Abstract
The environments we humans encounter daily are sources of exposure to diverse microbial communities, some of potential concern to human health. In this study, we used culture-independent technology to investigate the microbial composition of biofilms inside showerheads as ecological assemblages in the human indoor environment. Showers are an important interface for human interaction with microbes through inhalation of aerosols, and showerhead waters have been implicated in disease. Although opportunistic pathogens commonly are cultured from shower facilities, there is little knowledge of either their prevalence or the nature of other microorganisms that may be delivered during shower usage. To determine the composition of showerhead biofilms and waters, we analyzed rRNA gene sequences from 45 showerhead sites around the United States. We find that variable and complex, but specific, microbial assemblages occur inside showerheads. Particularly striking was the finding that sequences representative of non-tuberculous mycobacteria (NTM) and other opportunistic human pathogens are enriched to high levels in many showerhead biofilms, >100-fold above background water contents. We conclude that showerheads may present a significant potential exposure to aerosolized microbes, including documented opportunistic pathogens. The health risk associated with showerhead microbiota needs investigation in persons with compromised immune or pulmonary systems.
Keywords: bioaerosols, Mycobacterium avium complex, Non-tuberculous mycobacteria, public health, rRNA metagenomics
Shower usage provides a source for repeated exposure to microbes through aerosolization and/or direct contact. The inside of a showerhead is a specific niche that is moist, warm, dark, and frequently replenished with low-level nutrient resources and seed organisms. Biofilms form on interior showerhead surfaces and potentially expose the user to a cohort of unknown, aerosolized microorganisms. Shower aerosol particles can be sufficiently small to carry bacteria deep into the airways (1). Pulmonary disease and other health risks such as asthma, bronchitis, and hypersensitivity pneumonitis are associated with inhalation of both viable bacteria and inviable microorganisms or their components (2–4). It has been hypothesized that the rise in pulmonary infections by nontuberculous mycobacteria (NTM) over recent decades is linked to increased use of showers rather than baths (5). Immune-compromised populations are on the rise; thus, identification of anthropogenic reservoirs of potential pathogens is of public health concern (3, 6).
Previous microbiological studies of showerhead biofilms have used culture methodology to detect and identify microbes, and have focused primarily on Legionella pneumophilia (7, 8, 9) and Mycobacterium avium (10–12). These organisms commonly occur in municipal waters and several studies have traced both L. pneumophilia and M. avium infections in hospitalized patients to microbes in their home showers (9, 10, 12).
Despite implication as a potential source of disease, the microbial composition of the showerhead environment is poorly known. Characterization of natural microbial communities by use of culture techniques may drastically under-sample the actual numbers and diversity, because most microbes are not readily cultured with standard methods (13, 14). Consequently, we used culture-independent methodology based on ribosomal RNA gene sequences to identify the composition of assemblages of microbes associated with showerhead surfaces over a wide geographical area of the U.S. Many of these microbes are closely related to organisms common in water, but some microbes of potential public health concern are enriched to high levels by the showerhead environment.
Results
Samples and Processing.
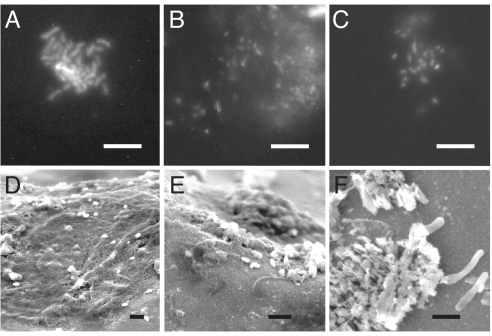
As described in Methods and Materials, biofilms were obtained by swab of interior surfaces of 45 showerheads from nine cities in the United States. Some sites were sampled on multiple occasions to assess the stability of the showerhead microbial assemblages. Water feeding into showerheads was sampled in parallel with the swabs at 12 sites. All swab samples examined by microscopy showed clear evidence of more or less dense microbiology. As illustrated in the micrographs in Fig. 1, microbes generally were clumped and embedded in extracellular material, consistent with biofilm morphology. The DNA yields from the swabs were highly variable, and DNA could not always be extracted.
Fig. 1.
Fluorescence and SEM images of showerhead biofilm. (A–C) Epifluorescence microscopy of biofilm samples stained with DAPI; scale bars, 10 μm. (D–F) SEM micrographs of increasing magnification of in situ showerhead biofilm on the inner surface of one water distributor (Scale bars, 2 μm.)
