Abstract
In Gram-negative pathogens, multidrug efflux pumps that provide clinically significant levels of antibiotic resistance function as three-component complexes. They are composed of the inner membrane transporters belonging to one of three superfamilies of proteins, RND, ABC, or MF; periplasmic proteins belonging to the membrane fusion protein (MFP) family; and outer membrane channels exemplified by the Escherichia coli TolC. The three-component complexes span the entire two-membrane envelope of Gram-negative bacteria and expel toxic molecules from the cytoplasmic membrane to the medium. The architecture of these complexes is expected to vary significantly because of the structural diversity of the inner membrane transporters. How the three-component pumps are assembled, their architecture, and their dynamics remain unclear. In this study, we reconstituted interactions and compared binding kinetics of the E. coli TolC with AcrA, MacA, and EmrA, the periplasmic MFPs that function in multidrug efflux with transporters from the RND, ABC, and MF superfamilies, respectively. By using surface plasmon resonance, we demonstrate that TolC interactions with MFPs are highly dynamic and sensitive to pH. The affinity of TolC to MFPs decreases in the order MacA > EmrA > AcrA. We further show that MFPs are prone to oligomerization, but differ dramatically from each other in oligomerization kinetics and stability of oligomers. The propensity of MFPs to oligomerize correlates with the stability of MFP–TolC complexes and structural features of inner membrane transporters. We propose that recruitment of TolC by various MFPs is determined not only by kinetics of MFP–TolC interactions but also by oligomerization kinetics of MFPs and pH.
Keywords: multidrug efflux transporters, protein–protein interactions, surface plasmon resonance, antibiotic resistance, gram-negative envelope
Drug resistance presents an ever-increasing threat to public health and encompasses all major microbial pathogens and antimicrobial drugs (1). Some pathogens have acquired resistance to multiple antibiotics and cause infections that are effectively untreatable. This problem is particularly serious with Gram-negative pathogens that have high levels of intrinsic resistance.
Current models postulate that high levels of intrinsic as well as acquired multidrug resistance of Gram-negative pathogens is the result of synergy between reduced uptake of drugs across the outer membrane (OM) and active drug efflux from the inner membrane (IM) (2–4). This synergy is possible because of the concerted action of three proteins: an IM transporter, a periplasmic membrane fusion protein (MFP), and an OM channel, such as Escherichia coli TolC. These three proteins form large, multicomponent assemblies that traverse both the IM and OM of Gram-negative bacteria. All components are absolutely essential for transport. Working together as a well-coordinated team, they achieve the direct extrusion of substrates across two membranes and into the medium.
Recent structural and functional studies highlighted key features of the multicomponent drug efflux pumps. The OM channels of these pumps are structurally conserved (5, 6). They exist as stable trimers, which are inserted into the OM via a β-barrel domain and extend deeply into periplasm by using their ≈100-Å-long, α-helical coiled-coil domain. Together, these domains form a tunnel-like structure that spans across the OM and halfway through the periplasm. Some bacteria, including E. coli and other Enterobacteriaceae species, contain a single OM channel TolC, which is shared by several transport complexes.
In contrast, the IM transporters that associate with the OM channels are structurally and functionally diverse. They can belong to any of the three major superfamilies of proteins: resistance-nodulation-cell division (RND), major facilitator (MF), and ATP-binding cassette (ABC) (7–10). The ABC transporters are driven by ATP hydrolysis, whereas drug efflux by RND and MF pumps is coupled with transport of protons. MF transporters are thought to function as monomers, whereas ABC and RND transporters are dimers and trimers, respectively. The transmembrane topologies of these transporters differ significantly as well.
The structural and functional fit between diverse IM transporters and OM channels is enabled by periplasmic MFPs (11). MFPs are structurally similar. They contain a short N-terminal hydrophilic region followed by a hydrophobic segment, which serves as a cleavable signal peptide or as a transmembrane α-helix. The periplasmic portion of MFPs is an elongated, sickle-shaped molecule comprising three domains: a β-barrel domain, a lipoyl domain centrally located, and a coiled-coil, α-helical hairpin at the other end of the molecule (12–15). Previous studies suggested that the β-barrel domain of MFPs is likely to interact with the IM transporters, whereas the α-helical hairpin establishes the contact with the OM channel (16, 17). This physical link created by MFPs makes possible transport of substrates across the two membranes without periplasmic intermediates. The mechanism of assembly and architecture of multicomponent drug efflux pumps remain unclear.
