Abstract
Bacteria have a complex internal organization with specific localization of many proteins and DNA, which dynamically move during the cell cycle and in response to changing environmental stimuli. Much less is known, however, about the localization and movements of RNA molecules. By modifying our previous RNA labeling system, we monitor the expression and localization of a model RNA transcript in live Escherichia coli cells. Our results reveal that the target RNA is not evenly distributed within the cell and localizes laterally along the long cell axis, in a pattern suggesting the existence of ordered helical RNA structures reminiscent of known bacterial cytoskeletal cellular elements.
Keywords: fluorescent protein, live E. coli cells, protein complementation, RNA visualization
Despite their relatively small dimensions, bacterial cells show a remarkable, rich internal subcellular organization that has captured the interest of researchers over the past decade (1–4). Many cytoplasmic and membrane proteins, particularly those involved in cell division, DNA replication, and chromosome segregation, have specific subcellular localizations that can change quickly over time in response to cell cycle progression, motility, and environmental cues. This dynamic and organized behavior is also true for bacterial chromosomal DNA. The use of GFP fusions and in situ fluorescence hybridization (FISH) have shown that every chromosomal locus has a defined subcellular address and is replicated and segregated into the new cell as part of an active and directed process (4, 5). Bacterial plasmids, both low and high copy, also have specific cellular addresses and segregate in a fashion that is unique for a given plasmid (6–8).
Little is known, however, about RNA dynamics in bacteria. With the advent of new methods to label RNA in live cells, the transcription kinetics, localization, and movement of RNA in the bacterium Escherichia coli has begun to be discerned only recently (9–13).
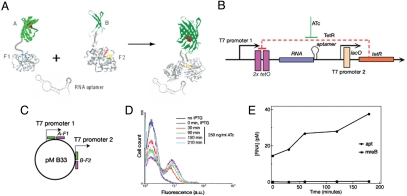
To understand RNA dynamics in live cells better, it would be useful to develop RNA labeling methods that would allow direct visualization and real-time quantitation of RNAs with low background levels. We recently reported a system based on protein complementation that uses binding of a split and inactive protein complex to a short interacting sequence on a target RNA. The marker protein re-associates and becomes fluorescent only upon binding to RNA (13), which makes this approach more desirable than alternative techniques relying on expression of full-size fluorescent proteins. Briefly, the method consists of fusing the N-terminal fragment of EGFP to the N-terminal domain of an RNA-binding protein, the eukaryotic initiation factor 4A (eIF4A), via a polypeptide linker. Similarly, the C-terminal fragment of EGFP is fused to the C-terminal domain of eIF4A. The target RNA is tagged at the 3′ end with an aptamer sequence known to bind eIF4A with high affinity (14) (Fig. 1A). Expression of the labeling components in E. coli cells generates a fluorescent signal only in the presence of the target RNA, caused by the reassociation of the two EGFP fragments and formation and maturation of the chromophore (13). This method offers two main advantages for live cell RNA imaging, namely low background fluorescence and small labeling complex.
Fig. 1.
RNA-inducible system to study transcription kinetics in live E. coli cells. (A) Experimental design. (B) A plasmid pMB133, expressing RNA tagged with the eIF4A-interacting aptamer under the control of TetR. TetR, expressed from the second T7 promoter, represses expression of RNA tagged with aptamer. Anhydrotetracycline (ATc) is required to relieve TetR repression and to induce RNA synthesis. (C) Plasmid expressing two protein fusions from the two T7/lacO promoters. (D) Flow cytometry analysis of E. coli cells transformed with the plasmids MB133 and MB33 after induction of RNA synthesis with anhydrotetracycline (ATc). (E) RNA concentration after induction with ATc as determined by competitive PCR (for details see ref. 13).
Here we present an extension of our previous RNA-labeling system, in which RNA induction and synthesis of the split proteins are separately controlled. Using this system we were able to image and quantify early stages of RNA synthesis at a single-cell level. Our results suggest that EGFP complexes formed in live E. coli cells are unstable or reversible, and that cell fluorescence reflects changes in target RNA concentration. Visualization of RNA in live cells revealed striking real-time spatiotemporal patterns of RNA not seen before in bacterial cells.
Results
RNA-Inducible System for Probing Spatiotemporal RNA Dynamics in Live Bacterial Cells.