To identify the microbial constituents of the showerhead biofilms, we amplified rRNA genes from sample DNAs by PCR, using nominally universal primers (515F-1391R), then cloned the amplicons and determined their sequences. Overall, >6,090 unique rRNA gene sequences were determined and used to identify phylogenetically the microbes associated with the sampled sites.
Composition of Showerhead Communities.
Ribosomal RNA gene sequences from natural microbial communities seldom are identical to previously encountered sequences. To relate these environmental sequences to named organisms, we binned the sequences into operational taxonomic units (OTUs) of greater than or equal to 97% identity, which corresponds approximately to the rRNA gene sequence variation seen in studied microbial species (15). Most of the sequences fell into species-level (≥97% identity) or genus-level (≥95% identity) bins with one or more named representative.
Fig. 2 summarizes the distribution of genera that comprised at least 0.5% of the total showerhead clones sequenced, grouped by municipality of origin (Dataset S1 shows all observed sequence types). The showerhead communities were comprised of multiple organisms, and the specific organisms varied from site to site. In general, however, compared to high-nutrient microbial communities (e.g., microbial mats, gut contents) the showerhead communities were relatively simple (2–29 sequence types per site) and collectively comprised limited phylogenetic diversity. Although representatives of many bacterial phyla were detected (33/≈70 known phyla), most of the sequences were diverse representatives of only three phyla: Actinobacteria, Proteobacteria, and Firmicutes (GreenGenes taxonomy) (16). Less than 1% of sequences analyzed were archaeal or eukaryotic. At the depth of sequence analysis performed, approximately 90 to a few hundred sequences per sample, full survey of the more rare organisms was not anticipated. Nonetheless we sampled the most abundant sequences [>2/3 of species predicted by Chao 1 estimation, (17)] and so collectively these analyses provide an overview of the kinds of microbes expected to occur in showerheads.
Fig. 2.
This heatmap-table summarizes the BLAST results for all showerhead swab libraries, pooled at the genus-level and grouped by municipality of origin. Genera representative by at least 0.5% of the total clones (>20 sequences) were included, for a total of 17 genera. Figure footnotes: *, “Other” is comprised of all genera representing less than 0.5% of total dataset; †, percent of total showerhead clones in study; ‡, showerhead fed by well water; §, signifies the first of multiple samples taken at the site as designated by the first three letters; ¶, signifies the second of multiple samples taken at the site; ‖, signifies the third of multiple samples taken at the site; −, signifies that clones representative of the genera were not detected in the sample.
Four sites were sampled 2–3 times over intervals of 2–12 months (see Dataset S2 for details) to test the temporal variability of the showerhead assemblages. In general, samples showed persistence of particular sequence types, although distributions varied between dates, perhaps an effect of patchiness in swab sampling (Fig. 2). In one case [showerhead BSK, a Denver Metro 1 showerhead sampled on three occasions (Fig. 2)], attempted cleaning with bleach solution resulted in a 3-fold increase in the load of M. gordonae, from approximately 25% of the assemblage sequences initially (BSK1Q) to 72% and 74% subsequently (BSK2Q and BSK3Q). Although anecdotal, this observation is interesting in light of the general resistance of mycobacteria to chlorine, which also may be one reason for the mycobacterial enrichment in municipal systems compared to well-water fed systems (discussed below).
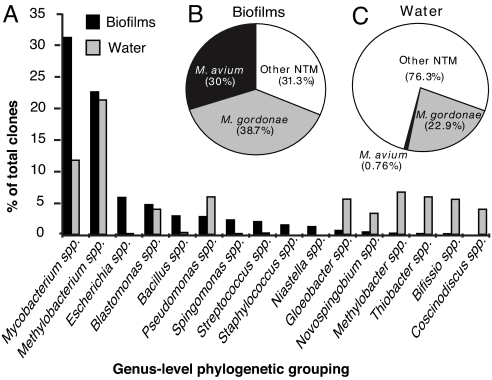
The overall distributions of abundant (>0.5% of total) genus-level OTUs in municipal water and showerhead biofilms are summarized in Fig. 3A. The biofilm assemblages were comprised of ubiquitous water and soil microbial groups, some known for biofilm formation. Surprising, however, was the abundance of sequences indicative of Mycobacterium spp. in showerhead biofilms compared to feedwaters. As summarized in Fig. 3B, the sequences of the dominant mycobacteria corresponded mainly to those of M. gordonae and M. avium, which comprised respectively 10.5% and 9.1% of the total municipal showerhead sequences, and were the most common sequences observed. Mycobacteria are known to occur at low levels in municipal waters, and were observed in the analyzed showerhead feed waters (Fig. 3C). However, libraries from showerhead biofilms were highly enriched in these organisms, >100-fold above background water contents. Moreover, sequences representative of M. avium, of particular note as an opportunistic pathogen, are enriched over those of M. gordonae in showerhead biofilms (Fig. 3B) compared to feed waters (Fig. 3C). The other minor microbial components that have been implicated in respiratory disease were all common water and soil organisms, including Pseudomonas spp. (3.8% of sequences), Sphingomonas spp. (2.7%), Staphylococcus spp. (2.0%), Streptococcus spp. (1%), Burkholderia spp. (0.8%), Neisseria spp. (0.6%), Acinetobacter spp. (0.6%), and Legionella spp. (0.1%) (Dataset S1 for details).