In this study, we investigated how MFPs interact with OM channels. For this purpose, we reconstituted interactions and compared binding kinetics of E. coli TolC with three MFPs: AcrA, MacA, and EmrA. These proteins function in drug efflux with transporters belonging to RND, ABC, and MF superfamilies, respectively (18–20). AcrA and its cognate transporter AcrB are constitutively expressed at high levels and provide intrinsic resistance to multiple structurally unrelated compounds (20). Recent studies showed that EmrAB is also expressed under laboratory conditions (21). However, its contribution to drug resistance is masked by the activity of AcrAB. Among EmrAB substrates are detergents, proton uncouplers, dyes, and some antibiotics. Finally, MacAB is regulated by PhoPQ system and is likely to be produced only under specific conditions (22). The overproduction of MacAB in ΔacrAB cells leads to elevated resistance to macrolide antibiotics. Although IM transporters vary significantly in size and topology, AcrA, EmrA, and MacA share a significant degree of structural homology. All three drug efflux complexes recruit the same OM channel TolC for their transport activities.
To investigate the mechanism of MFP–TolC interactions, we developed a real-time binding assay based on the surface plasmon resonance (SPR) technique. In this assay, AcrA, MacA, and EmrA were attached to the surface of a CM5 biochip, whereas TolC was present in solution and was free to bind the immobilized MFPs. We report that MFP–TolC interactions are highly dynamic and sensitive to pH. Despite structural similarity, MFPs differ dramatically from each other in oligomerization kinetics and stability of oligomers. We propose that MFP oligomerization and pH contribute to the assembly and stability of MFP–TolC complexes.
Results
MFPs Form Dynamic Oligomers.
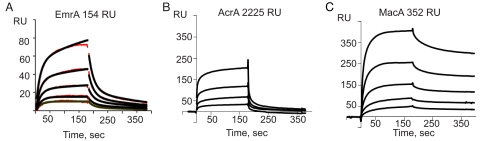
Previous structural studies showed that TolC exists as a stable trimer (6). However, the oligomeric state of MFPs and whether monomers or oligomers of MFPs interact with TolC remain unclear. To investigate oligomerization of MFPs, soluble periplasmic domains of EmrA, AcrA, and MacA were coupled to a CM5 biosensor chip surface, and the respective MFPs were injected in buffer containing 0.05% detergent dodecyl-β-d-maltoside (DDM). EmrA readily formed oligomers, as seen by the rapid increase in an SPR response upon injection of EmrA solution (Fig. 1A). When EmrA was injected at a concentration of 1.9 μM, ≈0.5 EmrA molecules from solution were bound per immobilized EmrA. At higher concentrations, EmrA formed multilayer arrays, as seen by a continuous linear increase in the amount of bound EmrA. This EmrA oligomerization on the surface was completely reversible, suggesting that the process is a concentration-dependent phenomenon different from the irreversible aggregation. Furthermore, injection of EmrA solution over the AcrA and MacA surfaces did not result in any significant response, demonstrating that EmrA oligomerization was due to specific protein–protein interactions rather than nonspecific association with the surface (Fig. S1).
Fig. 1.
Oligomerization of MFPs on the chip. (A) EmrA was immobilized at the density of 154 response units (RU), and soluble EmrA was injected in the Mes-DDM running buffer (pH 5.9) at concentrations of 0.375, 0.56, 0.84, 1.26, and 1.89 μM. The SPR sensorgrams (shown in red) were fitted with the ST model (black lines). (B) AcrA was immobilized at the density of 2,225 response units, and soluble AcrA was injected in the Mes-DDM running buffer (pH 5.8) at concentrations of 1.25, 2.5, 5.0, and 10.0 μM. (C) MacA was immobilized at the density of 352 response units, and soluble MacA was injected in the same buffer as AcrA at concentrations 0.375, 0.75, 1.5, 3.0, and 6.0 μM.
Quantitative analysis of SPR sensorgrams revealed a complex pattern of EmrA–EmrA interactions. Both the binding and dissociation phases of the sensorgrams could be approximated as double-exponential decays, indicating multiple events during both binding and dissociation. The simplest kinetic model that could provide a reasonable fit with experimental data was the sequential trimerization (ST) model, which postulates that two molecules of EmrA bind in sequence to a single immobilized EmrA (Fig. 1A). In support of this interpretation, we found by using chemical cross-linking that both dimers and trimers of EmrA are present in solution (Fig. 2B). We concluded, therefore, that the ST model is an adequate approximation of EmrA–EmrA interactions.