Our original RNA imaging system involved simultaneous expression of two proteins, each consisting of a fragment of a split fluorescent protein fused to a fragment of an RNA-binding protein, and the target RNA (13). All components of the system were expressed from the T7/lacO promoters and were induced simultaneously by the inducer IPTG. To be able to follow transcription and localization of RNA particles in real time, we decoupled the expression of the protein fusions from that of the target RNA by modifying the T7 promoter directing RNA expression. We inserted tetO sequences in the promoter region to place RNA synthesis under the control of the repressor molecule, TetR (see Fig. 1B and Supplementary procedures for details). This new plasmid, pMB133 (Fig. 1B), would encode a model RNA tagged with two copies of the aptamer in tandem.
We transformed E. coli cells with two plasmids: pMB133, encoding RNA and pMB33, which encodes two split fusion proteins that interact with the RNA-aptamer (13). To induce synthesis of the two fused proteins and the TetR protein, we allowed cells to grow at room temperature in Luria–Bertani (LB) medium supplemented with antibiotics and the inducer IPTG for 2 days and monitored fluorescence of the cells by flow cytometry. This was done to ensure that no fluorescent signal was coming from an unrepressed TetR-responsive T7 promoter. We grew cells at room temperature to slow cell growth and allow proper folding of proteins. Once the system was fully repressed (with no detectable fluorescence above cell background), we added the inducer of RNA-aptamer expression, anhydrotetracycline (ATc; 250 ng/ml), and measured fluorescence of the cells by flow cytometry every 30 min after addition of ATc (Fig. 1D). We observed an increase in average cell fluorescence and in the number of fluorescent cells over time. These results show that fluorescence was developed after the derepression of the target RNA synthesis. In parallel, we measured target RNA concentration using competitive polymerase chain reaction (PCR) as described (13). This experiment also revealed an increase in target RNA concentration as a result of induction with ATc (Fig. 1E). Thus, we can correlate an increase in number of fluorescent cells after ATc induction with an increase in target RNA concentration. Finally, we also observed an increase in fluorescence with increasing concentrations of the RNA inducer [supporting information (SI) Fig. S1]. These results confirmed that cell fluorescence is directly related to the synthesis of target RNA.
Transcription Kinetics in Individual E. coli Cells.
A possible issue when using fluorescent protein complementation, also referred to as bimolecular fluorescence complementation (BiFC), for kinetic measurements is the degree to which these complexes are reversible. For example, in vitro assays show that BiFC complexes may be irreversible under certain conditions (15). However, some BiFC complexes studied both in vitro and in vivo were found to be reversible (16–19).
We examined the stability of our fluorescent RNP complexes in live cells, taking advantage of the RNA-inducible system. If BiFC complexes formed upon induction of RNA synthesis are irreversible, they would not dissociate even when the molecules triggering their association, the target RNAs, are absent. Therefore, one would expect to observe steady fluorescence intensity, even when RNA is depleted. To test the stability of split fluorescent complexes in vivo, we followed RNA-dependent cell fluorescence over time in the absence of new RNA synthesis by flow cytometry (see Supplementary procedures, Fig. S2). We showed that removal of the inducer of RNA synthesis, ATc, causes a decrease in the fluorescent signal. These results suggest that BiFC complexes formed in the presence of RNA are unstable and/or can come apart once target RNA synthesis is impaired.
Evidence that the decrease in fluorescence is not due to dilution of the RNA caused by cell growth comes from time-lapse fluorescence microscopy of single cells. We grew cells in culture and induced protein components of the RNA labeling system as described in the previous section. We ensured that the culture was not fluorescent before induction with ATc. At this time, we resuspended cells in M9 minimal medium supplemented with antibiotics and IPTG as a way to reduce background fluorescence from components in the rich LB medium. We immobilized E. coli cells on microscope slides and imaged them by high-resolution time-lapse microscopy right after addition of ATc (see Materials and Methods).