Fig. 3.
Comparison of the diversity of abundant sequences from swab biofilm and water samples collected from sites supplied by treated municipal water, private well water supplied sites were excluded from this analysis. (A) A comparative histogram of the most abundant swab and water genera identified by BLAST. The total number of sequences for municipal biofilms was n = 3,454, for municipal water n = 1,146. (B) Pie chart of mycobacterial sequences (n = 1,051) identified in showerhead biofilm samples. (C) Pie chart of mycobacterial sequences (n = 131) from water samples.
Sequences indicative of M. avium, the most noteworthy potential pathogen detected, were identified in 20% of showerhead swabs overall, with an average density of 32% of the library when observed. Sequenced mycobacterial genomes contain only single copies of rRNA genes, so the frequency of mycobacterial rRNA genes in the assemblages represents a minimal contribution to the observed organismal abundances. The bacterial species M. avium is comprised of several extremely closely related subspecies, including M. avium hominissuis, M. avium avium, M. avium paratuberculosis and others, which are not discriminated by rRNA sequences (18). To verify that the sequences belonged to microbes of the M. avium complex to the exclusion of other NTMs, we amplified and sequenced several rRNA internal transcribed spacers (ITS), and compared those to known M. avium sequences from clinical and environmental isolates. ITS sequences are highly variable and consequently afford better differentiation of organisms represented by the sequences (18, 19). Showerhead M. avium sequences clustered phylogenetically with those of known environmental and clinical isolates of M. avium (Fig. S1). Twenty-eight of 49 (57%) of the ITS sequences analyzed were identical to those of clinical isolates from NTM disease. Clearly, showerhead biofilms pose an enriched exposure to this recognized opportunistic pathogen.
Although M. avium was commonly encountered, many samples were negative for this organism, either because the organism was not present or because it is less abundant than others and not detected because sequence analysis samples only the most abundant microbial species. To test the possibility that M. avium was present in samples that were negative by sequence analysis, we used quantitative PCR (Q-PCR) with M. avium-specific primers to screen DNAs from 32 biofilm and 14 water sources. Q-PCR identified M. avium DNA in 25 of 32 (78%) swab extracts tested, including 20 in which M. avium was not encountered in the rRNA gene libraries (Dataset S3). Although M. avium was encountered only rarely among 16S rRNA sequences determined from water samples (Fig. 3), Q-PCR detected that organism in 13 of 14 water samples tested from Denver and New York metropolitan systems (Dataset S3).
The opportunistic pathogen L. pneumophila, the cause of Legionnaire's Disease, receives much popular attention, but sequences indicative of that organism were encountered only rarely in this survey (only 3/≈6,000 sequences determined). L. pneumophila constitutes a broad relatedness group, however, so detection of a representative of the group does not indicate a pathogen. Because of the potential human health implication of this detection, we conducted quantitative PCR (QPCR) assays with a L. pneumophila-specific primer pair that targets a pathogenesis gene, the macrophage infectivity potentiator (mip) gene (20–24), to screen a subset of samples. Thirty-six samples (16 water and 20 swabs, representing 10 cities) were tested in duplicate reactions, including samples with positive L. pneumophila detection by sequence. The L. pneumophila mip gene was not detected in any sample at a sensitivity of 0.5 copies/μL of DNA extract.
Microbial Constituents of Shower Aerosols.
Showerhead biofilms and water are potential sources of aerosolized microorganisms. However, different microbes and biofilms have different qualities that can influence partitioning into aerosols. Indeed, we and others have shown that mycobacteria can be selectively aerosolized, possibly a consequence of their waxy, hydrophobic quality (3, 25). To determine the makeup of shower aerosol microbiology, we collected aerosols during 20-min unoccupied shower operations with three showerheads analyzed rRNA gene sequences and compared them with biofilm, water, and ambient bathroom air samples. Microbial constituents were reflective of feedwaters and not biofilm. It seems possible, however, that any initial pulse of biofilm components would have been extensively diluted by water delivered during the aerosol collection period, and so not detected.
