Abstract
p300 and CREB-binding protein (CBP) act as multifunctional regulators of p53 via acetylase and polyubiquitin ligase (E4) activities. Prior work in vitro has shown that the N-terminal 595 aa of p300 encode both generic ubiquitin ligase (E3) and p53-directed E4 functions. Analysis of p300 or CBP-deficient cells revealed that both coactivators were required for endogenous p53 polyubiquitination and the normally rapid turnover of p53 in unstressed cells. Unexpectedly, p300/CBP ubiquitin ligase activities were absent in nuclear extracts and exclusively cytoplasmic. Consistent with the cytoplasmic localization of its E3/E4 activity, CBP deficiency specifically stabilized cytoplasmic, but not nuclear p53. The N-terminal 616 aa of CBP, which includes the conserved Zn2+-binding C/H1-TAZ1 domain, was the minimal domain sufficient to destabilize p53 in vivo, and it included within an intrinsic E3 autoubiquitination activity and, in a two-step E4 assay, exhibited robust E4 activity for p53. Cytoplasmic compartmentalization of p300/CBP's ubiquitination function reconciles seemingly opposed functions and explains how a futile cycle is avoided—cytoplasmic p300/CBP E4 activities ubiquitinate and destabilize p53, while physically separate nuclear p300/CBP activities, such as p53 acetylation, activate p53.
p53, or a component of its tumor suppressor pathway, is mutated or dysregulated in nearly all human cancers (1). Depending on context, it can signal cells to arrest, senesce, or apoptose through both transcription-dependent as well as independent mechanisms (2, 3). Its activity is controlled posttranslationally by a multitude of covalent modifications, including phosphorylation, ubiquitination, sumoylation, neddylation, methylation, and acetylation (4, 5).
The ubiquitination and proteasome targeting of p53 by the RING E3 enzyme MDM2 has been considered to generally inhibit p53 function (6). However, the distinction between multiple monoubiquitination (MUM) and polyubiquitination of p53 adds additional complexity to its regulation. MUM has been reported to enhance p53 nuclear export, and in the absence of stress, p53 is very likely polyubiquitinated and then degraded by the proteasome in the cytoplasm (7, 8). In addition, recent evidence points to a potential positive role for p53 ubiquitination in its capacity as a nuclear transcription factor (4, 9, 10). Adding to the difficulty of understanding p53 ubiquitination, a multitude of ubiquitin ligases (E3s) have been identified for p53 in unstressed cells besides MDM2, most notably E4F1 (10), COP1 (11), and ARF-BP1 (12), making it unclear how each E3 contributes to overall p53 regulation in any given cell-type or environmental condition (13).
Whether p53 MUM vs. polyubiquitination is actively and specifically controlled in cells also remains unclear. In stressed cells, the induction of MDM2 expression by activated p53, can specifically increase the abundance of polyubiquitinated p53 adducts (8). In unstressed cells, specific polyubiquitin ligases (E4s), or ubiquitin (Ub) chain-extending factors, exist for p53—namely the coactivator p300 (14) and transcription factor YY1 (15). p300-dependent polyubiquitination of p53 in vitro required priming of p53 by MDM2-driven p53 MUM (16), although p300, and its paralog CREB-binding protein (CBP), both exhibited robust E3 autoubiquitination activity (14). YY1, by contrast, stabilized MDM2/p53 complexes, allowing MDM2 to act more processively toward chain elongation (15). A recognizable E3 or E4 domain could not be identified in p300/CBP, but the N-terminal 595 aa of p300 appeared to harbor both its E3 (autoubiquitination) and E4 activities (14). Recently, Zn-finger like domains have also been characterized as active E3 enzymes (10, 17), and p300 does harbor three conserved Zn2+-binding Cys-His-rich regions, one of which (C/H1-TAZ1) lies within its first 595 aa (18–20).
To better define the role of p300 or CBP in p53 stability regulation, p53 turnover was studied in p300- or CBP-deficient cells. p300 or CBP depletion in unstressed cells led to p53 stabilization, and p300 and CBP E3 activities were exclusively cytoplasmic, with detectable amounts of both proteins localized to the cytoplasm. CBP, like p300, encoded an E3 activity within its N terminus and exhibited E4 activity toward p53 in vitro. p300 and CBP therefore engage in compartmentalized regulation of p53, by cytoplasmic ubiquitination and presumed nuclear acetylation, that is required for the proper homeostasis of p53 in basal and stressed conditions.
Results
Regulation of p53 Abundance and Stability by p300 and CBP.