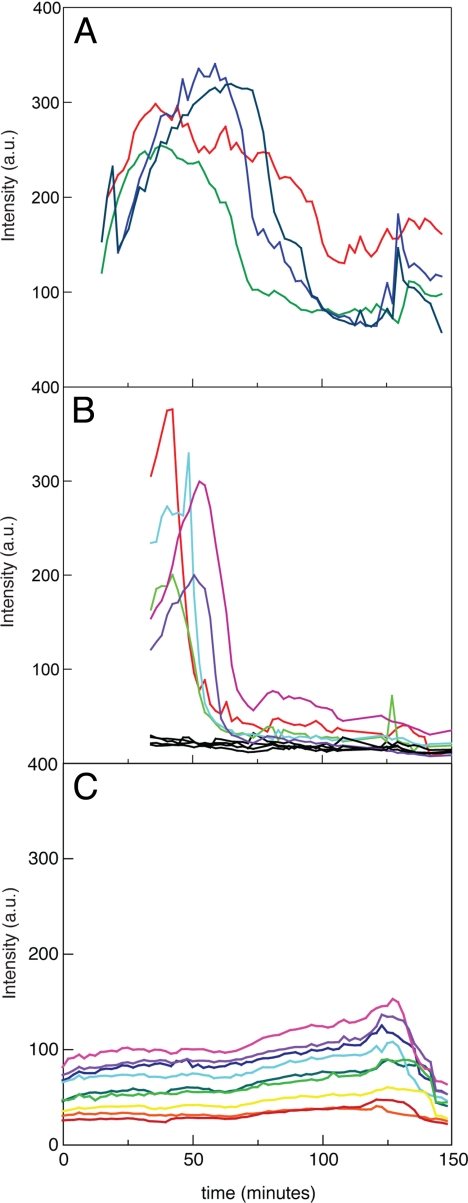
Fig. 2 A and B display two typical experiments showing changes in total fluorescence intensity in individual cells over 2.5 h after RNA induction. In all experiments, the majority of the cells displayed a rapid increase in cell fluorescence. In ≈50% of the cells, the total fluorescence intensity increased 5–8- fold over the background level, displaying a prominent peak 50–70 min after induction. The fluorescent signal then dropped, nearly reaching the uninduced level. In some cells, we observed a weak recovery of fluorescence at later stages of imaging. We also observed cells that showed a very slow and steady increase in fluorescence over the course of the experiment with no decline in the signal. Finally, we also found cells (≈50%) that showed no change in fluorescence over time. This cell population may represent cells not responsive to the inducer also observed by flow cytometry (Fig. 1D). The initial delay (15–20 min) in detecting cell fluorescence right after induction with ATc is not related to a delay in RNA synthesis or a limitation of our labeling system but, rather, a technical limitation resulting from the handling of the cells and the microscope. By automating this process, we should be able to detect the very early stages of transcription.
Fig. 2.
RNA kinetics in single E. coli cells upon induction with ATc. (A, B) Time course of total fluorescence in individual E. coli cells in two representative experiments. The t = 0 time is defined as the moment of ATc addition to cells. Measurements are taken at 2-min intervals. (C) Control measurements using cells with expression of full-size EGFP (13). The fluorescence excitation intensity in the control experiments is identical to the intensity used in experiments shown in (A) and (B), but the emission intensity in (C) is attenuated 10-fold.
As a control measurement, we used cells expressing full-size EGFP that were mounted on the microscope and imaged over a period of 2.5 h. Fig. 2C shows a representative set of time traces using identical illumination conditions as the ones used for imaging the split EGFP complexes. In this case, we generally observed a steady fluorescence trend or a slight increase after 70 min of observation. These results clearly eliminate the possibility of photo-bleaching caused by excessive illumination under the conditions used for imaging.
The fact that the total fluorescence intensity in ATc-induced cells displays a cyclical pattern (an increase, decrease, and in some cases a smaller increase) when the control cells display no evidence of photophysical degradation of the EGFP complexes, implies that these complexes are either degraded or dissociated during the course of the observation (we cannot discriminate these two possibilities). This result enables us to quantitatively characterize RNA transcription kinetics using our EGFP complementation system.
Estimation of Number of EGFP Molecules per Cell After RNA Induction.