Well-Water vs. Municipal-Supplied Showerhead Biofilms.
Most of the samples analyzed were supplied by municipal water distribution systems, but we included four homes supplied by private water wells. The microbial compositions of well-water biofilms were distinct from those associated with municipal waters, and no mycobacteria were detected. Three of the systems examined were supplied by individual water wells in Southwestern Colorado overlying the San Juan Basin's Fruitland coal formation. Oil and gas drilling have been implicated in increased methane and chemical release into the aquifers that overlie the coal beds (26). Both water and showerhead biofilm libraries from these homes showed an abundance of sequences closely related to bacterial genera such as Methylocystis spp. (10% of biofilm and water clones from these three sites), Methylobacteria spp. (8.1%), Methylomicrobium spp. (5.2%), and other close relatives of known methane and methanol metabolizing organisms (27). These results indicate that microbial analysis can provide insight into local groundwater geochemistry.
Discussion
In our daily lives, we humans move through a sea of microbial life that is seldom perceived except in the context of potential disease and decay. Indoor air typically has approximately 106 bacteria per m3; municipal tap water usually contains at least 107 bacteria per L. Little is known about the nature of these microbial populations, but they are expected to derive from both human traffic and microbial ecosystems that happen to be enriched by the character of the particular setting, in this case the showerhead biofilm ecotope.
The majority of showerhead microbiota encountered in our survey is composed of genus- or species-level relatedness groups that are commonly found in water and soil. The showerhead environment strongly enriches for microbes that are known to form biofilms in water systems, including Mycobacterium spp., Sphingomonas spp., Methylobacterium spp. and others (Fig. 3 and Dataset S1). Particularly, the enrichment and prevalence of mycobacteria were unexpected. Mycobacteria were detected in clone libraries or by QPCR in most showerheads fed by municipal water systems (Dataset S3).
The detection of significant loads of M. avium in many showerhead biofilms identifies a potential personal health concern. The reasons for the enrichment of mycobacteria are not clear. Mycobacteria readily form biofilms and, because of their generally waxy quality, may be particularly resistant to shear forces generated in shower operation (28–36). Furthermore, many species of biofilm-forming mycobacteria are chlorine-resistant, and thus potentially can be enriched by chlorine disinfection protocols used by many municipalities (28, 29, 34, 37, 38). Consistent with this, we only observed mycobacterial rRNA gene sequences in municipal water systems, not in untreated well water systems.
The occurrence of M. avium in showerhead biofilms raises the question of exposure. Does shower usage increase risk for NTM disease? At this time there are no epidemiological data with which to assess risk for NTM infections. However, M. avium and other NTM can cause pulmonary disease in healthy people, as well as those predisposed to pulmonary infection. In many centers, NTM now outnumber M. tuberculosis detections in clinical mycobacteriology labs (39, 40). Risk factors associated with NTM pulmonary infection include smoking, chronic lung disease, alcoholism, and pulmonary or immune genetic defects (5, 40). Diagnoses of disseminated NTM infections of the blood, lymph, bone, skin or other tissues have increased, especially in immune compromised populations such as HIV/AIDS and transplant patients (5, 40–42). Moreover, a few recent studies have shown a link between pulmonary M. avium infections and home showerhead water microbiology (10, 12). M. avium and other NTM infection rates are on the rise throughout the developed world (5, 39) and have been hypothesized to correlate with increased exposure to aerosolized microbes through increased use of showers rather than bathing (5). Thus, shower usage possibly is contraindicated for individuals with compromised immune or pulmonary systems, an issue that needs evaluation in these populations.
This study is a culture-independent molecular survey of the nature of showerhead microbiology. The finding that NTM are abundant and prevalent in showerhead biofilm assemblages points to one clear source of opportunistic pathogens known for pulmonary disease. Many home and public devices, such as humidifiers and evaporative cooling units, also produce moist aerosols that likely disperse microorganisms associated with the particular system. Little is known about the microbiology of such settings, which we commonly encounter in daily life. We conclude that there is need for further epidemiological investigations of potential sources of NTM infections, including showerheads. The methods we use here provide an experimental approach for such investigations.
Materials and Methods
Sample Collection.