Fig. 2.
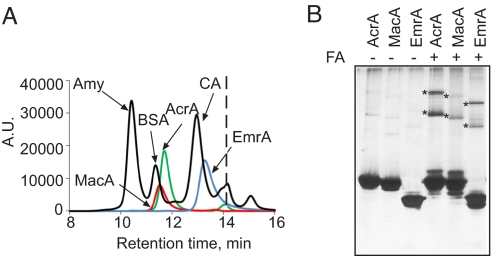
Oligomeric state of MFPs in solution. (A) LS/SEC analysis of MFPs. MFPs (0.2 mg/mL) and size standards (0.15 mg/mL) were analyzed in Mes-DDM running buffer at the flow rate 1.0 mL/min. As determined from the LS analysis, molecular masses of MFPs in peaks were 43.4 kDa for AcrA (blue), 44.4 kDa for MacA (red), and 35.5 kDa for EmrA (green). These values are in a good agreement with the masses of monomeric proteins predicted from their amino acid compositions: 40.6 kDa for AcrA, 39.0 kDa for MacA, and 38.1 kDa for EmrA. The vertical dashed line marks the included volume of the column. The molecular mass standards are: BSA (66 kDa), β-amylase (200 kDa), and carbonic anhydrase (29 kDa). (B) Formaldehyde cross-linking. Purified MFPs in Hepes-DDM buffer (pH 7.0) were incubated in the presence of 0.1% formaldehyde for 30 min at 37 °C. Proteins were separated by 10% SDS/PAGE and stained with silver nitrate. Asterisks mark dimers and trimers.
From the best-fit values for the dissociation rate constants, the lifetimes of EmrA dimers and trimers were found to be 17 s and 227 s, respectively. The equilibrium dissociation constants for the formation of EmrA dimer and trimer differ by ≈2-fold, with Kd1 = 4.22 μM and Kd2 = 1.87 μM, respectively. The higher affinity in the formation of trimers is consistent with the chemical cross-linking data, which show that EmrA trimers are the prevalent cross-linked species (Fig. 2B).
We observed a similar oligomerization with two other MFP proteins, AcrA and MacA. The two proteins, however, notably differed from EmrA in their affinity. When injected in concentrations up to 10.0 μM, AcrA did not show any detectable oligomerization on the surface, with an AcrA density of 75 response units. Weak AcrA binding was detected on the high-density (2,225 response units) AcrA surface (Fig. 1B). When AcrA was injected at a concentration of 10.0 μM, only 1 in 10 immobilized AcrAs could bind AcrA from solution. In contrast, MacA oligomerization was very efficient, even at low MacA concentration (Fig. 1C). When MacA was injected at a concentration of 6.0 μM, we calculated a stoichiometry of one MacA per one immobilized MacA.
Similar to EmrA, SPR sensorgrams for AcrA and MacA oligomerization could be approximated as double-exponential decays. Unlike with EmrA, however, these data could not be fitted to the ST model, suggesting an even more complex kinetic mechanism of the reaction. To compare stabilities of MFP oligomers, the dissociation phases of MacA and AcrA sensorgrams were fitted with a two-exponential rate equation. AcrA oligomers rapidly dissociated, with the lifetimes 7 s and 14 s for the fast- and slow-dissociating species, respectively. MacA oligomers, with a lifetime of >3 min, were notably more stable than those of EmrA and AcrA. This high stability of MacA oligomers significantly complicates quantitative analysis of MacA–MacA binding kinetics.
We next analyzed oligomerization of MFPs in solution. The size-exclusion chromatography and light-scattering analyses showed that all three MFPs are monomeric in solution (Fig. 2A). However, size-exclusion chromatography is not expected to reveal formation of short-lived oligomers. To confirm the propensity of MFPs for oligomerization, the proteins were treated with formaldehyde. In agreement with SPR experiments, both dimers and trimers were detected for all three cross-linked proteins (Fig. 2B).