To estimate the actual concentration of the assembled EGFP molecules in individual E. coli cells, we have performed the following experiments. First, using the confocal configuration (see Materials and Methods), the intensity of samples of purified EGFP (BioVision, catalog no. 499-1000) at different concentrations in PBS buffer (pH 7) was measured immediately after preparation. These measurements formed a calibration curve that was used to estimate the concentration of split EGFP in mounted E. coli cells expressing RNA. We did this by focusing the laser at the center of the fluorescent E. coli cells and measuring intensity in several trials. Here we took advantage of the fact that our confocal volume in z direction is close to the cell thickness (≈1 μm). Therefore, it is possible to assume that the volumes probed in homogeneous solution used for calibration and in the cell are similar. Then we switched our system to wide-field illumination and recorded images of the cells that were first analyzed in confocal format. In this way, we could translate the CCD camera images into concentration of complemented EGFP in each cell. This method was not perfect, but it provided a rough estimate of EGFP concentration in the cells. These measurements resulted in the determination of ≈500 complemented EGFP molecules in the cells expressing our RNA-inducible system (≈30 min after induction).
Spatiotemporal Patterns of Fluorescence Within Cells.
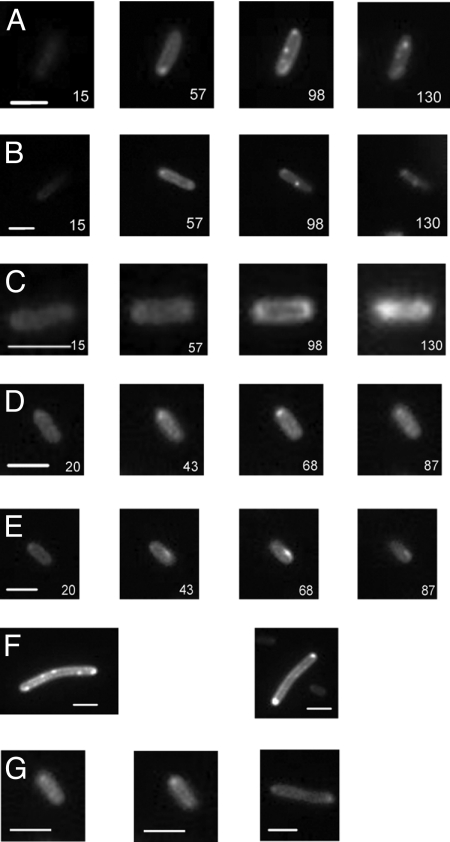
Time-lapse microscopy of cells expressing our inducible RNA labeling system revealed striking spatial changes in fluorescence over time. Generally, we could visualize fluorescence in cells under the microscope 15–20 min after induction of RNA synthesis. As time progressed, more cells became fluorescent and the fluorescence intensity in each cell increased. Three major trends in fluorescence distribution could be discriminated (Fig. 3 and Figs. S3-S7). First, RNA-dependent fluorescence was distributed laterally along the long axis of the cell body, whereas the central area of the cell was much dimmer (Fig. 3 A and B and Fig. S8). This trend was characteristic of most cells including dividing cells. Therefore, we think it is unlikely that this pattern is an artifact of our labeling method.
Fig. 3.
Time-lapse fluorescent images of live E. coli cells expressing RNA. Time after ATc induction is given in minutes in each image. Bars represent 2 μm. (A) In this E. coli cell, fluorescence is localized mainly along the edges of the cells (best visible in the time window 55–85 min). Focal points at the mid-cell and quarter-cell positions appear ≈100 min after RNA induction with ATc (see Fig. S3 for the full set of time-lapse images). (B) In this E. coli cell, the peripheral distribution of fluorescence is clearly visible in images in the time window 36–78 min. Mid-cell focal accumulation appears ≈75 min after RNA induction with ATc (see also Fig. S4). (C) A small E. coli cell showing nonuniform lateral fluorescence distribution. The dimmer fluorescence in the middle of the cell is best visible in the time window 85–109 min (see also Fig. S5). (D) A cell displaying polar accumulation of fluorescence (see also Fig. S6). (E) An E. coli cell displaying time-dependent fluorescent accumulation at a pole, which disappears later in the cycle and reappears at a different location (see also Fig. S7). (F) Examples of E. coli cells with fluorescence accumulations at the edges of the cell, likely indicators of helical structures, and the cells with visible diffuse helical structures slanted to the long cell axis.