Showerhead swab samples were collected between May 2006 and January 2008 from homes, apartment buildings, and public buildings in Colorado (Southwestern Colorado city #1, and four Denver-Metro Cities #1–4), Illinois (IL), Tennessee (TN), North Dakota (ND), and New York City (NYC). Sampling and site data are presented in online Dataset S2. A total of 52 samples from 45 sites were analyzed (see Dataset S2). Following removal and disassembly of the showerhead, sterile swabs were used to wipe biofilm from the inner surface. Swabs were stored in 70% ethanol until DNA extraction. Water samples were collected in new sterile 1-L Nalge bottles and stored at 4 °C until filtration (0–5 h). Water (≈1 L) was filtered through a 0.2-μL polycarbonate filter (Isopore™ Membrane Filters, Millipore) and the filter processed for DNA as described below with the addition of 500 μL chloroform to dissolve the filter.
Aerosol Sample Collection.
Before collection of air samples, the shower stall was washed with a 10% bleach solution and a new vinyl shower curtain was hung to minimize aerosols from dislodged biofilms of these surfaces. A specially designed OMNI 3000 air sampler (Evogen Inc.) with UV-sterilized contactor and virgin tubing and cartridges was used to collect shower aerosol samples. The OMNI sampled for 20 min at approximately 270 L/min at the outside periphery of the shower at approximately 2-m high (high enough to place the intake above the shower curtain), impinging into a sterile solution of 1× PBS and 0.005% Tween. Two 20-min air samples were collected: the ambient bathroom air, and then a sample with the shower running at a warm temperature. Aerosol samples were filtered and processed in the same manner as the water samples.
Additional Sample Details.
Dataset S4 shows all observed sequence types for water samples, and Dataset S5 shows a comparison of water, biofilm swab, and ambient bathroom air samples.
PCR Amplification of rDNA.
DNA was extracted from the cotton tips of the swabs (biofilm samples), or from polycarbonate filters (water and aerosol samples) using a bead beating protocol as previously described (43). DNA extracts were amplified with the universally conserved 16S rRNA primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1391R (5′-GACGGGCGGTGWGTRCA-3′) or bacterially conserved 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (44) and 338R (5′-CTGCTGCCTCCCGTAGGAGT-3′) (45). PCR Reactions were conducted at 94 °C for 2 min, followed by 30 cycles at 94 °C for 20 s, 52 °C for 20 s, and 65 °C for 1:30 min, followed by a 65 °C elongation step for 10 min. Each 25-μL reaction contained 10 μL Eppendorf 2.5× HotMasterMix (Eppendorf), 10 μL water, 0.05% BSA (Sigma-Aldrich), 100 ng of each oligonucleotide primer, and 1–5 ng of DNA template. Triplicate PCR products were pooled before purification and cloning. Individual PCR-amplified rRNA genes were isolated by cloning with Topo-TA as per manufacturer's instructions (Invitrogen) and collected into libraries of 96 randomly chosen clones. DNA preparation and sequencing was performed as previously described (46).
ITS Gene Sequencing.
Samples identified to contain M. avium were used as template for Mycobacterium spp.-specific amplification of the 16S-23S rRNA ITS sequence using Myco1121F primer (5′-CATGTTGCCAGCRGGTAATGCCGGG-3′) or Myco 1432F primer (5′-GAAGCCRGTGGCCTAACC-3′) and universal 23S rRNA primer 130R (5′-GGGTTBCCCCATTCGG-3′) (44). Myco1121F and Myco1432F were designed using the ARB software package to identify regions conserved across the non-tuberculosis mycobacterial sequences of interest, and synthesized by IDT (Integrated DNA Technologies). PCR, cloning and sequencing was performed as described above. Phylogenetic analysis was performed by comparison of unknown ITS gene sequences to genes of known Mycobacteria in an ARB (http://www.mpibremen.de/ARB.html) database (47).
Phylogenetic Analysis.
Sequences initially were compared to other known small subunit rRNA (SSU rRNA) gene sequences in the National Center for Biotechnology Information (NCBI) database through use of the Basic Local Alignment Search Tool (BLAST) (48) using the program XplorSeq (49). A total of 6,090 swab and water 16S, and 52 MAC ITS DNA sequences generated in this study have been deposited in GenBank with accession numbers EU629353–EU635442, and EU697021–EU697072. 16s sequences with low bit scores (<500) or shorter than 300 base pairs were excluded from the analysis; of 6,090 total sequences, 5,745 were used.
Quantitative PCR.
