM is a general procedure for testing haplotype-disease association, we will focus on evaluating the method under the CD/RV scenario.
SIMULATION SETTINGS
A total of four settings of haplotype-disease association were considered. In the first three settings, we assumed that the haplotypes arise from five SNPs in a haplotype block. Each haplotype is coded as a series of zeros and ones, with 1 denoting the minor allele of a SNP. The three settings differ in their degrees of haplotype diversity, with 6, 9, and 12 haplotypes for setting 1, 2, and 3, respectively (Table I). Regardless of the degrees of haplotype diversity, the baseline disease prevalence, that is, the disease penetrance probability given a non-associated haplotype, was set to be 10%. Two rare haplotypes, 10100 and 11011, with frequencies of 0.005 and 0.03, were assumed to increase the odds of getting the disease by 4 and 2 folds (i.e., OR = 4 and 2), respectively. In the fourth setting, we investigated the interacting effect between two haplotype blocks that are in linkage equilibrium. This setting also led to a larger number of haplotypes over the two blocks jointly. Four haplotypes within each block were considered, and Haplotype 1100 (with a frequency of 0.05) in either block is assumed to be the only disease associated haplotype. The ORs were set to be 2 if the 1100 haplotype was present in only one of the two blocks. If the haplotypes in both blocks were 1100, then the OR was assumed to be 6 (Table II). Each of the four settings were investigated with three different sample sizes: 200, 400, or 1,000, with equal numbers of cases and controls.
TABLE I.
Three settings of disease association with haplotypes in a single block
| Setting 1 | Setting 2 | Setting 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Hap | Freq | OR | Hap | Freq | OR | Hap | Freq | OR |
| 10100 | 0.005 | 4 | 10100 | 0.005 | 4 | 10100 | 0.005 | 4 |
| 11011 | 0.030 | 2 | 11011 | 0.030 | 2 | 11011 | 0.030 | 2 |
| 11111 | 0.100 | 1 | 11111 | 0.100 | 1 | 11101 | 0.080 | 1 |
| 11100 | 0.155 | 1 | 11100 | 0.080 | 1 | 11110 | 0.130 | 1 |
| 01100 | 0.290 | 1 | 10000 | 0.080 | 1 | 10010 | 0.070 | 1 |
| 10011 | 0.420 | 1 | 11101 | 0.085 | 1 | 11111 | 0.100 | 1 |
| 01010 | 0.050 | 1 | 01000 | 0.020 | 1 | |||
| 01100 | 0.250 | 1 | 01011 | 0.050 | 1 | |||
| 10011 | 0.320 | 1 | 00111 | 0.070 | 1 | |||
| 01101 | 0.060 | 1 | ||||||
| 01110 | 0.140 | 1 | ||||||
| 10001 | 0.245 | 1 | ||||||
TABLE II.
The odds ratios (ORs) of a setting of disease association with inter-acting effects of haplotypes from two blocks
| Block 1 | ||||||
|---|---|---|---|---|---|---|
| Freq | 0.05 | 0.10 | 0.30 | 0.55 | ||
| Freq | Hap | 1100 | 1011 | 0001 | 1111 | |
| 0.05 | 1100 | 6 | 2 | 2 | 2 | |
| 0.10 | 1011 | 2 | 1 | 1 | 1 | |
| 0.30 | 0001 | 2 | 1 | 1 | 1 | |
| 0.55 | 1111 | 2 | 1 | 1 | 1 | |
DATA GENERATION AND ANALYSIS PROCEDURE
Data for R = 500 replicates were generated under each of the four settings and three sample sizes. Each replicate was analyzed using both hapassoc and rGLM, and powers were calculated. The following four steps provide the details of the data generation and analysis procedure.
1. Generate genotype data
Generate phased haplotype pairs using the frequencies given in Table I (Table II for setting 4) assuming HWE. Assign the individual to be a case or a control based on the sampled haplotype pair according to the probabilities derived from the logistic regression model. Continue sampling until the desired sample size is met. Remove the phase information so that only unphased SNP genotypes are used in the subsequent analysis.