Second, a considerable number of cells displayed accumulations of fluorescence at or near cell poles (≈40–60% of cells) as seen before in our previous study (13). The polar accumulations were widely variable among different cells. In some cells, fluorescence first appeared at the poles, whereas in other cells accumulations at the poles formed only 30–50 min after ATc induction, when other parts of the cell were already fluorescent. In addition, in some cells these accumulations were dynamic, meaning that spots appeared, then disappeared or appeared later at a different location (Fig. 3E and Movie S1). In a smaller number of cells (5%), which usually were elongated rods, we observed focal accumulations of fluorescence at the middle and, on some occasions, at ¼ and ¾ positions of the cell (Fig. 3 A and B; Fig. S3, Fig. S4, and Fig. S9). These accumulations became visible 80–100 min after RNA induction. They differed from the polar accumulations in that they were much more localized and less dynamic, as they did not change their position with time (Fig. 3 A and B and Movie S2).
An inspection of circumferential fluorescence distribution revealed a clear pattern. In many cells, the presence of small fluorescent dots (clusters) on the cell edges was discernable (Fig. 3F). Peripheral fluorescent clusters might be indicators of helical structures because at the cell edges the helix is closest to being perpendicular to the image plane and therefore brightest (20, 21). Indeed, in other cells the presence of bands slanted relative to the long axis of the cell was recognizable (Fig. 3G). This pattern was reminiscent of many helical patterns present in bacterial cells (20, 22–28). Note, however, that these RNA-related patterns were more diffuse than most helical patterns revealed by protein labeling.
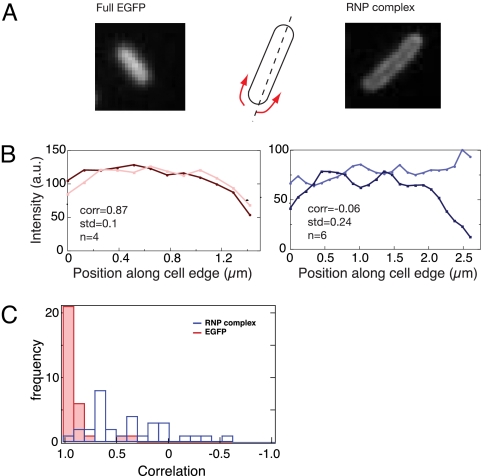
To characterize these structures better, we analyzed fluorescence patterns along the circumference of the cells. Approximating the cells to capped rods, we extracted the fluorescence intensities along the two sides of the E. coli cell edges (as explained in Supplementary procedures). Fig. 4 A and B display such an analysis applied to two cells: a cell expressing full-size EGFP (left), and a cell expressing the RNP complex (right) (see Fig. S10 for more examples of RNA-expressing cells). The extracted intensity curves of the two cells are clearly different: whereas the curves associated with the two sides of the cell expressing full-size EGFP generally display similar patterns, the curves of the two sides of the cell with RNP complex appear to be strikingly out of phase.
Fig. 4.
Lateral fluorescence distribution in bacterial cells expressing RNP complexes is principally different from the random distribution of EGFP in E. coli cells. (A) Examples of cells expressing full-size EGFP (Left) and RNP complex (Right), and schematic illustration of the lateral fluorescence profiling approximating the cell as rods. (B) Typical examples of lateral fluorescence profiles in cells expressing EGFP (Left) and RNP complex (Right). The two lines in each panel represent the fluorescence intensity on the left and right sides of the rods. Insets show the correlation coefficient and standard deviation, where n is the number of the replicate measurements of one cell (for more examples, see Fig. S10). (C) Histogram of the correlation coefficients for 30 cells expressing EGFP (blue) and 31 cells expressing RNP (red) measured as in B, showing clear differences between the full-size EGFP-expressing cells and the RNP-expressing cells.
We reasoned that if fluorescence is distributed randomly along the rod-shaped cell, the fluorescence patterns on the two edges of the rod should be similar. Therefore, the correlation between the two functions should be close to 1. The correlation should also be close to 1 if there is a nonrandom but symmetrical longitudinal distribution (for example, the ring-type distribution perpendicular to the long axis). If the distribution is not random and not symmetrical, the correlation coefficient between the two functions should be <1.