Thus, all three MFPs are prone to oligomerization, albeit with different kinetics. EmrA and AcrA oligomers are highly dynamic, whereas MacA oligomers are more stable. EmrA oligomers are formed with the low micromolar affinities. In contrast, oligomerization of AcrA requires very high protein concentrations and is likely to depend on additional factors at physiological conditions. MFPs can be arranged by the affinity and stability of their oligomers in the order MacA > EmrA > AcrA.
Complex Kinetics of MFP–TolC Interactions.
To analyze MFP–TolC interactions, TolC was injected over MFP surfaces in buffers of different pH and composition. We found little TolC binding at pH 7.0 (see Fig. 4A). In contrast, injection of 0.04 μM to 2.0 μM TolC at pH 5.8 and 150 mM NaCl resulted in a significant binding to all three MFPs (Fig. 3). All three MFPs could be saturated with TolC and demonstrated very similar, low-micromolar range affinities toward TolC (Fig. S2). Quantitative analysis of the curves showed complex reactions. The association and dissociation phases could be well-approximated by using double-exponential rate equations, indicating multiple reaction events.
Fig. 4.
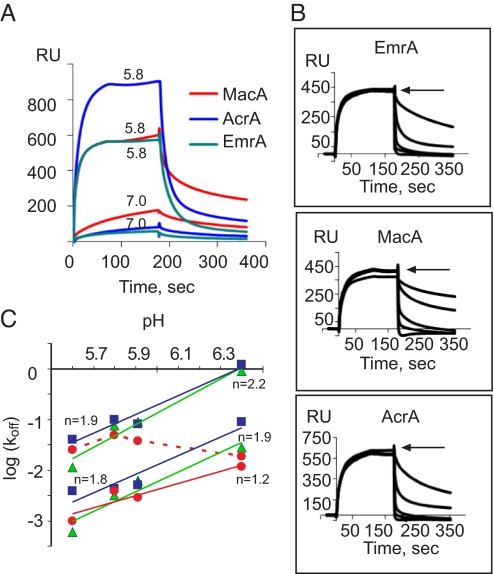
TolC–MFP interactions are modulated by pH. (A) Assembly of TolC–MFP complexes at pH 5.8 and pH 7.0. MFPs were immobilized to densities of 2,225 response units of AcrA (blue), 1,553 response units of EmrA (green), and 2,663 response units of MacA (red), and the chip was equilibrated in running buffers of the indicated pH. TolC in the respective running buffer was injected at a concentration of 0.5 μM. (B) Effect of pH on the dissociation rate of TolC–MFP complexes. TolC–MFP complexes were preassembled in the Mes-DDM buffer (pH 5.8) by injecting 0.25 μM TolC for 3 min. The dissociation (arrow) was initiated by injecting buffers of pH levels 5.6 (the slowest dissociation on all sensorgrams), 5.8, 6.3, 6.5, 7.0, and 7.7, respectively. (C) The slow and fast dissociation rate constants for EmrA (green squares), AcrA (blue triangles), and MacA (red circles) were determined by using a two-exponential rate equation and were plotted as a function of pH. The linear fit and slopes (n) are shown. The fast MacA–TolC complexes are not sensitive to pH (red dashed line).
Fig. 3.
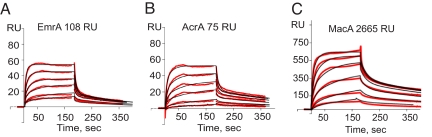
Kinetics of MFP–TolC interactions. (A) EmrA was immobilized at the density of 108 response units. TolC was injected in the Mes-DDM running buffer (pH 5.8) at doubling concentrations, increasing from 0.125 to 2.0 μM. (B) AcrA was immobilized at the density of 75 response units. The experiment was done as in A. (C) MacA was immobilized at the density of 2,665 response units. TolC was injected in the same buffer as in A and B but at doubling concentrations increasing from 0.04 to 1.28 μM. The red lines correspond to the experimental data and the black lines to the fit by using the HL model. Residuals and standard deviations for each fit are shown in Table S1.
The best fit of MFP–TolC binding curves was obtained for the model that assumed the presence of two populations of ligand on the surface with fast and slow dissociation rates [heterogeneous ligand (HL)]. Based on the HL model, the lifetimes for the two EmrA–TolC complexes were 200 ± 30 s and 10 ± 2 s, and the affinities of TolC to two EmrA populations were Kd1 = 0.08 μM and Kd2 = 1.27 μM for the slow and fast complexes, respectively (Fig. 3A and Table S1). At the higher surface densities, the affinity of fast EmrA–TolC complexes increased to Kd2 = 0.33 μM.