2. Calculate the test statistics
(a) Given the unphased genotypes, obtain the test statistics using hapassoc and rGLM, denoted as Tr and Sr. To calculate the statistic under rGLM, a series of the tuning parameter λ, {λmax = 10, 0.81λmax, …, 0.819λmax, 0}, with geometric decay, are considered.
(b) Randomly permute the cases and control statuses of all individuals in the sample and recalculate the statistics, denoted as Tr,1 and Sr,1 for hapassoc and rGLM, respectively.
3. Repeat steps 1–2 for R = 500 times
4. Calculate the powers
Obtain the power under hapassoc by finding the p-value, , for each replicate based on formula (8) using {T1,1, T2,1, …, TR,1} as a sample from the null distribution. For a significance level α, . Similarly, find the p-values, , and calculate the power for the analyses based on rGLM as using {S1,1, S2,1, …, SR,1} as a sample from the null distribution.
RESULTS
ONE HAPLOTYPE BLOCK
Because of the existence of rare haplotypes, hapassoc had difficulty finding estimates for the regression coefficients, especially when the sample size was relatively small (Table III, Column NC). When n=200 (100 cases and 100 controls), at least half of the samples failed to converge for all three settings; the larger the number of haplotypes, the higher the rate of non-convergence. As the sample size increases, the non-convergence rate decreases (for each of the three settings, respectively) because the rare haplotypes were being observed more frequently and thus the coefficient estimates were more stable.
TABLE III.
Comparisons of powers between results obtained from hapassoc and rGLM
| rGLM | hapassoc | ||||
|---|---|---|---|---|---|
| n | Setting | Power | Power | NCa | Power.Cb |
| 200 | 1 | 24.0 | 10.2 | 50.0 | 20.4 |
| 2 | 19.8 | 8.0 | 57.2 | 18.7 | |
| 3 | 16.0 | 4.4 | 63.8 | 12.2 | |
| 400 | 1 | 54.0 | 36.4 | 21.6 | 46.4 |
| 2 | 29.4 | 21.2 | 29.4 | 30.0 | |
| 3 | 23.0 | 13.8 | 38.6 | 22.5 | |
| 1,000 | 1 | 86.4 | 83.6 | 2.2 | 85.5 |
| 2 | 69.0 | 68.6 | 7.0 | 73.8 | |
| 3 | 69.4 | 68.6 | 4.6 | 71.9 | |
NC denotes the non-convergence rates (%) of the analyses using hapassoc.
Power.C gives the powers of hapassoc based only on the replicates that converged.
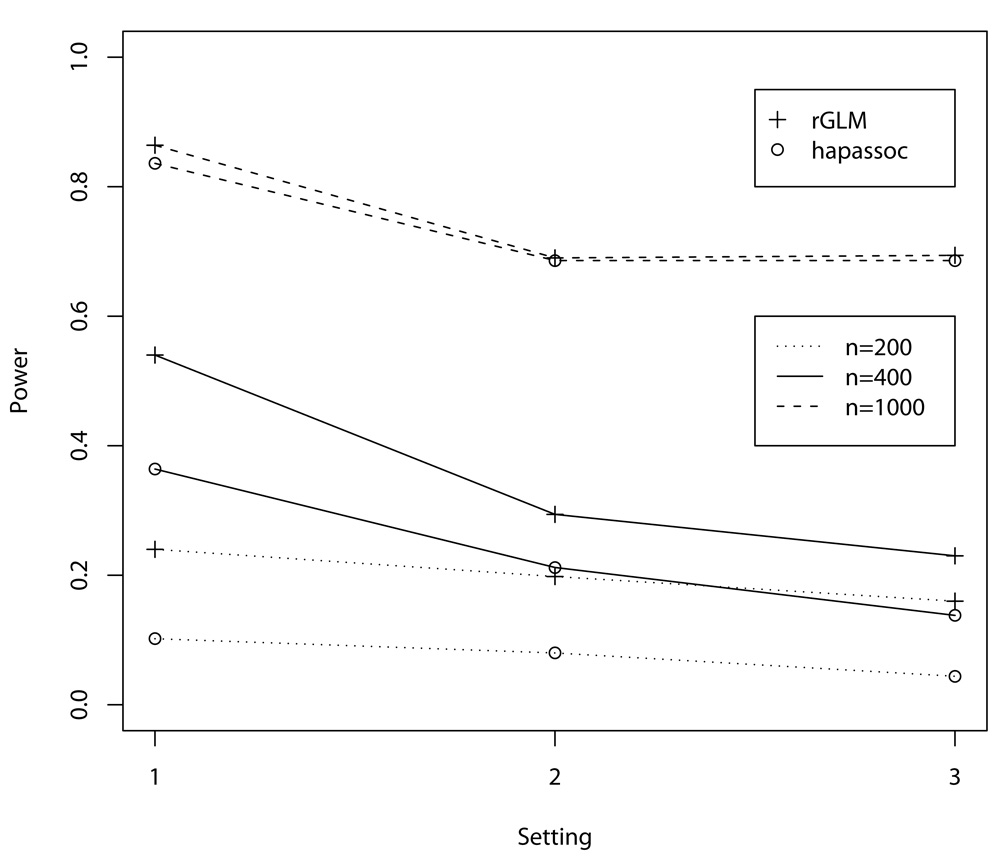