To validate this assumption, we performed the analysis displayed in Fig. 4 for 30 cells expressing full-size EGFP as well as 31 cells expressing the fluorescent RNP complex. The correlation results are shown in Fig. 4C. We found that in the cells expressing full-size EGFP, the fluorescence distribution profiles were very similar on both edges of the cell and that the correlation between the two functions was close to 1 with very slight variability (0.90, R2 = 0.05, 95% confidence). In contrast, in cells expressing fluorescent RNP complexes, fluorescence patterns for the two cell sides differed dramatically and often appeared to be out of phase. In these cells, the correlation value was much lower (0.37, R2 = 0.13, 95% confidence) and, in several cases, an anticorrelation was seen (negative correlation values). The cells that expressed fusion proteins in the absence of target RNA and displayed a small background fluorescence that could be detected by the improved imaging method showed a distribution of fluorescence similar to that of cells expressing full-size EGFP. Thus, the observed patterns were unique to the cells expressing the target RNA.
This analysis shows that distribution of RNA-based fluorescence in E. coli cells qualitatively differs from the uniform distribution of full-size EGFP. These data suggest that the lateral distribution observed for the labeled RNA is not random and is nonsymmetrical relative to the long cell axis. This is consistent with the visual evidence suggesting orderly helical structures slanted to the long axis in E. coli cells expressing RNA (Fig. 3 F and G; see also Movies S1–S3).
Discussion
We report on an inducible transcription system combined with fluorescent protein complementation for real-time visualization of RNA in live bacterial cells. Using this system, the rapid onset of transcription as well as the subsequent distribution of RNA was directly visualized in single cells. Noncomplemented proteins expressed in the cell produce low background fluorescence, and high fluorescent signal is triggered only in the presence of the aptamer-tagged RNA. In our previous RNA detection system, protein fusions and RNA were induced simultaneously (13). Therefore, it was difficult to study real-time transcription kinetics because of the lag period necessary for fluorescent protein maturation.
This study using RNA-inducible system alleviated this obstacle and revealed a pulsing transcription dynamics that agrees with previous results on RNA dynamics in live E. coli cells (11, 12). Using the MS2 coat protein-based RNA labeling method and fluorescence correlation spectroscopy (FCS), Le et al. also showed a pulsating pattern of transcription after measuring RNA concentration. In this study, the efflux pumps AcrAB-TolC, which expel ATc inducer out of the cell, were responsible for this pulsating transcriptional activity (11, 12). Our results are consistent with this interpretation. Thus, our data show that our RNA labeling system, based on complementation of split fluorescent proteins, is capable of detecting real-time RNA dynamics in bacteria.
Time-lapse fluorescent imaging of cells expressing short RNA revealed striking fluorescence patterns, which we did not observe before (13). RNA was localized mostly at the periphery of the cells along the long cell axis (Fig. 3 and Figs. S3-S7). We also observed dynamic accumulations of fluorescence at the poles, and the presence of focal points at the mid (Fig. S9), and quarter points of the cell in larger more elongated rods. Finally, we obtained evidence suggesting the presence of ordered helical structures underlying RNA localizations (Figs. 3 and 4, Movies S1-S3). Such RNA patterns were not detected in other studies aimed at RNA visualization in live cells (9, 10), including our earlier study which used simultaneous induction of proteins and RNA (13).
These differences can be explained by several factors. First, in our previous system (in which proteins and RNA were induced simultaneously), we needed to grow and express the labeled RNP complex overnight to be able to obtain an RNA-dependent fluorescent signal. Because of this long incubation time, the fluorescent signal was usually 5-fold higher than the signal obtained with our inducible system. Brighter and stronger cell fluorescence could make the study of finer details of RNA localization difficult to see. Also, the application of an advanced imaging technique in this study, coupled with the ability to control the onset of transcription, enabled us to visualize RNA only a few minutes after the start of transcription, which was not possible in our previous system.
The RNA patterns observed in this study are in line with the common knowledge of bacterial cell morphology and plasmid localization. The lateral RNA localization could be caused by the presence of a dense chromosomal nucleoid in the center of the cell (29, 30), which is thus less accessible for RNA localization and movement than the periphery of the cell. The polar accumulations of fluorescence in a large number of cells likely reflect localization of the corresponding plasmids and high concentrations of nascent RNA transcripts at the sites of plasmid location. In this study, target RNA was transcribed from a high copy number plasmid pETDuet1 with a ColE1 origin of replication. ColE1-containing plasmids form clusters at the cell poles, which migrate to the mid- and quarter points of the cell during plasmid segregation (31). Appearance of fluorescent focal points at the mid- and quarter-points in some cells (Fig. 3 A and B and Fig. S9) is consistent with this mechanism.