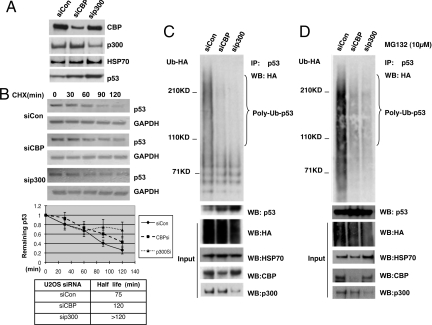
To assess the physiologic contributions of p300 and CBP to p53 regulation in unstressed cells, p300 and CBP were transiently silenced in U2OS cells with specific siRNA duplexes, followed by analysis of cell lysates for p300, CBP, and p53 levels, as well as determination of p53 half-life (t1/2) by cycloheximide decay (Fig. 1 A and B). p300 siRNA has been reported to increase p53 abundance but the mechanism was not determined further (21). p300 and CBP siRNAs both caused increases in steady-state p53 abundance (Fig. 1A) and p53 half-life (from 75 min to >2 h and 2 h, respectively; Fig. 1B) compared to that seen in control siRNA-treated cells. Similar gains in p53 abundance were also seen after stable CBP knockdown by two distinct shRNA sequences (CBP-shA, CBP-shB), and CBP-shA also induced p53 stabilization (Fig. S1A). Demonstrating that these results were not cell type-specific or due to the use of a cancer cell line, siRNA depletion of CBP or p300 in nontransformed MCF10A human breast epithelial cells similarly stabilized p53 from t1/2 = 75 min with control siRNA to >2 h for both p300 and CBP siRNAs (Fig. S1B).
Fig. 1.
CBP and p300 regulate p53 stability and polyubiquitination. (A and B) p53 abundance and stability in U2OS cells after CBP or p300 siRNA treatment. (A) U2OS cells transiently transfected with the indicated siRNAs for 72 h were harvested for immunoblotting with anti-CBP, -p300, -hsp70 (loading control), and -p53 antibodies. (B) (Top) U2OS cells transfected with the indicated siRNAs for 72 h were treated with cycloheximide, and lysates were harvested at the indicated times for analysis by p53 immunoblot. (Bottom) p53 levels were quantitated on a blot scanner (LiCOR), and half-life calculated based on the decay of normalized (to GAPDH loading control) p53 levels to 50% of their original level. Values are an average of three independent experiments. Error bars, ±1 standard deviation (S.D.). (C and D) CBP and p300 both drive p53 polyubiquitination in vivo. U2OS cells were treated with the indicated siRNAs for 48 h and transfected with HA-Ub expression vector 24 h before harvest. MG132 (10 μM) was added for 4 h before harvest as indicated. Cell lysates were IP'd with anti-p53 Ab, followed by anti-HA or anti-p53 immunoblot. PolyUb indicates those p53 species >100 kDa that are larger than the largest MUM species as determined by use of non-chain-forming Ub moieties (14). Input lysates were blotted with the indicated antibodies.
Indirect explanations for the increased p53 abundance and stability induced by p300/CBP depletion, such as increased p53 mRNA expression, decreased MDM2 expression, or induction of genotoxic stress (the latter two of which might stabilize p53) were investigated. RT-PCR analysis showed that p53 mRNA levels were actually decreased, not increased, in CBP-shA as compared with control-sh cells (Fig. S1C), and MDM2 protein levels were increased or unchanged, not decreased, in CBP-shA or CBP-shB cells as compared with control-sh cells (Fig. S1A). The possibility that CBP depletion might stimulate a p53-stabilizing stress response (22) was investigated by mock or doxorubicin (Dox)-treatment (as positive control for stress stimulus) of control-sh or CBP-shA cells, followed by immunoblotting of lysates for p53 serine 15 (S15) phosphorylation, a marker of p53 activation in response to stress (Fig. S1D) (23, 24). The untreated cell lines had an equivalent and very low level of serine 15 phosphorylation, and both showed robust induction of phosphorylation after treatment with Dox for 2 h (Fig. S1D). Thus, the lack of basally induced p53 S15 phosphorylation in unstressed CBP-depleted cells does not support a nonspecific stress mechanism as the reason for the observed p53 stabilization.
CBP Is Required for p53 Polyubiquitination In Vivo.
Given that p300 can act as an E4 for p53 in vitro (14), the abundance of p53-Ub conjugates was analyzed in U2OS cells expressing HA-tagged Ub after control, CBP, or p300 siRNA treatment, and in the presence or absence of MG132 proteasome inhibitor. Lysates of siRNA-treated cells were analyzed by p53 IP under stringent conditions, followed by HA immunoblot (Fig. 1 C and D). Polyubiquitinated p53-Ub conjugates were detected in a p53 IP of lysate from control siRNA-treated cells, as evidenced by the broadly distributed HA immunoreactive species (Fig. 1C, first lane). Based on the pattern of p53-Ub conjugates seen in vitro when non-chain-forming Ub moieties are used, those p53-Ub conjugates <100 kDa represent largely mono-Ub and MUM species, while those >100 kDa represent polyubiquitinated species (14). p53 IPs from lysates of p300 or CBP siRNA-treated cells, by contrast, had significantly reduced levels of polyubiquitinated p53 (Fig. 1C, second and third lanes). Similar results were seen after proteasome inhibition, except for a stronger polyubiquitinated signal in the p53 IP from control siRNA-treated cells (Fig. 1D). Similar results were also seen when the p53 ubiquitination pattern was compared between control-sh and CBP-shA cell lines expressing HA-Ub (Fig. S2A). In this case, the relative abundance of polyubiquitinated vs. mono-Ub/MUM p53 was quantitated showing an inverse correlation between the two forms when CBP was silenced, with polyubiquitination decreasing and mono-Ub/MUM forms increasing (Fig. S2A). p300 and CBP are therefore both required to maintain physiologic levels of polyubiquitinated p53 conjugates in unstressed U2OS cells.