The rate and equilibrium constants derived from the kinetic analysis of AcrA–TolC interactions are comparable to those obtained for EmrA–TolC complexes (Fig. 3B and Table S1). The affinity of TolC toward AcrA is somewhat lower than EmrA, with Kd1 = 0.11 μM and Kd2 = 2.1 μM for the slow and fast complexes, respectively.
TolC binding to MacA was the most reproducible at the high surface density of the immobilized MacA. Although the HL model provided the best fit for this protein, notable deviations from the model could be seen as well (Fig. 3C). Quantification of the model revealed that the affinity and stability of MacA–TolC complexes are the highest among the three MFPs used in these studies, with both the fast and slow Kds in the nanomolar range (Table S1).
Structural reasons for the existence of the two populations of MFPs are unclear. Such heterogeneity could arise, for example, if the immobilized MFPs were cross-linked to the surface in various conformations or oligomeric states that have two distinct affinities for TolC. To address this issue, we first confirmed that in our system, the α-helical hairpin of AcrA was the site of TolC binding (17). In agreement with the in vivo studies, the immobilized AcrA mutant lacking the essential α-hairpin (AcrAΔCC) did not bind TolC (Fig. S3). We next examined interactions of TolC with the immobilized AcrAS362C. This mutant contains only one cysteine residue, Cys-362, which is located at its C terminus away from the α-hairpin (23). Immobilization of AcrAS362C onto the CM5 chip by using a thiol-coupling procedure produced a surface with homogeneously oriented AcrAs. We found that this protein interacted with TolC with the affinity and kinetics similar to those for the randomly oriented AcrA (Fig. S4). We conclude, therefore, that the observed heterogeneity of AcrA–TolC interactions in our SPR experiments (and perhaps, by extension, EmrA–TolC and MacA–TolC) was not due to the coupling reaction or the orientation of the immobilized proteins, but reflects the intrinsic property of the protein.
MFP–TolC Interactions Are Modulated by pH.
TolC binding to MFPs was inhibited by 250 mM NaCl but was unresponsive to the presence of antibiotics, the substrates of MFP-dependent pumps. Surprisingly, MFP–TolC complexes were highly sensitive to pH of the buffer. As shown in Fig. 3, TolC interacted with all three MFPs at pH 5.8. At pH 7.0, the efficiency of TolC binding decreased for all three proteins, with AcrA affected in the most dramatic fashion (Fig. 4A).
We next investigated the effect of pH on dissociation of MFP–TolC complexes. In this experiment, the binding step was carried out at pH 5.8, whereas the dissociation was initiated by injecting buffers with pH levels ranging from 5.6 to 7.7 (Fig. 4B). The MFP–TolC complexes were most stable at pH 5.6 but rapidly disintegrated at pH levels >6.0. AcrA–TolC interactions were the most sensitive to rising pH: 6 s after injection of the buffer with pH 6.4, only 10% of TolCs remained bound to the AcrA surface, whereas 30% of TolCs were still bound to MacA (Fig. 4B).
The dissociation phases were fitted with a two-exponential rate equation. In the case of EmrA and AcrA, both the slow- and the fast-dissociation steps were affected by pH (Fig. 4C), suggesting that interactions in both complexes are mediated by similar contacts. In contrast, only slow-dissociating MacA–TolC complexes were sensitive to pH, whereas the interactions in the fast complexes were not affected by protonation/deprotonation events. To estimate the number of ionizable residues involved in the pH-dependent affinity change, we plotted the dissociation rate constants as a function of pH. The slope of this plot gives the net number of protons that are released upon formation of the transition state during TolC dissociation from MFPs (Methods and refs. 24 and 25). The slopes of the transitions between pH 5.6 and pH 6.4 for the fast and slow EmrA–TolC and AcrA–TolC complexes ranged between 1.8 and 2.2, suggesting that two protons are released during dissociation of each complex (Fig. 4C). Interestingly, the slope of the transitions for the slow-dissociating MacA–TolC complex was 1.2. Thus, only one proton is involved in dissociation of this complex.
