Abstract
Mammalian mitochondrial (mt) tRNAs, which are required for mitochondrial protein synthesis, are all encoded in the mitochondrial genome, while mt aminoacyl-tRNA synthetases (aaRSs) are encoded in the nuclear genome. However, no mitochondrial homolog of glutaminyl-tRNA synthetase (GlnRS) has been identified in mammalian genomes, implying that Gln-tRNAGln is synthesized via an indirect pathway in the mammalian mitochondria. We demonstrate here that human mt glutamyl-tRNA synthetase (mtGluRS) efficiently misaminoacylates mt tRNAGln to form Glu-tRNAGln. In addition, we have identified a human homolog of the Glu-tRNAGln amidotransferase, the hGatCAB heterotrimer. When any of the hGatCAB subunits were inactivated by siRNA-mediated knock down in human cells, the Glu-charged form of tRNAGln accumulated and defects in respiration could be observed. We successfully reconstituted in vitro Gln-tRNAGln formation catalyzed by the recombinant mtGluRS and hGatCAB. The misaminoacylated form of tRNAGln has a weak binding affinity to the mt elongation factor Tu (mtEF-Tu), indicating that the misaminoacylated form of tRNAGln is rejected from the translational apparatus to maintain the accuracy of mitochondrial protein synthesis.
Keywords: EF-Tu, Glu-tRNAGln amidotransferase, glutamyl-tRNA synthetase, mitochondrial tRNA
Translational accuracy is required to properly decipher the genetic code to form proteins. The fidelity of protein synthesis largely depends on the formation of correct aminoacyl-tRNAs (aa-tRNAs) by the corresponding aminoacyl-tRNA synthetases (aaRSs). In the classical model, each species of aaRS strictly discriminates one amino acid among the 20 canonical amino acids, as well as its cognate tRNA isoacceptor, from non-cognate tRNA species. However, recent genomic analyses have revealed that the full complement of 20 aaRSs is used only in eukaryotic cytoplasm and a minority of bacteria, whereas a majority of the bacterial and all of the known archaeal genomes do not encode a glutaminyl-tRNA synthetase (GlnRS) (1). In addition, an asparaginyl-tRNA synthetase (AsnRS) is not encoded in most bacterial and archaeal genomes (1). In these organisms, the amide-amino acid (Gln or Asn) on its cognate tRNA is synthesized via an indirect pathway (2). In most bacteria and archaea, in which GlnRS is absent, tRNAGln is first misaminoacylated with Glu catalyzed by a non-discriminating GluRS (ND-GluRS) that can aminoacylate both tRNAGlu and tRNAGln. The Glu moiety on the Glu-tRNAGln is then transamidated by a glutamyl-tRNAGln amidotransferase (Glu-AdT) in the presence of ATP and Gln as an amide donor to synthesize Gln-tRNAGln. Similarly, in the case of Asn-tRNAAsn formation in organisms lacking AsnRS, Asn-tRNAAsn is synthesized by a non-discriminating aspartyl-tRNA synthetase and an aspartyl-tRNAAsn amidotransferase (Asp-AdT).
Two types of tRNA-dependent amidotransferase exist, the heterotrimeric GatCAB (3) and heterodimeric GatDE (1). Bacterial GatCAB functions as a Glu-AdT or an Asp-AdT in a species-specific manner. In some bacteria lacking both AsnRS and GlnRS, GatCAB has a dual activity as both a Glu-AdT and an Asp-AdT (2). As each subunit of GatCAB is encoded only in archaeal genomes lacking a gene for AsnRS, archaeal GatCAB seems to function as an Asp-AdT. GatDE is only found in archaea, and serves as a Glu-AdT (2).
GatB and GatE are paralogous subunits having a tRNA-dependent kinase activity, while GatA and GatD are the glutaminase subunits of the AdT. For transamidation of Glu on the tRNAs, Glu-AdT catalyzes three sequential reactions. First, the GatB (or GatE) subunit uses ATP to phosphorylate the Glu moiety on the Glu-tRNAGln, forming a γ-phosphoryl-Glu-tRNAGln as an activated intermediate. Second, the GatA (or GatD) subunit catalyzes the liberation of ammonia from Gln as an amide donor. Finally, Glu-AdT amidates the activated intermediate using the liberated ammonia to form Gln-tRNAGln.