CBP N-Terminal Sequences Destabilize p53.
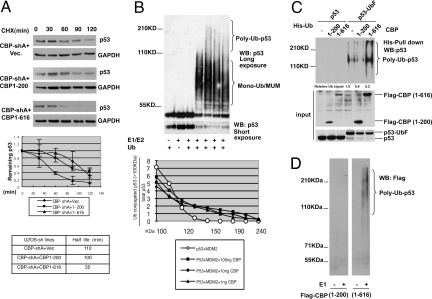
To map the portion of CBP responsible for p53 destabilization, vector control, full-length or truncated CBP cDNAs were expressed as “rescue” alleles in CBP-deficient CBP-shA cells, and p53 stability was measured by cycloheximide-decay analysis. Full-length CBP* (mutated in the shRNA target sequence) expression in CBP-shA cells restored p53 half-life to the normal 60 min from the 120 min observed in CBP-shA cells (Fig. S2B). Given that the p300 N terminus has been proposed as necessary and sufficient for E4 activity (residues 1–595) (14), the homologous portion of the CBP N terminus was analyzed in detail for its ability to rescue p53 instability in CBP-shA cells (Fig. 2A). CBP (1–616) and (1–200), which do and do not, respectively, include the C/H1-TAZ1 domain (residues 348–432), were expressed in CBP-shA cells (Fig. S2C). p53 was destabilized from a t1/2 of 110 min seen in CBP-shA cells to 55 min in cells expressing CBP (1—616) (Fig. 2A), while the p53 t1/2 remained similar to the value in CBP-shA cells when CBP (1–200) was expressed (100 min) (Fig. 2A). Thus, the CBP N terminus is sufficient to destabilize p53, and the sequence between residues 200 and 616, including the C/H1-TAZ1 domain, is required for this activity.
Fig. 2.
The CBP N terminus regulates p53 polyubiquitination and stability. (A) CBP N-terminus regulates p53 stability. (Top) CBP-shA cells were transfected with vector or the indicated CBP rescue alleles, treated with cycloheximide 48 h after transfection, and lysates analyzed by p53 immunoblot. (Bottom) p53 levels were quantitated by densitometry, and half-life calculated based on decay of normalized (to GAPDH loading control) p53 levels to 50% of their original level. Values are an average of three independent experiments. Error bars, ±1 S.D. (B) Purified recombinant CBP protein catalyzes p53 polyubiquitination. (Top) Purified recombinant p53-MDM2 protein complexes were preincubated with ubiquitin reaction components at 37 °C for 30 min. Purified FLAG-CBP (1, 10, or 100 ng) was then added, followed by further incubation for 1 h at 37 °C. Reactions were analyzed by p53 immunoblot. A shorter exposure (middle) shows the unmodified p53 protein level. (Bottom) The ratio of high molecular weight (>100 kDa) p53 species: Total p53 abundance was plotted at 20-kDa intervals for the indicated conditions. (C) The CBP N terminus is an active E4 in vivo. H1299 cells were transfected with His-Ub vector, p53 or p53-UbF expression vectors, along with the indicated CBP plasmids or empty vector. (Top) Forty-eight hours after transfection, lysates were purified on Ni-NTA beads, and eluted products immunoblotted with anti-p53 antibody. (Bottom) Input lysates were immunoblotted with anti-FLAG and p53 antibodies. The abundance of high molecular weight p53-Ub conjugates was determined by scanning densitometry and normalized to total amount of p53UbF in the input lysate. Experiment shown is representative of three independent experiments. (D) Sequential E4 assay. Purified FLAG-MDM2/HA-p53 complexes were exposed to E1, E2, Ub, and ATP to generate mono-Ub/MUM p53. HA-p53-Ub conjugates were purified by anti-HA IP and stringent washing, and then exposed to E2 and ATP a second time with or without E1, along with FLAG-Ub and affinity purified CBP (1–616) or (1–200). After further washes, HA-p53-Ub conjugates were analyzed by anti-FLAG immunoblot.
The CBP N Terminus Exhibits E4 Activity In Vitro and In Vivo.
Given the suggestion from the in vivo data that CBP and p300 both exhibit an E4 function in vivo, we verified that CBP, like p300 (14), could act directly as an E4 in a reconstituted in vitro p53 ubiquitination reaction using purified components (Fig. 2B). p53-MUM conjugates were prepared using purified E1, ubch5a (E2), Ub, and limiting quantities of purified recombinant MDM2 (14). When purified FLAG-CBP was added to this reaction, there was a dose-dependent increase in the abundance and the maximum molecular weight of high molecular weight species of p53, consistent with polyubiquitination (polyubiquitin conjugates are assumed at a molecular weight of p53 species >100 kDa) (14) (Fig. 2B). Control reactions lacking MDM2 revealed no ubiquitination activity of p300 or CBP toward native p53 (Fig. S3A, right), and eliminating p53 from the reaction eliminated all signal on the p53 immunoblots (Fig. S3A, left). To quantitate the E4 effect of CBP, the ratio of p53 signal in the high molecular weight range (>100 kDa) of the gel to total p53 signal was calculated at 20-kDa intervals between 100 kDa and the top of the gel, and plotted vs. mean molecular weight (Fig. 2B, bottom). The clear separation of the control (MDM2-only) curve from the CBP titration curves reveals that CBP had a clearly quantifiable impact on Ub content and, by inference, Ub chain length of p53-Ub conjugates.