Discussion
The interaction between an MFP and TolC is a critical step in various transport reactions that take place across the two-membrane envelope of E. coli. We developed a real-time binding assay to study this interaction. By using SPR technique, we successfully reconstituted interactions between TolC and three MFPs—AcrA, EmrA, and MacA—that function in multidrug efflux. We identified three key features of MFP–TolC interactions. First, the assembly of MFP–TolC complexes is a highly dynamic process. From the dissociation rate constants, we estimate that the lifetimes of MFP–TolC complexes are seconds to minutes. However, these short-lived complexes are formed with high affinity. The Kds for MFP–TolC complexes were found to be in the low nanomolar and micromolar range (Table S1). Previously, isothermal calorimetry (ITC) was used to determine the affinity of AcrA–TolC interactions in solution (26). The low- and high-affinity complexes with Kds of 13–17 μM and 1.9–3.1 μM were detected as well. The lower affinity of AcrA–TolC in the ITC experiments could be explained by the high pH 7.4 of the buffer that was used in those experiments. As shown in Fig. 2, at a pH >6.0, the affinity of TolC to AcrA was dramatically reduced (Fig. 4).
Second, TolC forms high- and low-affinity complexes with MFPs. Even at low densities of ligands, the SPR sensorgrams are best fitted with the HL model, which predicts interactions of TolC with two populations of the immobilized ligand (Fig. 3 and Table S1). Importantly, we did not find such surface heterogeneity in oligomerization experiments, although experiments were carried out on the same surfaces. Thus, these two populations of MFPs differ only in their interactions with TolC. Furthermore, AcrA immobilized onto the surface in a specific orientation demonstrated similar heterogeneity in TolC binding (Fig. S4). Thus, this heterogeneity was unlikely due to steric occlusion of the TolC-binding site on some of the proteins. Given the high propensity to oligomerization, these two populations are probably MFP monomers and oligomers.
The properties of oligomers vary significantly among MFPs. MacA forms relatively stable oligomers with lifetimes exceeding 3 min. EmrA oligomers are formed with low-micromolar affinity and are highly dynamic. AcrA oligomerization requires nonphysiological protein concentrations. These results are in agreement with the hexameric assembly of MacA and dimerization of AcrA in crystals as well as with oligomerization of EmrA in solution (12, 15, 18). Interestingly, the stability of MFP oligomers correlates with the stability of MFP–TolC complexes: MacA forms the most stable complexes with TolC, followed by EmrA and then AcrA. It is tempting to propose that MFP oligomerization affects MFP–TolC interactions.
The differences between MFPs could play an important role during assembly of the tripartite complexes in vivo. AcrA and its cognate transporter AcrB are constitutively expressed in relatively large quantities, with ≈6,000 copies of AcrA per cell (27). With the volume of periplasm estimated to be 0.065 fL (28), this amount of AcrA is translated into ≈150 μM concentration of protein. For MacAB and EmrAB to successfully compete with AcrAB for binding TolC, these MFPs should either be produced in the same amounts as AcrAB or have higher affinity toward TolC, which would give them an advantage at the relatively low copy number. Our studies suggest that this advantage could come from the ability of MacA and EmrA to form oligomers that establish more stable interactions with TolC.
The dynamic nature and variations in MFP oligomerization befit the structural particularities of respective IM transporters. ABC and MF transporters do not have large periplasmic domains and are unlikely to contact TolC directly. Therefore, MFPs functioning with ABC- and MF-type transporters are envisioned to form a sheath surrounding the periplasmic tip of OMF (29). Our data support this idea: both EmrA and MacA form oligomers that could seal the gap between the transporter and TolC. In contrast, the periplasmic domains of RND pumps, such as AcrB, protrude deep into periplasm and have the geometry corresponding to the periplasmic entrance of TolC (30). Thus, in the AcrAB–TolC system, AcrA is not needed to seal the gap between IM and OM, and the substrate can pass from AcrB directly to TolC without escaping into periplasm. In agreement, we found that AcrA has a very low propensity to oligomerization.