Mitochondria have own translational apparatus that synthesizes proteins encoded in mitochondrial (mt) genome. While with the exception of some protozoans the required rRNA and a full set of tRNAs are encoded in the mtDNA (4), all other factors in the mt translational apparatus are encoded in nuclear genome, translated in the cytoplasm, and then imported into the mitochondria. However, according to eukaryotic genomic analyses, to date, it is known that mitochondrial ortholog of GlnRS is not encoded in nuclear genome (4). This fact is consistent with the observation that no GlnRS activity could be detected in mitochondria purified from some eukaryotic species (5), implying that mitochondrial Gln-tRNAGln formation proceeds via the indirect pathway. In Saccharomyces cerevisiae, it is known that gatA and gatB orthologs encoded in nuclear genome are involved in mitochondrial function (6–8), although gatC ortholog was not identified. These observations strongly indicated that Gln-tRNAGln is formed via the indirect pathway in yeast mitochondria. However, it was reported that yeast mtGluRS is not an ND-GluRS that does not generate Glu-tRNAGln, obligatory intermediate of the indirect pathway (9, 10). These contradictory facts complicated the issue of Gln-tRNAGln formation in yeast mitochondria. Recent study has reported that cytoplasmic GluRS is imported into yeast mitochondria, and acts as an ND-GluRS that glutamylates mt tRNAGln to synthesize Glu-tRNAGln (9). In addition, they have found a new type heterotrimeric amidotransferase GatFAB that converts Glu-tRNAGln into Gln-tRNAGln (9). GatF is a fungi-specific subunit of Glu-AdT. In higher plants, dual targeted GluRS and GatCAB are responsible for both mitochondrial and chloroplastic Gln-tRNAGln formation in the respective organelles (11). Thus, mitochondrial Gln-tRNAGln formation is likely species-specific, which might make it difficult to predict what pathway and factors generate mitochondrial Gln-tRNAGln for mitochondrial translation in a given eukaryote.
Mammalian mitochondria have a translational apparatus that synthesizes 13 proteins encoded in the mitochondrial (mt) genome. While two rRNAs and a full set of tRNAs are encoded in the mtDNA, all other translational factors including aaRSs are encoded in the nuclear genome. To our knowledge, 11 species of mammalian mt aaRSs have been characterized (12). Among them, only two aaRSs, mtLysRS and mtGlyRS, share the same genes as the cytoplasmic LysRS and GlyRS, respectively. The other aaRSs which are mitochondria-specific have been bioinformatically predicted as bacterial homologs in mammalian genomes (12). However, no mitochondrial homolog of GlnRS has been predicted in a mammalian genome, implying that Gln-tRNAGln is synthesized via the indirect pathway in mammalian mitochondria. This speculation is supported by the observation that mitochondrial extracts from mouse liver do not exhibit GlnRS activity (5). It was recently reported that cytoplasmic tRNAGln is imported into human mitochondria (13), indicating an involvement of this tRNA in the biogenesis of Gln-tRNAGln in human mitochondria. However, fate of the imported tRNA and the enzymatic pathway for generating mitochondrial Gln-tRNAGln were unknown. In this study, we aimed to clarify the pathway and mechanism of Gln-tRNAGln formation in human mitochondria by characterizing the mtGluRS and a human homolog of GatCAB in vitro as well as in vivo.
Results
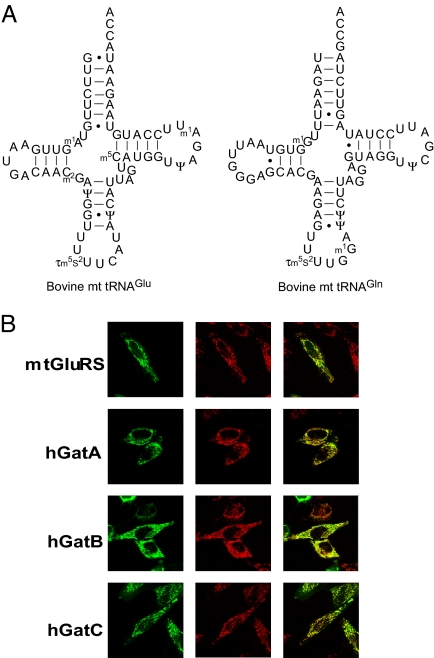
Post-Transcriptional Modifications of Mammalian Mitochondrial tRNAGlu and tRNAGln Isolated from Bovine Liver.
In E. coli, the wobble modification of tRNAGlu is essential for glutamylation by GluRS (14). Therefore, to study Gln-tRNAGln formation in mammalian mitochondria, we needed to examine an aminoacylation of native mt tRNAs which are fully modified, instead of using in vitro transcribed tRNAs with no modification. We isolated mt tRNAs for Glu and Gln from bovine liver using chaplet column chromatography (15). The primary structures, including the posttranscriptional modifications, of the isolated tRNAs were determined by mass spectrometric analysis (16, 17) (Fig. S1 and Tables S1 and S2). As shown in Fig. 1A, both tRNAs contain modified bases at several positions. At the wobble positions, the tRNAs commonly contain 5-taurinomethyl-2-thiouridine (τm5s2U), which is a mammalian mitochondria-specific uridine derivative we previously identified at the wobble position in mt tRNALys (18). In addition, we also isolated mt tRNAs for Glu and Gln from human placenta (19), and confirmed the presence of τm5s2U at the wobble positions.
Fig. 1.