Our results also suggest that RNA localization at the periphery of the cell is not random; rather, it appears to form structures reminiscent of helices. This finding raises many intriguing questions about RNA localization and processing in bacteria. Recent studies showed that RNase E and RNA helicase, two components of the bacterial degradosome, form helical filamentous structures that coil around the length of the cell (27, 28). These structures are independent of the MreB and MinD cytoskeletal structures and may play important roles in cell growth, division, and chromosome segregation. They may also serve to compartmentalize RNA processing and RNA degradation (27, 28). It is tempting to speculate that the RNA network that we are revealing may be related to the same structures.
This work establishes a new RNA labeling system with tight control of RNA synthesis and provides a noninvasive method for RNA visualization in live bacterial cells over long time periods. The system is also capable of capturing changes in RNA concentrations in single cells, as the RNP complexes formed are dynamic and reversible. The method still has its limitations. First, the target RNA needs to be genetically modified for the labeling to occur. Second, it remains to be seen whether a chromosomally expressed RNA target can also be detected using this technology.
Nevertheless, this could be a powerful approach for studies exploring the biological roles of different RNAs in bacteria as well as for biophysical studies on the movement of RNA in live cells. System biology studies will also benefit from an alternative method to label and quantify RNA molecules in live bacterial cells.
Materials and Methods
Construction of plasmid pMB33, which expresses two protein fusions each containing, a fragment of EGFP and of the eukaryotic initiation factor 4A protein, has been described earlier (13). Construction of the plasmid pMB133 with regulated expression of RNA is described in Supplementary procedures.
Bacterial Growth and Induction.
E. coli cells BL21(DE3) were transformed with the plasmids pMB133 and pMB33 for expression of the RNA labeling system. Cells were grown in LB medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol with vigorous shaking at 37 °C for 3–4 h. For split proteins and TetR expression, cells were diluted 1:150 in fresh medium supplemented with 1 mM IPTG and grown overnight with shaking at 25 °C. To achieve a complete repression of RNA synthesis, next-day cultures were diluted in fresh LB medium plus antibiotics and IPTG and were allowed to continue growing at 25 °C for another day. At the time of RNA induction, the OD600 of the cultures was between 0.4 and 0.6. Expression of the aptamer-tagged RNA was turned on by adding 250 ng/ml anhydrotetracycline (ATc) to half of the culture, whereas the other half remained untreated. These untreated cells served to determine background fluorescence coming from a leaky T7 regulated promoter. Before microscopy studies, untreated ATc cells were washed and resuspended in M9 media supplied with antibiotics and IPTG to reduce fluorescence coming from components in the LB medium.
Flow cytometry was performed as described earlier (13).
Microscopy and Image Analysis.
Cells were immobilized on number 1 cover slips, which were washed with piranha solution (30% H2O2:70% H2SO4) and subsequently coated with (3-aminopropyl)-triethoxysilane to promote adhesion of E. coli cells. Suspension of bacterial cells in M9 medium with IPTG was applied on the treated cover glass and incubated for 5 min. Unbound cells were next removed by rinsing the binding surface several times with M9 until only bound cells remained. M9 medium with IPTG and ATC was then added to the immobilized cells. Analysis was performed at room temperature. Under these conditions the cells remained alive and dividing, although the cell cycle was substantially longer. Cell doubling time determined in cell culture at 25 °C was 120 min.
The cells were mounted on a custom-modified inverted microscope (Zeiss 200M) fitted and imaged using a 100X/1.40 oil immersion objective. White light was used to confirm an adequate E. coli coverage on the cover glass and proper immobilization, before the fluorescence imaging started. Excitation was performed using a solid-state 488-nm laser beam (Coherent, Sapphire), which was attenuated using a quarter wave-plate/polarizing cube combination, and shaped into the backport of the microscope forming nearly-flat illumination field. At the imaging arm, scattered laser light and Raman emission were effectively blocked using appropriate filters (Semrock). Imaging was performed using a back-illuminated Electron-Multiplying CCD (iXon 897, Andor), cooled to −100 °C, and hooked to the laser shutter. Camera acquisition was timed using an external pulse generator, set for 0.2-s exposure time and 120-s intervals between exposures. Images were saved to disk using Solis (Andor) software, and the total cell fluorescence was analyzed using custom code written in MatLab.