To demonstrate that the CBP N terminus could also act as an E4 for p53-mono-Ub or MUM conjugates in vivo, p53 or p53-Ub fusion (Ub cDNA fused in frame to the p53 C terminus; p53-UbF) protein cDNAs were expressed in cells, to determine if p53-UbF could serve as a model for p53-monoUb or MUM conjugates, and act as a direct substrate for the CBP E4 activity. His-Ub and CBP (1–200) and (1–616) polypeptides were expressed along with p53 or p53-UbF in H1299 cells, followed by Ni-NTA pulldown of lysates and p53 immunoblotting. Analysis of lysates used for Ni-NTA pulldown by FLAG and p53 immunoblot revealed even expression of CBP (1–200) and CBP (1–616) (Fig. 2C, middle), as well as equivalent expression of p53 and p53-UbF (Fig. 2C, bottom). Coexpression of the CBP N terminus with native p53, which is not even multiply monoubiquitinated in H1299 cells (Fig. 2C, top, first lane) had no effect, as ubiquitinated p53 remained undetectable (Fig. 2C, top, second and third lanes). Coexpression of CBP (1–616) with p53-UbF resulted in robust induction of high molecular weight Ub conjugates of the p53-Ub fusion, as compared with the effects of control (empty vector) or CBP (1–200) coexpression, both of which were associated with a similar background level of p53UbF ubiquitination signal (Fig. 2C). The effect of the CBP N terminus on the ubiquitination status of the p53-Ub fusion specifically demonstrates that the first 616 aa of CBP are sufficient to polyubiquitinate p53-monoUb or MUM conjugates in vivo as well as in vitro.
CBP E4 Activity Is Separable from MDM2.
To definitively delineate the independent contribution of CBP to p53 polyubiquitination, a two-step in vitro E4 assay was developed. In the first step, p53-MUM conjugates were prepared by incubating with MDM2 in a similar fashion as seen in Fig. 2B, fourth lane, using purified FLAG-MDM2/HA-p53 complex from insect cells. The reaction products were immunopurified on anti-HA-conjugated beads under stringent (RIPA buffer) conditions to remove MDM2. CBP (1–616) or (1–200) polypeptides were then added to the immobilized HA-p53-Ub conjugates with fresh E1, E2, ATP, and FLAG-ubiquitin, followed by washing of the beads and blotting of the reaction products with anti-FLAG antibody. As is seen in Fig. 2D, CBP (1–616), but not (1–200), clearly catalyzed the synthesis of polyubiquitin chains when p53 had been previously mono- or multiply monoubiquitinated by MDM2. Moreover, no FLAG-MDM2 signal was seen among the reaction products (absence of signal at 90-kDa section of blot; Fig. 2D), confirming that MDM2 had been effectively removed before the addition of CBP polypeptides. Thus, CBP (1–616) encodes an MDM2-independent p53-directed E4 function.
The CBP N Terminus Encodes an Intrinsic E3 Activity.
The E4 effect of CBP would be most consistent with an intrinsic MDM2-independent ubiquitin ligase activity that can only recognize a monoubiquitinated or multiply monoubiquitinated substrate, as has been observed for p300 (14). We investigated whether sequences arising from the CBP N terminus, which demonstrate E4 activity (Fig. 2 C and D) and promote p53 instability in vivo (Fig. 2A), encode an intrinsic E3 activity that could be clearly separated from MDM2. Prokaryotically synthesized, purified, GST-CBP (1–616), (1–451), and (452–721) were incubated with Ub, E1, E2 (ubch5a), and ATP in an autoubiquitination reaction, and the reaction products detected by anti-Ub immunoblot. Titration of GST-CBP (1–616) or (1–451) with standard ubiquitin reaction components, revealed robust E3 activity (Fig. S3C). GST-CBP (452–721), however, was inactive as an E3 (Fig. S3C). Thus, the CBP polypeptide that promotes p53 degradation in vivo and p53 polyubiquitination in vivo and in vitro, also encodes an intrinsic, MDM2-independent, E3 ubiquitin ligase domain. The core sequences responsible for this activity appear to lie within at least the N-terminal 451 aa of CBP, and include the C/H1-TAZ1 domain.
CBP Promotes p53 Degradation in the Cytoplasm.
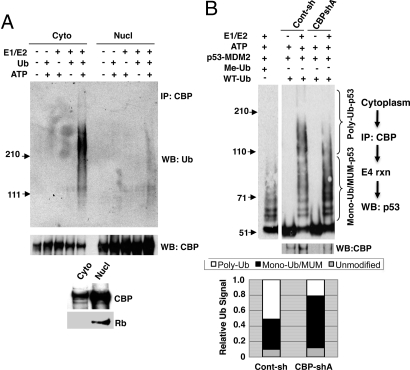
Since p300/CBP are considered to be nuclear coactivators (25), but p53 and MDM2 both shuttle between nucleus and cytoplasm (26), the cellular localization of CBP's regulation of p53 ubiquitination and degradation was explored further. Levels of unmodified p53 were proportionately increased in both nuclear and cytoplasmic fractions in CBP-shA vs. control cells, and controls for nuclear (Rb) and cytoplasmic (actin) proteins revealed that there was little cross-contamination of the fractions (Fig. 3A). Mono-Ub/MUM p53 was found predominantly in the cytoplasm, and levels of this species in the cytoplasm were significantly increased in CBP-shA vs. control cells, either due to the overall increase in total p53, or a loss of an (E4) activity required to convert p53-mono-Ub or p53-MUM conjugates to polyubiquitinated forms (Fig. 3A).