The large number of non-convergence replicates using hapassoc reduces its power to detect association between the rare haplotypes and disease, whereas rGLM did not encounter any problem with convergence and thus has greater power, especially when the sample sizes are small to moderate (Figure 1, n = 200 and 400). As can be seen from the figure, analysis using hapassoc when the disease associated haplotypes are rare can be vastly under-power. For example, the analysis using rGLM with a sample size of n = 200 (scenarios 2 and 3) can lead to as much power as an analysis using hapassoc with the sample size being double (n = 400). When the sample size is large (n = 1, 000), the non-convergence rates are in the single digits (Table III) and as such the powers for detecting associations using rGLM or hapassoc are comparable, as expected. The fact that hapassoc and rGLM achieve similar results without the problem of non-convergence is also demonstrated in the last column (Power.C) of Table III, where the power calculations were based on only those replicates that converged. The differences in the two procedures are rather small, and may be explained by random variability.
Fig. 1.
Power comparisons between the results from hapassoc and rGLM. The X-axis denotes the three one-haplotype-block settings, which correspond to 6, 9, and 12 haplotypes, respectively. All simulations were based on 500 replicates.
Since the goal of an association study is not simply to determine whether there is association, but also what haplotype(s) are involved, it is of interest also to ascertain whether the associated haplotypes in the simulation models are contained in the final model resulting from the analyses. For rGLM, the haplotypes that had non-zero coefficient estimates were taken to be the associated haplotypes because those that were not implicated received a coefficient of zero and dropped out of the model. For hapassoc, none of the coefficient estimates would be zero, and therefore we constructed 95% confidence intervals for the coefficients of the two associated haplotypes. Table IV shows the percentages of replicates including each of the two rare haplotypes (the β1 and the β2 columns), both (the β1& β2 column), or at least one (the β1/β2 column) in the final models estimated by hapassoc or rGLM. The results show that rGLM identified the associated haplotypes more often than hapassoc. It is of particular interest to note that, for n = 1, 000, despite comparable results for detecting association (Figure 1), hapassoc had much more difficulty identifying both rare haplotypes in the same run. These results attest to the advantage of employing the underlying principle of rGLM to reduce the degrees of freedom. Although an omnibus test using hapassoc can detect association, the large number of parameters that need to be estimated would lead to large standard errors, thus reducing its ability for precise estimation of the coefficients of rare, associated, haplotypes.
TABLE IV.