Quantitation of EGFP molecules in single cells, RNA concentration determination and statistical analysis of fluorescence distribution in single cells is described in Supplementary procedures.
Supplementary Material
Acknowledgments.
We thank James Deshler and Olga Lomovskaya for insightful discussions; Steven Colburn and Sergey Broude for help with correlation analysis; Yingjie Sun for help in image analysis; and Dan Dwyer, James Collins, and Lawrence Rothfield for critical reading of the manuscript and helpful suggestions. This work was supported by the SPRiNG Award (to N.E.B.) and Biosurfaces Initiative Award from BU (to A.M. and N.E.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907495106/DCSupplemental.
References
- 1.Collier J, Shapiro L. Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol. 2007;18:333–340. doi: 10.1016/j.copbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen RB, Wang SC, Shapiro L. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat Rev Mol Cell Biol. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- 3.Møller-Jensen J, Löwe J. Increasing complexity of the bacterial cytoskeleton. Curr Opin Cell Biol. 2005;17:75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Thanbichler M, Viollier PH, Shapiro L. The structure and function of the bacterial chromosome. Curr Opin Genet Dev. 2005;15:153–162. doi: 10.1016/j.gde.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Thanbichler M, Shapiro L. Chromosome organization and segregation in bacteria. J Struct Biol. 2006;156:292–303. doi: 10.1016/j.jsb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Gordon GS, et al. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 7.Pogliano J, Ho TQ, Zhong Z, Helinski DR. Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:4486–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho TQ, Zhong Z, Aung S, Pogliano J. Compatible bacterial plasmids are targeted to independent cellular locations in Escherichia coli. EMBO J. 2002;21:1864–1872. doi: 10.1093/emboj/21.7.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci USA. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Le TT, et al. Real-time RNA profiling within a single bacterium. Proc Natl Acad Sci USA. 2005;102:9160–9164. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guet CC, et al. Minimally invasive determination of mRNA concentration in single living bacteria. Nucleic Acids Res. 2008;36:e73. doi: 10.1093/nar/gkn329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat Methods. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 14.Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA. 2003;9:394–407. doi: 10.1261/rna.2161303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magliery TJ, et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: Scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt C, et al. Mechanisms of proinflammatory cytokine-induced biphasic NF-kappaB activation. Mol Cell. 2003;12:1287–1300. doi: 10.1016/s1097-2765(03)00390-3. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y, Rebecchi M, Scarlata S. Phospholipase Cbeta2 binds to and inhibits phospholipase Cdelta1. J Biol Chem. 2005;280:1438–1447. doi: 10.1074/jbc.M407593200. [DOI] [PubMed] [Google Scholar]
- 18.Demidov VV, et al. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proc Natl Acad Sci USA. 2006;103:2052–2056. doi: 10.1073/pnas.0511078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderie I, Schmid A. In vivo visualization of actin dynamics and actin interactions by BiFC. Cell Biol Int. 2007;31:1131–1135. doi: 10.1016/j.cellbi.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 20.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 21.Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- 22.Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 23.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vats P, Rothfield L. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc Natl Acad Sci USA. 2007;104:17795–17800. doi: 10.1073/pnas.0708739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vats P, Shih YL, Rothfield L. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol Microbiol. 2009;72:170–182. doi: 10.1111/j.1365-2958.2009.06632.x. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh AS, Young KD. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 2005;187:1913–1922. doi: 10.1128/JB.187.6.1913-1922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taghbalout A, Rothfield L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci USA. 2007;104:667–1672. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taghbalout A, Rothfield L. RNaseE and RNA helicase B play central roles in the cytoskeletal organization of the RNA degradosome. J Biol Chem. 2008;283:13850–13855. doi: 10.1074/jbc.M709118200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiraga S, et al. Chromosome partitioning in Escherichia coli: Novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao S, Helinski DR, Toukdarian A. Localization of the naturally occurring plasmid ColE1 at the cell pole. J Bacteriol. 2007;189:1946–1953. doi: 10.1128/JB.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.