Fig. 3.
Subcellular localization of CBP/p300 E3 activity. (A) p53 localization in CBP-deficient cells. Nuclear or cytoplasmic fractions of control or CBP-shA cells were analyzed by immunoblotting with Rb (nuclear marker), actin (cytoplasmic marker), and p53 antibodies. (B) CBP regulation of p53 turnover in the cytoplasm and nucleus. Control and CBP-shA U2OS cells treated with cycloheximide were fractionated into nuclear and cytoplasmic fractions at the indicated time points. (Top three panels) The fractions were immunoblotted for p53, PARP (nuclear marker), and actin (cytoplasmic marker). (Lower left) Subcellular fractions (time = 0) from control and CBP-shA cells were immunoblotted with anti-CBP, anti-PARP, anti-α-tubulin, and anti-p53 antibodies. (Lower right) Determination of nuclear and cytoplasmic p53 half-life from Cont-sh and CBP-shA cells. Result is the average half-life from four separate experiments. * indicates significant P = 0.01 for difference in cytoplasmic p53 t1/2 between Cont-sh and CBP-shA. (C) Subcellular localization of p300 and CBP. U2OS cells were fractionated into nuclear and cytoplasmic fractions, and the fractions were immunoblotted for CBP, p300, Rb (nuclear marker), and actin (cytoplasmic marker).
To determine if the differential ubiquitination of nuclear and cytoplasmic p53 correlated with different rates of turnover, nuclear and cytoplasmic p53 half-life was measured by cycloheximide decay in control and CBP-shA cells (Fig. 3B and Fig. S3D). PARP (nuclear marker) and α-tubulin (cytoplasmic marker) immunoblots of the fractions from each time point revealed negligible cross-contamination (Fig. 3B). Surprisingly, CBP silencing led to stabilization of the normally rapidly degraded cytoplasmic p53 (t1/2 increase from 50 to 85 min, P = 0.01), but had no significant effect on the slower turnover of nuclear p53 (t1/2 increase from 95 to 100 min, P = 0.63; Fig. 3B and Fig. S3D). Given that p300/CBP are considered to be nuclear proteins (27, 28), but the major effect of CBP depletion was on cytoplasmic p53 half-life, the possibility that cytoplasmic pools of CBP or p300 might influence p53 metabolism was explored.
CBP/p300 Are Partly Cytoplasmic.
CBP and p300 are localized in PML oncogenic domains (PODs) and senescence-associated nuclear bodies, respectively (27, 29). To gain a more precise understanding of p300 and CBP subcellular localization, their abundance in cytoplasmic and nuclear fractions of U2OS cells was determined (Fig. 3C). The nuclear:cytoplasmic abundance ratios of Rb and actin, as determined by densitometry, revealed that the upper limit of cross-contamination of one fraction into the other was no greater than 10% in either direction (Fig. 3C). CBP and p300 were predominantly nuclear, as expected, but a substantial fraction of CBP was cytoplasmic, whereas a small but reproducible fraction of p300 was also observed in the cytoplasmic fraction (Fig. 3C). These fractionation results were further confirmed by immunofluorescent staining of U2OS cells for CBP and p300, where a clear cytoplasmic signal was detected for CBP, but p300 appeared predominantly nuclear (Fig. S3E). The specificity of the CBP immunofluorescence signal was confirmed by the near complete loss of CBP staining seen in CBP-shA cells (Fig. S3E).
CBP/p300 E3 Activities Are Exclusively Cytoplasmic.
To determine if nuclear and cytoplasmic CBP or p300 molecules have any differential capacities to influence p53 ubiquitination, nuclear and cytoplasmic CBP/p300 were assayed for E3 autoubiquitination activity. Nuclear and cytoplasmic fractions of U2OS cells were IP'd with anti-CBP or p300 antibody, and the IPs were assayed for E3 activity by the addition of ubiquitin, E1, E2 (ubch5a), and ATP, followed by Ub, CBP, or p300 immunoblotting (Fig. 4A and Fig. S4A). Dropout control reactions lacking E1/E2, Ub, or ATP were also performed to demonstrate that any Ub chains observed were the result of bona fide in vitro E3 activity and not contamination by cellular Ub chains binding nonspecifically to the IPs (Fig. 4A and Fig. S4A). CBP and p300 were observed in the cytoplasm and nuclear fractions as expected, and controls indicated little cross-contamination (Fig. 4A, bottom, and Fig. S4A, bottom). Surprisingly, CBP and p300 derived from cytoplasm demonstrated robust E3 autoubiquitination activity dependent on the simultaneous addition of E1/E2, Ub, and ATP, whereas nuclear-derived CBP/p300 exhibited much lower or undetectable activity (Fig. 4A and Fig. S4A). CBP/p300 immunoblotting of the reactions revealed that the lack of E3 activity associated with nuclear CBP/p300 could not be explained by lesser abundance (Fig. 4A and Fig. S4A). The lack of nuclear CBP autoubiquitination activity could also not be explained by nonspecific inhibition by extraction conditions or inhibitory factors within the nuclear fraction, as immunopurified nuclear MDM2 exhibited easily detected autoubiquitination activity, which if anything, had a higher specific activity than cytoplasmic MDM2, based on the relative abundance of MDM2 in the two fractions (Fig. S4B).