Percentages of replicates that identified the two rare haplotypes as the associated ones
| rGLM | hapassoc | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Setting | β1 | β2 | β1&β2 | β1/β2 | β1 | β2 | β1&β2 | β1/β2 |
| 200 | 1 | 24.4 | 39.8 | 22.6 | 41.6 | 0.0 | 2.2 | 0.0 | 2.2 |
| 2 | 13.6 | 31.0 | 11.6 | 33.0 | 0.0 | 0.6 | 0.0 | 0.6 | |
| 3 | 11.0 | 27.8 | 9.2 | 29.6 | 0.0 | 0.4 | 0.0 | 0.4 | |
| 400 | 1 | 45.4 | 62.6 | 43.2 | 64.8 | 12.4 | 33.2 | 5.4 | 40.2 |
| 2 | 33.6 | 52.4 | 30.2 | 55.8 | 10.2 | 27.6 | 5.6 | 32.2 | |
| 3 | 24.8 | 46.6 | 22.4 | 49.0 | 11.0 | 22.2 | 5.6 | 27.6 | |
| 1000 | 1 | 82.4 | 92.8 | 81.6 | 93.6 | 28.4 | 58.2 | 18.6 | 68.0 |
| 2 | 69.6 | 90.8 | 68.6 | 91.8 | 16.4 | 36.6 | 7.8 | 45.2 | |
| 3 | 68.6 | 89.6 | 66.2 | 92.0 | 12.2 | 41.0 | 5.2 | 48.0 | |
TWO HAPLOTYPE BLOCKS
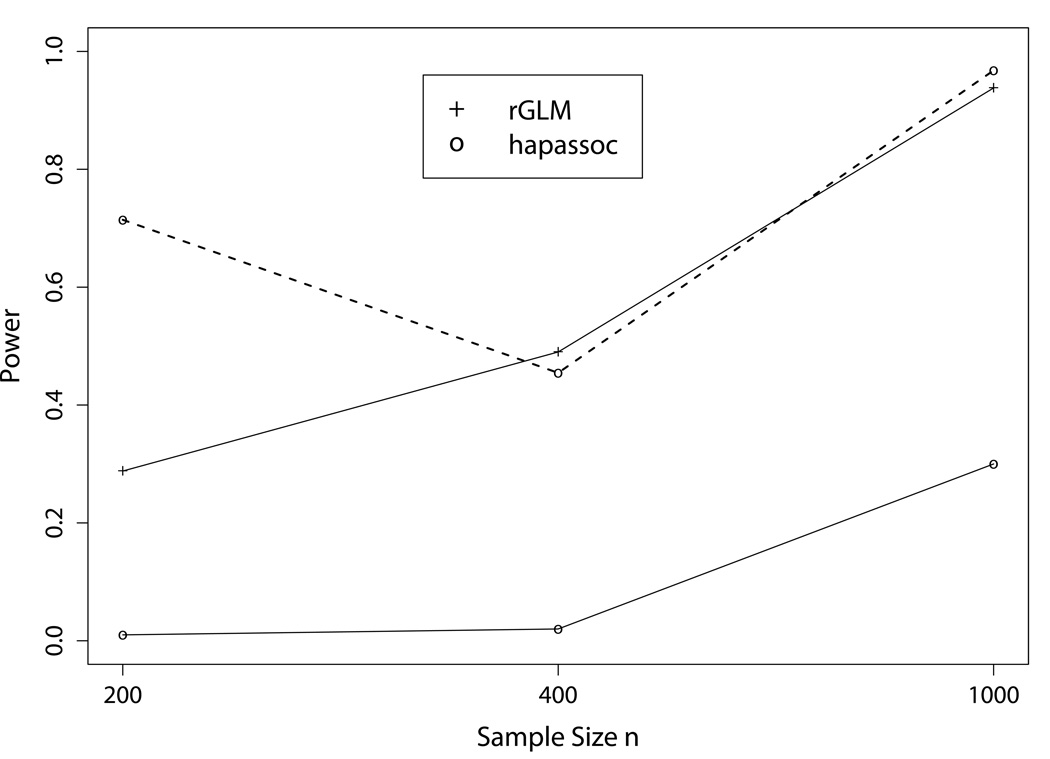
When the interacting effects between two haplotype blocks were considered, there were a total of 16 haplotype combinations, with many having small frequencies. As such, hapassoc had much greater difficulty converging, even when the sample size was 1,000. The non-convergence rates were 98.6, 95.6, and 69%, for n = 200, 400, and 1,000, respectively. The two solid curves in Figure 2 show that rGLM has much higher power than hapassoc, even when n = 1, 000, unlike the results from the one-haplotype-block settings. When the powers for hapassoc were computed only based on those replicates that converged, they (dashed line; for n = 400 and 1,000) were comparable to rGLM, as in the one haplotype block settings. For n = 200, the result was based on only 7 replicates (i.e., hapassoc was only able to obtain results in 7 out of 500 replicates), and thus the vast difference in the powers between rGLM and hapassoc could be due to random variation given the very small sample size. In terms of identification of associated haplotypes, rGLM included at least one of the seven associated rare haplotypes with reasonable rates (Table V). Their counter parts for hapassoc were almost all zeros. Even for n = 1, 000, only 11% of the runs identified one or more associated haplotypes.