Quantitative PCR was conducted using MAC-specific, Legionellae-specific and L. pneumophila mip (L.p.-mip) gene SYBR Green (ABI Biosciences) assays, to determine the prevalence of M. avium, Legionellae, and L. pneumophila throughout the sample set, including those samples in which the species were not represented in the 16S library. Q-PCR was performed on a DNA Engine Opticon System (MJ Research) in 25-μL reaction volumes composed of 12.5 μL SYBR Green, 25 ng each primer, 0.4 μL 10× BSA, 8.6 μL water, and 1.5 μL DNA, the Legionellae and L.p.-mip reaction mixtures excluded BSA. Sample DNAs were diluted 1:5 before amplification for the MAC assays, and full concentration for the L.p.-mip assay.
MAV-specific analysis was conducted with bacteria-specific primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (44) and MAV-specific primer MAV199R (5′-ACCAGAAGACATGCGTCTTG-3′) (Degroote MA, and NRP). For the MAV-assay, a deletion plasmid (MAP- 8FΔ) was constructed with a 40 base pair deletion in positions 28–68 at the 5′ end of the M. avium subsp. paratuberculosis 16S rRNA gene (Degroote, MA, and NRP). The plasmid was amplified, and cloned into TOPO-4 vector, then screened by agarose gel electrophoresis to verify the deletion. Sequencing of the insert was performed to verify that the correct region was amplified. E. coli cells containing the cloned deletion plasmid grew overnight in 2× yeast-tryptone broth containing 0.1 mM ampicillin, and were purified with a QiaFilter Plasmid Maxi Kit (QIAGEN). Plasmid concentration was quantified with spectrofluorometry and serially diluted from 107 to 1 copies. The MAV-specific QPCR assay included an initial denaturation step of 94 °C for 10 min was followed by 45 cycles of 94 °C for 15 s, 60 °C for 45 s, a fluorescence read, then 1 s at 80 °C, and a second plate read.
Legionella-specific primers were Leg448F (5′-GAGGGTTGATAGGTTAAGAGC-3′) (50) and Leg880R (5′-GGTCAACTTATCGCGTTTGCT-3′) (51). L. pneumophila primers were Lp-mip-PT69 (5′- GCA TTG GTG CCG ATT TGG- 3′) and Lp-mip-PT70 (5′- GYT TTG CCA TCA AAT CTT TCT (52). Standards for the Legionella and L.p.−mip assay were generated with DNA extracted from a plate scrape of L. pneumophila subsp. pneumophila strain Philadelphia-1, ATCC 33152. The genes were amplified, cloned into TOPO-4 vector, purified, and quantified as described above. The Legionella-specific QPCR cycling was as follows 94 °C for 10 min, 45 cycles of 94 °C for 15 s, 52 °C for 15 s, and 65 °C for 30 s, a fluorescence read, then 1 s at 80 °C, and a second plate read. L.p.-mip-specific QPCR cycling was as above except the annealing temperature was 58 °C and the second plate read was at 75 °C.
Duplicate Q-PCR reactions were performed on each sample and for each primer set tested. Copy numbers were adjusted to account for the 1:5 dilution (when applicable) and baseline and blank subtracted. Samples with >0.5 copies/μL of DNA extract were considered “positive.” A melting curve was used in all assays to ensure specificity of amplification. Data from the second plate reads were used for quantitation.
Microscopy.
Fluorescence and SEM microscopy were used to visualize showerhead biofilm microbes. Fluorescence microscopy entailed wiping the inner surface of a showerhead with a sterile swab, then rolling the swab into 100 μL of 1× TE on a glass slide. Slides were then heat fixed and stained with 10 μg 4,6-diamidino-2-phenylindole (DAPI; Sigma). Epiflourescence microscopy was performed on a Nikon Eclipse E600 microscope (Nikon Instruments Inc.). SEM was carried out on several biofilms in situ on the showerhead surface. Preparation for SEM entailed disassembly and fragmentation of the plastic showerhead distributor, fixation in a 2% glutaraldehyde solution in sodium cacodylate buffer for 1 h, then soaking in a 1% osmium tetroxide, 20% acetone solution for 30 min. Samples were desiccated by ethanol series dehydration (15 min in each 30% and 70%, and 45 min in 100% ethanol), affixed to microscope stubs with double-sided carbon conductive tape and colloidial silver liquid, then sputter coated with approximately 5 nm of gold/palladium using a Cressington 108Auto Sputter Coater (Cressington Scientific Instruments). Microscopy was performed on a JEOL JSM-6480 LV-SEM (JEOL).
Supplementary Material
Acknowledgments.