Finally, the assembly of MFP–TolC complexes is modulated by pH. The effect of pH is dramatic, with almost no detectable association at pH >7.0. In contrast, complexes are readily assembled and highly stable at pH <6.0 (Fig. 4C). This result suggested that ionization of amino acid residues in MFP, TolC, or both is needed for interactions. The previously identified AcrA–TolC interface, which includes the N-terminal helix of AcrA α-hairpin and the intermolecular groove between the inner and the outer entrance coils of TolC H7/H8/H3, does not contain histidine residues (pKa ≈ 6.2) (17). However, several ionizable groups are present that could contribute to this pH dependence. Alternatively, a conformational change induced by pH, such as the one previously reported for AcrA (12, 31), could play a role in formation of MFP–TolC complexes. At normal physiological conditions, the pH of periplasm is estimated to be ≈1.5 units (pH ≈ 6.0) lower than that of the cytoplasm (pH ≈ 7.5) (28). These conditions promote high-affinity interactions between various MFPs and TolC.
This analysis also predicts that increase in pH due to dissipation of proton-motive force or alkaline pH of growth medium would adversely affect the MFP–TolC interactions and, as a result, the activity of tripartite transporters. The effect of pH on antimicrobial activity of drugs is well-characterized (32). Intriguingly, the activity of many macrolides, quinolones, and aminoglycosides is higher at alkaline pH, especially in Gram-negative pathogens. This marked pH effect is often explained by the degree of ionization of these compounds. Our studies suggest that changes in activity of multidrug efflux pumps could also contribute to the pH-dependent changes in the antimicrobial activity of drugs.
Methods
Overproduction and Purification of Proteins.
Construction and purification of soluble MFP variants containing the C-terminal six-histidine (6His) affinity tags have been reported (18, 33, 34) and are described in SI Materials and Methods. To construct AcrA lacking the α-helical hairpin (AcrAΔCC), amino acid residues Q102–V173 of AcrA were replaced with an LLGTQ sequence by PCR using pET-AcrA plasmid as a template. An AcrA mutant containing a single S362C substitution has been reported (23, 31).
TolC containing the C-terminal 6His-tag was purified from E. coli ET101 cells harboring pTolCHis plasmid (6, 33). DDM detergent was used during solubilization and purification steps.
Light-Scattering and Size-Exclusion Chromatography (LS/SEC).
MFPs and TolC were analyzed by HPLC (Shimadzu) using YMC-Pack Diol-300 size exclusion column at 0.2 to 2.0 mg/mL protein concentrations. Protein samples were injected in the buffer containing 20 mM Mes-KOH (pH 5.8), 0.05% DDM, and 150 mM NaCl at the flow rate of 1 mL/min. The LS and refraction index (RI) of eluted proteins were measured by using a PD2010 light scattering detector (Precision Detectors) and Waters 2414 detector, respectively. The molecular mass of the proteins was calculated from the LS and RI data by using Discovery software (Precision Detectors), as described in ref. 35. BSA (66 kDa), β-amylase (200 kDa), and carbonic anhydrase (29 kDa) were used as molecular weight standards.
Surface Plasmon Resonance.
A BIAcore 3000 biosensor system was used to characterize MFP–TolC binding interactions. MFPs and AcrAΔCC were immobilized onto a CM5 biosensor chip by using the amino coupling procedure, whereas the thiol coupling was used to immobilize AcrAS362C (BIAcore). Surface activation and coupling procedures are described in SI Materials and Methods.
To develop an SPR-based binding assay, MFPs were coupled to a CM5 biosensor chip surface to various densities, as indicated in the figures. A coupling density of 2,000 response units (RU) corresponds to 2 ng/mm2, or approximately three molecules of protein per 104Å2 (100 Å × 100 Å). Most of the experiments were carried out in the Mes-DDM running buffer (pH 5.8). For pH-dependence experiments, 20 mM Mes-KOH (pH 5.8) was replaced with either 20 mM Hepes-KOH (pH 7.0–7.5) or 20 mM Tris·HCl (pH 8.2). After each experiment, all surfaces were regenerated by a quick injection of 20 mM CHAPS prepared in buffer containing 20 mM Tris·HCl (pH 8.2) and 150 mM NaCl. All experiments were done at least in duplicates. Unless otherwise noted, the flow rate was maintained at 30 μL/min. Further increase in the flow rate did not affect the on and off rates of binding reactions.
All presented sensograms show only normalized data, which are the differences between response curves for the MFP-coupled and MFP-free surfaces.
Kinetic Analysis of TolC–MFP and MFP–MFP Interactions.