Fig. 2.
Power comparisons between the results from hapassoc and rGLM for the fourth setting with interacting effects between haplotypes from two blocks. The dashed curve shows the powers for hapassoc based on 7, 22, and 155 replicates, the numbers of replicates (out of 500) that converged, for n = 200, 400, and 1,000, respectively.
TABLE V.
Percentages of replicates that identify a variable number of the associated haplotypes
| n = 200 | n = 400 | n = 1000 | ||||
|---|---|---|---|---|---|---|
| #βi | rGLM | hapassoc | rGLM | hapassoc | rGLM | hapassoc |
| 1 | 16.6 | 0.0 | 13.6 | 0.2 | 1.6 | 8.6 |
| 2 | 14.4 | 0.0 | 14.8 | 0.0 | 6.8 | 1.4 |
| 3 | 11.6 | 0.0 | 18.4 | 0.0 | 11.4 | 1.0 |
| 4 | 7.4 | 0.0 | 16.8 | 0.0 | 21.0 | 0.0 |
| 5 | 4.2 | 0.0 | 10.4 | 0.0 | 25.0 | 0.0 |
| 6 | 2.0 | 0.0 | 5.8 | 0.0 | 21.2 | 0.0 |
| 7 | 1.4 | 0.0 | 2.2 | 0.0 | 12.4 | 0.0 |
| Total | 57.6 | 0.0 | 82.0 | 0.2 | 99.4 | 11.0 |
DISCUSSION
It has been a decade-long notion, foreseen by Risch and Merikangas [1996], that genetic association analysis is much more promising in mapping common genetic diseases compared to linkage mapping, which is more suitable for mapping rare diseases caused by rare variants with high penetrances. Whole genome association studies have indeed surged recently in popularity as technological advances have made large-scale genotyping more feasible and as new exciting results offer tremendous hope and optimism [Chanock and Hunter, 2008; Hung et al., 2008; Thorgeirsson et al., 2008; Amos, et al., 2008]. The standard logic of WGAS rests upon the CD/CV hypothesis. However, it has been argued that rare variants are valuable for mapping common diseases [Purcell et al., 2007; Gorlov et al., 2008] and as such, powerful statistical methods for detecting CD/RV associations need to be developed.
The method proposed in this paper is applicable to both the CD/CV and CD/RV scenarios based on the GLM framework. We borrow a dimension-reduction method from the data mining and statistical learning literature, but use it for the purpose of weeding out haplotypes that are not associated with the disease so that the associated haplotypes, especially those that are rare, can stand out and be accounted for more precisely. By using high-dimensional data analysis techniques, which are frequently employed in microarray analyses, interacting effects among haplotypes in different blocks can also be investigated without much concern about the sample size being overwhelmed by the number of haplotype combinations.
The results from our simulation study show that, for detecting association under CD/RV, standard GLM method such as that implemented in hapassoc may fail to provide coefficient estimates for the (rare) associated haplotypes, especially if inter-block interaction effects are considered, resulting in a great loss of power compared to our rGLM method. In addition to power consideration, the ability to identify the associated haplotypes is just as important. Our results indicate that rGLM can uncover the associated variants much more frequently than can hapassoc. However, our comparison may be unfair because the analysis strategies are different: an ordinary GLM method requires the confidence interval estimates for the coefficients (to see whether they include zero because none of the coefficient estimates them-selves would be zero); whereas all haplotypes with non-zero coefficient estimates are taken to be associated under the regularized GLM (because the estimated unassociated haplotypes have a coefficient of zero). As a general analysis strategy, if one is interested in estimating the odds ratios of the associated haplotypes, a follow-up step could be performed using the ordinary GLM by pooling all the haplotypes with zero estimated coefficients from rGLM as a reference category. Confidence interval estimates for the coefficients can then be constructed.