Fig. 4.
p300/CBP E3 activities are predominantly cytoplasmic. (A) (Top and Middle) Nuclear and cytoplasmic fractions of U2OS cells were immunoprecipitated with anti-CBP antibody, and the washed IPs incubated with E1/E2, Ub, and ATP as indicated, followed by anti-Ub and CBP immunoblotting of the reactions. (Bottom) Immunoblot of CBP and Rb (nuclear marker) in nuclear and cytoplasmic fractions. (B) E4 activity of cytoplasmic CBP. p53/MDM2 complexes (insect-cell-derived) were incubated with E1/E2, Ub, or methyl-Ub, ATP and CBP IPs from control-sh or CBP-shA cytoplasmic fractions as indicated, followed by anti-p53 (Top) or anti-CBP (Middle) immunoblotting of the reactions. Migration positions for native, monoubiquitinated/MUM and polyubiquitinated p53 species are indicated. (Bottom) Relative abundances of native, mono-Ub/MUM, and polyubiquitinated p53 species were quantitated by densitometry.
To further assess the specific dependence of the observed E3 activity on the actual presence of CBP or p300 in the reaction, cytoplasmic fractions of control, CBP-shA, and p300-sh cells were IP'd with anti-CBP or anti-p300 antibodies, as appropriate, and the IPs assayed for E3 activity with or without added E1 and E2 (Fig. S4 C and D). There was no significant cross-contamination of nuclear and cytoplasmic markers (Fig. S4 C and D, bottom), and the IPs were also probed for p300 and CBP to assure that the levels of p300/CBP in the IPs reflected the respective shRNA knockdowns for each (Fig. S4 C and D, middle). CBP and p300 IPs from control cell cytoplasm were both highly enriched for E3 autoubiquitination activity dependent on added E1/E2 (Fig. S4 C and D, upper). However, CBP and p300 IPs from both CBP-shA and p300-sh cytoplasm were substantially depleted of E3 activity (Fig. S4 C and D, upper), confirming that the E3 activity seen in the p300 and CBP IPs from control cell cytoplasm did depend on the specific presence of p300 or CBP proteins in the respective reactions.
Cytoplasmic CBP Is an Active p53 E4.
The specific localization of E3 autoubiquitination activity of CBP/p300 to the cytoplasm suggests that their E4 activity, likewise resides in the cytoplasm. To confirm that cytoplasmic CBP was an active E4 for p53, immunopurified cytoplasmic CBP derived from control-sh or CBP-shA cells was exposed to MDM2/p53 complexes in the presence or absence of E1/E2, along with Ub and ATP (Fig. 4B). To identify the migration positions of p53-mono-Ub/MUM species, p53/MDM2 complexes were incubated with E1/E2, methyl-Ub (nonconjugatable), and ATP (Fig. 4B, first lane). CBP IP'd from control-sh cells clearly catalyzed the formation of polyubiquitinated p53 (p53-polyubiquitin conjugates ≈50% of total p53 species; Fig. 4B), and this activity was significantly reduced in proportion to the reduction in CBP abundance when a CBP IP from CBP-shA cells was used as the source for cytoplasmic CBP (p53-polyubiquitin conjugates ≈20% of total p53 species; Fig. 4B). Thus, cytoplasmic CBP is an active E4 for previously mono/multiply monoubiquitinated p53.
Discussion
p300 and its paralog CBP have been identified as physiological regulators of p53 ubiquitination and stability. Depletion of CBP or p300 resulted in p53 stabilization, and CBP and p300 were both required for physiologic p53 polyubiquitination. The CBP N terminus (1–616) was found to encode a p53 destabilization function in vivo, and residues 1–616 and 1–451, respectively, constitute minimal functional E4 and E3 domains within CBP. CBP and p300 E3 activities were only observed from cytoplasmic fractions, and p53-mono-Ub and MUM conjugates accumulated in the cytoplasm of CBP silenced cells. The degradation of cytoplasmic p53 was specifically altered by CBP silencing, consistent with the cytoplasmic localization of CBP E4 function in vivo.
The N terminus of p300 harbors its E3/E4 activity (14), and the same region of CBP encodes its E3/E4 activity and shares significant areas of sequence conservation (18, 30). Within this region of p300/CBP is a cys-his-rich region, but no signatures for RING, HECT, U-box, or PHD domains can be found. Based on findings with the A20, Rabex-5, and E4F1 proteins (10, 17, 31), a generic Zn2+-binding region (cys- or cys-his-rich) can encode an active E3, with no clear sequence or structural homology to the Zn2+-binding RING E3 domain. Perhaps more directly relevant, the evolutionarily related HAT P/CAF was recently shown to encode a noncanonical E3 that targets MDM2 (21). In this case, the protein domain responsible for the activity contains no structural motifs found in known E3 proteins (21).