We thank Dr. Marcel Behr for advice on M. avium taxonomy, Thomas Giddings for his invaluable assistance with the SEM, the students of Spring 2006 MCDB 4110 at the University of Colorado at Boulder for their initial efforts on this research, Ray Grunzinger for the collection of many showerhead swab samples, and the multiple people and institutions that allowed us access to their showerheads. This work was supported by grants from the Alfred P. Sloan Foundation and the National Institute of Occupational Safety and Health to N.R.P.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU629353–EU635442 and EU697021–EU697072).
This article contains supporting information online at www.pnas.org/cgi/content/full/0908446106/DCSupplemental.
References
- 1.Zhou Y, Benson JM, Irvin C, Irshad H, Cheng YS. Particle size distribution and inhalation dose of shower water under selected operating conditions. Inhal Toxicol. 2007;19:333–342. doi: 10.1080/08958370601144241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorn J. The inflammatory response in humans after inhalation of bacterial endotoxin: A review. Inflamm Res. 2001;50:254–261. doi: 10.1007/s000110050751. [DOI] [PubMed] [Google Scholar]
- 3.Falkinham JO., 3rd Mycobacterial aerosols and respiratory disease. Emerg Infect Dis. 2003;9:763–767. doi: 10.3201/eid0907.02-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marras TK, et al. Hypersensitivity pneumonitis reaction to Mycobacterium avium in household water. Chest. 2005;127:664–671. doi: 10.1378/chest.127.2.664. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien DP, Currie BJ, Krause VL. Nontuberculous mycobacterial disease in northern Australia: A case series and review of the literature. Clin Infect Dis. 2000;31:958–967. doi: 10.1086/318136. [DOI] [PubMed] [Google Scholar]
- 6.Exner M, et al. Prevention and control of health care-associated waterborne infections in health care facilities. Am J Infect Control. 2005;33:S26–40. doi: 10.1016/j.ajic.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Bollin GE, Plouffe JF, Para MF, Hackman B. Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets. Appl Environ Microbiol. 1985;50:1128–1131. doi: 10.1128/aem.50.5.1128-1131.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alary M, Joly JR. Risk factors for contamination of domestic hot water systems by legionellae. Appl Environ Microbiol. 1991;57:2360–2367. doi: 10.1128/aem.57.8.2360-2367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedro-Botet ML, Stout JE, Yu VL. Legionnaires' disease contracted from patient homes: the coming of the third plague? Eur J Clin Microbiol Infect Dis. 2002;21:699–705. doi: 10.1007/s10096-002-0813-2. [DOI] [PubMed] [Google Scholar]
- 10.Falkinham JO, Iseman MD, Haas P, Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6:209–213. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- 11.Shin JH, et al. Prevalence of non-tuberculous mycobacteria in a hospital environment. J Hosp Infect. 2007;65:143–148. doi: 10.1016/j.jhin.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Nishiuchi Y, et al. The recovery of Mycobacterium avium-intracellulare complex (MAC) from the residential bathrooms of patients with pulmonary MAC. Clin Infect Dis. 2007;45:347–351. doi: 10.1086/519383. [DOI] [PubMed] [Google Scholar]
- 13.Jannasch HW, Jones GE. Bacterial populations in sea water as determined by different methods of enumeration. Limnol Oceanogr. 1959;4:128–139. [Google Scholar]
- 14.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 15.Stackebrandt E, Goebel BM. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 16.DeSantis TZ, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao A, Lee SM. Estimating the number of classes via sample coverage. J Am Stat Assoc. 1992;87:210–217. [Google Scholar]
- 18.Turenne CY, Wallace R, Jr, Behr MA. Mycobacterium avium in the postgenomic era. Clin Microbiol Rev. 2007;20:205–229. doi: 10.1128/CMR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth A, et al. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S–23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cianciotto NP, Eisenstein BI, Mody CH, Toews GB, Engleberg NC. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cianciotto NP, Fields BS. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cianciotto NP, Stamos JK, Kamp DW. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr Microbiol. 1995;30:247–250. doi: 10.1007/BF00293641. [DOI] [PubMed] [Google Scholar]
- 23.Helbig JH, et al. The PPIase active site of Legionella pneumophila Mip protein is involved in the infection of eukaryotic host cells. Biol Chem. 2003;384:125–137. doi: 10.1515/BC.2003.013. [DOI] [PubMed] [Google Scholar]