The results from our simulation study are consistent with what we observed in our analysis of a dataset on multiple sclerosis (unpublished data). Using hapassoc, we encountered non-convergence for 15% of the tests performed due to the existence of rare haplotypes. Using the proposed rGLM, non-convergence was no longer an issue. Furthermore, we were able to identify a number of significant results that have haplotype frequencies as low as 0.0014.
A number of assumptions and other specific issues in the analysis procedure deserves further discussion. First, haplotype frequencies are estimated from the pooled sample of cases and controls. This may lead to biased estimates of haplotype effects, especially if the cases are represented in a higher proportion. Such an effect is minimal, though, unless there is substantial haplotype ambiguity from the genotype data [Stram et al., 2003]. However, if concerns arise, then one can easily modify the model in (1) to specify separate frequency parameters for the cases and the controls. To reduce the number of parameters in the model, we assume that the system of haplotypes is in HWE. If there is sufficient indication of departure from HWE, then we can instead employ a model to account for such departure, for example, by remodeling (3) using a within-population inbreeding coefficient parameter that allows for excess/reduction of homozygosity [Weir, 1996]. Furthermore, if HWE holds in the controls but is violated in the cases, then separate modeling and estimation of the frequencies can be entertained. For the selection of the penalty term and the amount of penalty (the λ parameter), we chose to work with LASSO and set the maximum penalty to be 10. The latter is due to our observation that the optimum amount of penalization is usually much smaller than 10, and therefore there is no need to go beyond that. For the former, there are other forms of penalty except LASSO, including SCAD [Fan and Li, 2001] and ridge [Horel and Kennard, 1970], but we note that employing a ridge-like penalty might not lead to better results than an unpenalized approach [Souverein et al., 2006].
The likelihood as depicted in (1) may be more specifically referred to as prospective because it models the probability of a disease outcome conditional on the underlying genetic variant and other covariates. This framework is most commonly adopted by statistical methods for genetic analysis under the case-control design [e.g., Schaid et al., 2002; Burkett et al., 2006] due to its modeling simplicity and computational feasibility. Theoretical work justifying such an approach exists [Prentice and Pyke, 1979], although the needed assumption may not be easily satisfied in genetic association studies. A retrospective likelihood formulation, in which the basic probability is for the genetic makeup and other covariates conditional on the disease status, has also been proposed [Satten and Epstein, 2004]. However, a number of assumptions are needed for the validity of the proposed approach, including that the disease is rare. As such, careful evaluation of whether such a formulation is appropriate for the CD/CV and/or CD/RV settings is warranted.
ACKNOWLEDGEMENTS
This work was supported in part by National Institutes of Health grant NS046696, National Science Foundation grant DMS-0112050, and the Biomedical Research and Technology Transfer grant from the State of Ohio Tech 05–062. We would like to thank Dr. Yang Liu and Dr. Wolfgang Sadee for sharing their unpublished multiple sclerosis data with us which motivated this study, and Dr. Dustin Potter for a critical read of the manuscript.