Subcellular fractionation revealed that p300/CBP E3 activities are limited to the cytoplasm and presumably account for the activity that converts mono-Ub/MUM p53 conjugates to polyubiquitinated unstable p53 in the cytoplasm. The mechanism of localization of p300/CBP E3 activity to the cytoplasm cannot yet be explained. Either a cytoplasmic-specific modification of p300/CBP or p300/CBP-interacting factor activates the activity or an interacting factor or modification in the nucleus represses it. The spatial separation of p300/CBP's E3/E4 activity from their nuclear HAT functions is intriguing and solves the seeming paradox of how these two presumed opposing regulatory functions exist within the same molecules (32).
This work raises a number of questions critical to p53, ubiquitin/proteasome, and cancer biology. How the plethora of p53 E3s, and now E4s, are coordinated to achieve p53 homeostasis remains unclear, although it is likely that the subcellular localization of these enzymes will be of critical importance to understanding their roles. The biochemical mechanism by which CBP/p300 recognizes ubiquitinated, and not native, p53 for further ubiquitination is also presently not understood. The simplest explanation for the preferred recognition of ubiquitinated substrate would be the existence of a Ub recognition motif within the p300/CBP N-termini—as is seen in the Rabex-5 atypical E3 (31), although no such domain is readily recognized in p300/CBP by homology search. Finally, the implications of these findings for cancer biology and therapy are potentially significant. As MDM2 is now a bona fide clinically relevant drug target for cancer therapy (33), other enzymes in the p53 stability regulation pathway, such as the p300/CBP E4 domain, might also be effectively targeted in human cancers.
Materials and Methods
Cell Culture and Plasmids.
U2OS cells were grown in DMEM supplemented with 10% FBS and antibiotics. Cells were treated, where noted, with 100 μg/mL cycloheximide, 2 μM Dox, or 10 μM MG-132 (Sigma). Plasmid transfection was done with Fugene 6 (Roche), and siRNA transfections used Oligofectamine (Invitrogen). pRSV-CBPmyc (34) and pcDNA-UbHA (9) have been described. To generate pN8.Flag-CBP (1–616) and -CBP (1–200), CBP fragments were PCR-amplified from pRSV-CBPmyc and cloned into pN8.Flag vector using BamHI and EcoR1. An shRNA-resistant CBP allele (pRSV-CBP*myc) was generated by silent mutation of the shRNA target sequence (AACTCCAATAGC mutated to AATAGTAACTCT; CBP residues 190–193) within pRSV-CBPmyc. pGEX-CBP (1–616) and (1–200) were generated by cloning the indicated PCR-amplified fragments into pGEX-2tk. pCDNA3-p53-UbF, which has the Ub cDNA cloned in-frame to the 3′ end of the p53 cDNA, was the generous gift of Christine Blattner, and MT107 His-Ub expression vector has been described (35).
Western Blotting, Immunofluorescence, and Immunoprecipitation.
For western blot analyses, cells were lysed in cold NETN240 buffer (20 mM HEPES, pH 7.4, 2 mM MgCl2, 10 μM ZnCl2, 240 mM NaCl, 0.2% Triton-X 100), supplemented with complete EDTA-free tablets (Roche). For immunoprecipitation of whole cell lysates, cells were lysed in cold RIPA buffer [50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10 mM MgCl2, 10 μM ZnCl2, 1% Triton X-100, 0.5% DOC supplemented with fresh 5 mM NEM and complete EDTA-free tablets (Roche)]. Immunoprecipitations from whole cell lysates were performed in the lysis buffer overnight, followed by capture with Protein A agarose (Upstate) and five washes in lysis buffer. Western blot signals were quantified after visualization of primary antibody by HRP-conjugated secondary antibody and enhanced chemiluminescence or by fluorescent-labeled secondary antibody and detection by Odyssey blot scanner (LiCor), using ImageJ National Institutes of Health (NIH) software.
In Vitro E3 Assays.
To determine CBP/p300 or MDM2 E3 ligase activity, CBP, p300, or MDM2 were immunoprecipitated from 0.7 mg cytoplasm or 0.2 mg nuclear fractions diluted with high salt buffer [10 mM HEPES (pH 7.5), 150 mM NaCl, 150 mM KCl, 1 mM MgCl2, 0.5% Triton X-100, supplemented with fresh 5 mM NEM and protease inhibitors) using A-22, N-15, or D-7 antibodies followed by protein A Sepharose. The IPs were washed in high salt buffer three times followed by two washes in Ub buffer (25 mM HEPES, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.05% Triton X-100, 0.5 mM DTT, 3 mM Mg-ATP). The washed and equilibrated IPs were then incubated with 100 ng E1 (rabbit; Boston Biochem), 25 ng E2 (UbcH5a, human recombinant; Boston Biochem), and 5 μg Ub (human recombinant; Boston Biochem) for 60 min at 37 °C. Reactions were then stopped by the addition of sample buffer, followed by SDS-PAGE and immunoblotting. E3 assays using purified GST-CBP polypeptides were performed as described (14). GST-CBP (1–616), (1–451), and (452–721) (36) were expressed in BL21 cells and purified using glutathione-Sepharose (GE Healthcare). Purified GST-CBP (0.1, 0.2 or 0.5 μg) (1–616), (1–451), or (452–721) proteins were added to ubiquitin reaction components as indicated and incubated at 37 °C for 60 min, followed by analysis of the reaction products with anti-Ub antibody.