Data were analyzed with BiaEvaluation version 6 (BIAcore) and Microsoft Excel software. In all examined cases, both the association and dissociation phases of SPR sensorgrams could be approximated as double-exponential decays. Such kinetics rules out simple reaction mechanisms and predicts at least two distinct events during both protein binding and dissociation. The best global fit was obtained by using the ST model for EmrA–EmrA interactions and HL for MFP–TolC interactions. Schematic representation and equations relevant to the HL and ST models are given in SI Materials and Methods.
The dissociation phases were analyzed by using a two-exponential dissociation rate equation:
where k−1 and k−2 represent slow and fast rate constants, and Rslow and Rfast correspond to the fraction of complexes dissociating with each rate constant.
The pH dependence of the dissociation rates was analyzed according to the following kinetic scheme:
where MFP·TolC* is the transition state during dissociation of MFP·TolC complex; K* is the equilibrium constant for the formation of the transition state; kf is the rate constant for dissociation of MFP·TolC* complex; and n is the net number of protons that are released from the complex upon formation of transition state. Given this scheme, the rate constant for dissociation of MFP·TolC, koff, can be evaluated according to the following equation:
and the number of the released protons can be determined from the slope of the pH-dependence of koff plotted in the double-logarithmic coordinates.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant AI052293 (to H.I.Z.) and by an Oklahoma Center for Advancement of Science and Technology grant (to V.V.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906601106/DCSupplemental.
References
- 1.Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med. 2004;10(Suppl):S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat Rev Drug Discov. 2007;6:56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Preventing drug access to targets: Cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol. 2001;12:215–223. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- 4.Zgurskaya HI, Krishnamoorthy G, Tikhonova EB, Lau SY, Stratton KL. Mechanism of antibiotic efflux in Gram-negative bacteria. Front Biosci. 2003;8:s862–s873. doi: 10.2741/1134. [DOI] [PubMed] [Google Scholar]
- 5.Akama H, et al. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: Dual modes of membrane anchoring and occluded cavity end. J Biol Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 6.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 7.Paulsen IT, Chen J, Nelson KE, Saier MH., Jr Comparative genomics of microbial drug efflux systems. J Mol Microbiol Biotechnol. 2001;3:145–150. [PubMed] [Google Scholar]
- 8.Saier MH, Jr, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 9.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 10.Saier MH, Jr, et al. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 11.Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta. 2009;1794:794–807. doi: 10.1016/j.bbapap.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akama H, et al. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 14.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yum S, et al. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol. 2009;387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 16.Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: Mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J Bacteriol. 2008;190:691–698. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobedanz S, et al. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc Natl Acad Sci USA. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borges-Walmsley MI, et al. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J Biol Chem. 2003;278:12903–12912. doi: 10.1074/jbc.M209457200. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183:5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, et al. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 21.Elkins CA, Mullis LB. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob Agents Chemother. 2007;51:923–929. doi: 10.1128/AAC.01048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 23.Ge Q, Yamada Y, Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC. J Bacteriol. 2009;191:4365–4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W. H. Freeman and Company; 1999. [Google Scholar]
- 25.Sprague ER, Martin WL, Bjorkman PJ. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J Biol Chem. 2004;279:14184–14193. doi: 10.1074/jbc.M313281200. [DOI] [PubMed] [Google Scholar]
- 26.Touze T, et al. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- 27.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 28.Boos W, Lucht JM. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC: ASM Press; 1996. [Google Scholar]
- 29.Federici L, et al. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 A resolution. J Biol Chem. 2005;280:15307–15314. doi: 10.1074/jbc.M500401200. [DOI] [PubMed] [Google Scholar]
- 30.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 31.Ip H, Stratton K, Zgurskaya H, Liu J. pH-induced conformational changes of AcrA, the membrane fusion protein of Escherichia coli multidrug efflux system. J Biol Chem. 2003;278:50474–50482. doi: 10.1074/jbc.M305152200. [DOI] [PubMed] [Google Scholar]
- 32.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in Laboratory Medicine. 4th Ed. Baltimore: Williams and Wilkins; 1996. pp. 52–111. [Google Scholar]
- 33.Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: The periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol Microbiol. 2007;63:895–910. doi: 10.1111/j.1365-2958.2006.05549.x. [DOI] [PubMed] [Google Scholar]
- 34.Zgurskaya HI, Nikaido H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol. 1999;285:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]
- 35.Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J Biol Chem. 2006;281:34208–34217. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.