Footnotes
WEB RESOURCES
The URLs for software packages are as follows: hapassoc, http://stat-db.stat.sfu.ca:8080/statgen/research/hapassoc rGLM, http://www.stat.osu.edu/~statgen/SOFTWARE/rGLMR, http://www.r-project.org/
REFERENCES
- Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second International Symposium on Information Theory; Akademia Kaido: Budapest; 1973. pp. 267–281. [Google Scholar]
- Akey JM, Jin L, Xiong M. Haplotypes vs. single-marker linkage disequilibrium tests: what do we gain? Eur J Hum Genet. 2001;9:292–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature Genet. 2008 doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Knapp M. A Powerful Strategy to Account for Multiple Testing in the Context of Haplotype Analysis. Am J Hum Genet. 2004;75:561–570. doi: 10.1086/424390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett K, Graham J, McNeney B. hapassoc: Software for likelihood inference of trait associations with SNP haplotypes and other attributes. Journal of Statistical Software. 2006;16:1–19. [Google Scholar]
- Burkett K, McNeney B, Graham J. A note on inference of trait associations with SNP haplotypes and other attributes in generalized linear models. Hum Hered. 2004;57:200–206. doi: 10.1159/000081447. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Hunter DJ. Genomics: When the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- Durrant C, Zondervan KT, Cardon LR, Hunt S, Deloukas P, Morris AP. Linkage disequilibrium mapping via cladistic analysis of single-nucleotide polymorphism haplotypes. Am J Hum Genet. 2004;75:35–43. doi: 10.1086/422174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin D, Cohen A, Essioux L, Chumakov I, Blumenfeld M, Cohen D, Schork NJ. Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer’s disease. Genome Res. 2001;11:143–151. doi: 10.1101/gr.148401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Li R. Variable selection via nonconcavee penalized likelihood and its oracle properties. J Am Stat Assoc. 2001;96:1348–1360. [Google Scholar]
- Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horel AE, Kennard RW. Ridge regression: biased estimation for non-orthogonal problems. Technometrics. 1970;12:55–67. [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Li Y, Sung WK, Liu J. Association Mapping via Regularized Regression Analysis of SNP Haplotypes in Variable-sized Sliding Windows. Am J Hum Genet. 2007;80:705–715. doi: 10.1086/513205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor J, Marjoram P, Thomas D. Fine-scale mapping of disease genes with multiple mutations via spatial clustering techniques. Am J Hum Genet. 2003;73:1368–1384. doi: 10.1086/380415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23:221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- Park MY, Hastie T. L1 regularization path algorithm for generalized linear models. J R Statist Soc B. 2007;69:659–677. [Google Scholar]
- Prentice RL, Pyke R. Logistic disease incidence model and case-control studies. Biometrika. 1979;66:403–411. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Rosenberg PS, Che A, Chen BE. Multiple hypothesis testing strategies for genetic case-control association studies. Stat Med. 2006;25:3134–3149. doi: 10.1002/sim.2407. [DOI] [PubMed] [Google Scholar]
- Satten GA, Epstein MP. Comparison of prospective and retrospective methods for haplotype inference in case-control studies. Genet Epidemiol. 2004;27:192–201. doi: 10.1002/gepi.20020. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltman H, Roeder K, Devlin B. Evolutionary-based association analysis using haplotype data. Genet Epidemiol. 2003;25:48–58. doi: 10.1002/gepi.10246. [DOI] [PubMed] [Google Scholar]
- Souverein OW, Zwinderman AH, Michael WT, Tanck MWT. Estimating haplotype effects on dichotomous outcome for unphased genotype data using a weighted penalized log-likelihood approach. Hum Hered. 2006;61:104–110. doi: 10.1159/000093476. [DOI] [PubMed] [Google Scholar]
- Stone M. Cross-validatory and assessment of statistical predictions (with discussion) J Roy Statist Soc B. 1974;36:111–147. [Google Scholar]
- Stram DO, Haiman CA, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Pike ML. Choosing haplotype-tagging SNPs based on unphased genotype data from a preliminary sample of unrelated subjects with an example from the multiethnic cohort study. Hum Hered. 2003;55:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58:267–288. [Google Scholar]
- Tzeng JY. Evolutionary-based grouping of haplotypes in association analysis. Genet Epidemiol. 2005;28:220–231. doi: 10.1002/gepi.20063. [DOI] [PubMed] [Google Scholar]
- van de Geer SA. High-dimensional generalized linear models and the lasso. Ann Stat. 2008;36:614–645. [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Human Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- Zhao JH, Curtis D, Sham PC. Model-free analysis and permutation tests for allelic associations. Hum Hered. 2000;50:133–139. doi: 10.1159/000022901. [DOI] [PubMed] [Google Scholar]
- Zou H, Hastie T, Tibshirani R. On the “Degrees of Freedom” of the Lasso. Ann Stat. 2007;35:2173–2192. [Google Scholar]