One-Step E4 Assay.
To determine CBP E4 ligase activity, insect-cell-derived human p53 and FLAG-MDM2 were purified as a complex using FLAG M2 (Sigma) agarose as described (14), then incubated with ubiquitin reaction components (100 ng E1, rabbit; Boston Biochem), 25 ng E2 (UbcH5a, human recombinant; Boston Biochem), and 5 ng Ub (human recombinant; Boston Biochem) at 37 °C for 30 min. FLAG-CBP (1, 10, or 100 μg) immunopurified from baculovirus-infected Sf9 insect cells (with anti-FLAG resin/FLAG peptide elution) (35) or CBP IP'd from U2OS cytoplasmic fraction (see In Vitro E3 Assay section) was then added, followed by further incubation of the reaction at 37 °C for 60 min and p53 immunoblot of the reaction products.
Two-Step E4 Assay.
To determine E4 ligase activity of CBP, a two-step ubiquitination reaction was performed. First, insect-cell-derived p53 was ubiquitinated by MDM2 as described above. To remove MDM2 and nontagged ubiquitin, p53 was then immunoprecipitated and washed three times with RIPA buffer, two times with PBS, and two times with Ub buffer. Immobilized p53 was then mixed with second ubiquitination reaction components [100 ng E1, 25 ng UbcH5a, 5 μg Flag-tagged Ub (Boston Biochem)] along with affinity-purified Flag-CBP (1–200) or (1–616) polypeptides obtained from transfected U2OS cells and incubated at 37 °C for 60 min. To remove CBP autoubiquitination products, beads were washed three times with RIPA buffer. p53 was eluted by boiling with NuPAGE LDS sample buffer, separated by SDS-PAGE, and visualized by immunoblotting with anti-Flag antibodies.
Additional Materials and Methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank C.M. Chiang (UT Southwestern, Dallas, TX) for generously providing FLAG-CBP baculovirus, C. Blattner (Forschungszentrum Karlsruhe, Karlsruhe, Germany) for providing pCDNA3.p53-UbF, and members of the Altieri and Grossman labs for helpful discussions. S.G. was supported by a Kimmel Scholar Award, and R01CA107532 from the National Cancer Institute (NCI).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904305106/DCSupplemental.
References
- 1.Michael D, Oren M. The p53 and Mdm2 families in cancer. Curr Opin Genet Dev. 2002;12:53–59. doi: 10.1016/s0959-437x(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 2.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Slee EA, O'Connor DJ, Lu X. To die or not to die: How does p53 decide? Oncogene. 2004;23:2809–2818. doi: 10.1038/sj.onc.1207516. [DOI] [PubMed] [Google Scholar]
- 4.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 5.Morgunkova A, Barlev NA. Lysine methylation goes global. Cell Cycle. 2006;5:1308–1312. doi: 10.4161/cc.5.12.2820. [DOI] [PubMed] [Google Scholar]
- 6.Brooks CL, Gu W. Ubiquitination, phosphorylation, and acetylation: The molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 7.Brooks CL, Li M, Gu W. Monoubiquitination: The signal for p53 nuclear export? Cell Cycle. 2004;3:436–438. [PubMed] [Google Scholar]
- 8.Li M, et al. Mono- versus polyubiquitination: Differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 9.Kaur M, Pop M, Shi D, Brignone C, Grossman SR. hHR23B is required for genotoxic-specific activation of p53 and apoptosis. Oncogene. 2006;26:1231–1237. doi: 10.1038/sj.onc.1209865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Cam L, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman SR, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 15.Sui G, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Lai Z, et al. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J Biol Chem. 2001;276:31357–31367. doi: 10.1074/jbc.M011517200. [DOI] [PubMed] [Google Scholar]
- 17.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 18.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 19.Freedman SJ, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Guzman RN, Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry. 2005;44:490–497. doi: 10.1021/bi048161t. [DOI] [PubMed] [Google Scholar]
- 21.Linares LK, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol. 2007;9:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- 22.Oren M, et al. Regulation of p53: Intricate loops and delicate balances. Ann N Y Acad Sci. 2002;973:374–383. doi: 10.1111/j.1749-6632.2002.tb04669.x. [DOI] [PubMed] [Google Scholar]
- 23.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 24.Siliciano JD, et al. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 26.Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaMorte VJ, Dyck JA, Ochs RL, Evans RM. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckner R, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 29.Pedeux R, et al. ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol. 2005;25:6639–6648. doi: 10.1128/MCB.25.15.6639-6648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 31.Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- 32.Grossman SR. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem. 2001;268:2773–2778. doi: 10.1046/j.1432-1327.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 33.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.