- 24.Swanson MS, Hammer BK. Legionella pneumophila pathogenesis: A fateful journey from amoebae to macrophages. Annu Rev Microbiol. 2000;54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- 25.Angenent LT, Kelley ST, St Amand A, Pace NR, Hernandez MT. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc Natl Acad Sci USA. 2005;102:4860–4865. doi: 10.1073/pnas.0501235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckstrom JA, Boyer DG. Aquifer-protection considerations of coalbed methane development in the San Juan basin. SPE Form Eval. 1993;8:71–79. [Google Scholar]
- 27.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steed KA, Falkinham JO., 3rd Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl Environ Microbiol. 2006;72:4007–4011. doi: 10.1128/AEM.02573-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norton CD, LeChevallier MW, Falkinham JO., 3rd Survival of Mycobacterium avium in a model distribution system. Water Res. 2004;38:1457–1466. doi: 10.1016/j.watres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Lehtola MJ, et al. Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Appl Environ Microbiol. 2007;73:2854–2859. doi: 10.1128/AEM.02916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams MM, et al. Structural analysis of biofilm formation by rapidly and slowly growing nontuberculosis mycobacteria. Appl Environ Microbiol. 2009;75:2091–2098. doi: 10.1128/AEM.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaerewijck MJ, Huys G, Palomino JC, Swings J, Portaels F. Mycobacteria in drinking water distribution systems: Ecology and significance for human health. FEMS Microbiol Rev. 2005;29:911–934. doi: 10.1016/j.femsre.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Le Dantec C, et al. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl Environ Microbiol. 2002;68:5318–5325. doi: 10.1128/AEM.68.11.5318-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilborn ED, et al. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl Environ Microbiol. 2006;72:5864–5869. doi: 10.1128/AEM.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falkinham JO. The changing pattern of nontuberculous mycobacterial disease. Can J Infect Dis. 2003;14:281–286. doi: 10.1155/2003/323058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szewzyk U, Szewzyk R, Manz W, Schleifer KH. Microbiological safety of drinking water. Annu Rev Microbiol. 2000;54:81–127. doi: 10.1146/annurev.micro.54.1.81. [DOI] [PubMed] [Google Scholar]
- 37.Hilborn ED, et al. Molecular comparison of Mycobacterium avium isolates from clinical and environmental sources. Appl Environ Microbiol. 2008;74:4966–4968. doi: 10.1128/AEM.02900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falkinham JO, 3rd, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in drinking water distribution systems. Appl Environ Microbiol. 2001;67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Field SK, Fisher D, Cowie RL. Mycobacterium avium complex pulmonary disease in patients without HIV infection. Chest. 2004;126:566–581. doi: 10.1378/chest.126.2.566. [DOI] [PubMed] [Google Scholar]
- 40.Heifets L. Mycobacterial infections caused by nontuberculosis mycobacteria. Semin Respir Crit Care Med. 2004;25:283–295. doi: 10.1055/s-2004-829501. [DOI] [PubMed] [Google Scholar]
- 41.Suffys P, et al. Detection of mixed infections with Mycobacterium lentiflavum and Mycobacterium avium by molecular genotyping methods. J Med Microbiol. 2006;55:127–131. doi: 10.1099/jmm.0.46218-0. [DOI] [PubMed] [Google Scholar]
- 42.Khan K, Wang J, Marras TK. Nontuberculous mycobacterial sensitization in the United States: National trends over three decades. Am J Respir Crit Care Med. 2007;176:306–313. doi: 10.1164/rccm.200702-201OC. [DOI] [PubMed] [Google Scholar]
- 43.Dalby AB, Frank DN, St Amand AL, Bendele AM, Pace NR. Culture-independent analysis of indomethacin-induced alterations in the rat gastrointestinal microbiota. Appl Environ Microbiol. 2006;72:6707–6715. doi: 10.1128/AEM.00378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane DJ. 16S/23S rRNA sequencing. In: Stackenbrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester, England: John Wiley & Sons; 1991. pp. 115–176. [Google Scholar]
- 45.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Frank DN. XplorSeq: A software environment for integrated management and phylogenetic analysis of metagenomic sequence data. BMC Bioinformatics. 2008;9:420. doi: 10.1186/1471-2105-9-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto H, et al. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl Environ Microbiol. 1997;63:2489–2494. doi: 10.1128/aem.63.7.2489-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang B, et al. Specific detection of viable Legionella cells by combined use of photoactivated ethidium monoazide and PCR/real-time PCR. Appl Environ Microbiol. 2008;75:147–153. doi: 10.1128/AEM.00604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wellinghausen N, Frost C, Marre R. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl Environ Microbiol. 2001;67:3985–3993. doi: 10.1128/AEM.67.9.3985-3993.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.