Primary structures of mitochondrial tRNAGlu and tRNAGln, and subcellular localization of mtGluRS and hGatCAB. (A) Nucleotide sequences of bovine mt tRNAGlu (Left) and mt tRNAGln (Right). Modified nucleosides: τm5s2U, 5-taurinomethyl-2-thiouridine; Ψ, pseudouridine; m1A, 1-methyladenosine; m2G, N2-methylguanosine; m1G, 1-methylguanine; m5C, 5-methylcytidine. (B) Subcellular localization of mtGluRS and three subunits of hGatCAB. Confocal fluorescence microscopic image of HeLa cells transfected with pEGFP-mtGluRS (top row), pEGFP-hGatA (second row), pEGFP-hGatB (third row) and pEGFP-hGatC (bottom row). Left panels show EGFP fluorescence. Middle images show mitochondria stained by MitoTracker Red. The right column shows the merged images.
Human Mitochondrial GluRS Is a Non-Discriminating GluRS.
Since most mitochondrial proteins are homologous to their bacterial counterparts, we searched for a gene having sequence similarity to bacterial GluRS in the non-redundant NCBI database, and retrieved a candidate gene for the human ortholog of the bacterial GluRS (protein ID = NP_001077083). This gene had been previously predicted to be a GluRS by other group (12). Sequence alignment of the putative human mtGluRS with other putative mitochondrial and bacterial GluRS proteins (Fig. S2) revealed that the mtGluRS has an extended N-terminal sequence, which is predicted to be a mitochondria-targeting signal with a high probability value (0.994) by MitoProt (http://ihg.gsf.de/ihg/mitoprot.html) (20), strongly suggesting that the putative mtGluRS is indeed localized in mitochondria. To examine the subcellular localization of the mtGluRS, full length mtGluRS with EGFP fused to its C terminus was transiently expressed in HeLa cells. As observed by confocal microscopy (Fig. 1B, Top), mtGluRS-EGFP was clearly localized to mitochondria.
As we were unable to identify a candidate gene for a human ortholog of the bacterial GlnRS, consistent with previous reports (12), we hypothesized that Gln-tRNAGln is synthesized via the indirect pathway in human mitochondria. Thus, mtGluRS may be a non-discriminating GluRS (ND-GluRS), which can aminoacylate both mt tRNAGlu and mt tRNAGln as natural substrates.
We therefore examined the aminoacylation ability of mtGluRS. The cDNA of mtGluRS lacking the N-terminal mitochondria-targeting sequence was cloned into an expression vector. The N-terminal His-tagged mtGluRS was recombinantly expressed in E. coli, and purified to homogeneity. To investigate whether the recombinant mtGluRS is an ND-GluRS, we carried out glutamylation of mt tRNAs for Glu and Gln isolated from bovine liver, as well as an in vitro transcribed mt tRNAGlu as a control. The mtGluRS efficiently glutamylated both mt tRNAGln and mt tRNAGlu (Table S3), demonstrating that mtGluRS is an ND-GluRS. Since the in vitro transcribed mt tRNAGlu was not glutamylated, it was found that posttranscriptional modifications in mt tRNAGlu are necessary for glutamylation. As reported for bacterial GluRS, mtGluRS presumably recognizes the wobble modifications of the mt tRNAs for Glu and Gln. We also confirmed that human mt tRNAs for Glu and Gln isolated from placenta were efficiently glutamylated by mtGluRS, and our observations are, therefore, not due to the heterologous combination of human mtGluRS and bovine mt tRNAs.
The kinetic parameters of glutamylation of the bovine mt tRNAs for Glu and Gln by the recombinant mtGluRS were determined (Table S3). Although the Km value (1.7 μM) for the mt tRNAGln was ≈6 fold higher than that (0.27 μM) for mt tRNAGlu, the kcat/Km of mt tRNAGln was only 2-fold lower than that of mt tRNAGlu. The ND-GluRS from Thermosynechococcus elongatus exhibits a Km value for glutamylation of tRNAGln that is approximately 5-fold higher than that of tRNAGlu (21). Therefore, the higher Km value for glutamylation of tRNAGln may be a common feature of ND-GluRS.
Identification and Characterization of Human Glu-tRNAGln Amidotransferase.
We next searched for human orthologs of bacterial gatA, gatB, and gatC from the human non-redundant NCBI database, and retrieved candidate genes for each: NM_018292 (NP_060762) as a gatA-like gene, NM_004564 (NP_004555) as a gatB-like gene, and NM_176818 (NP_789788) as a gatC-like gene (named hGatA, hGatB and hGatC, respectively). The sequence alignment of each of the human homologs with bacterial and mitochondrial counterparts revealed that hGatB and hGatC possess N-terminal mitochondria-targeting sequences, while hGatA does not (Fig. S3). The MitoProt values for mitochondria-targeting of hGatA (0.18), hGatB (0.82), and hGatC (0.16) are not indicative as to whether all of the homologs are actually localized in mitochondria and form a heterotrimeric amidotransferase. To determine the subcellular localization of each subunit, the full-length cDNA of each protein fused to EGFP at its C terminus was transiently expressed in HeLa cells. As shown in Fig. 1B (Lower), all three proteins are clearly localized in mitochondria.
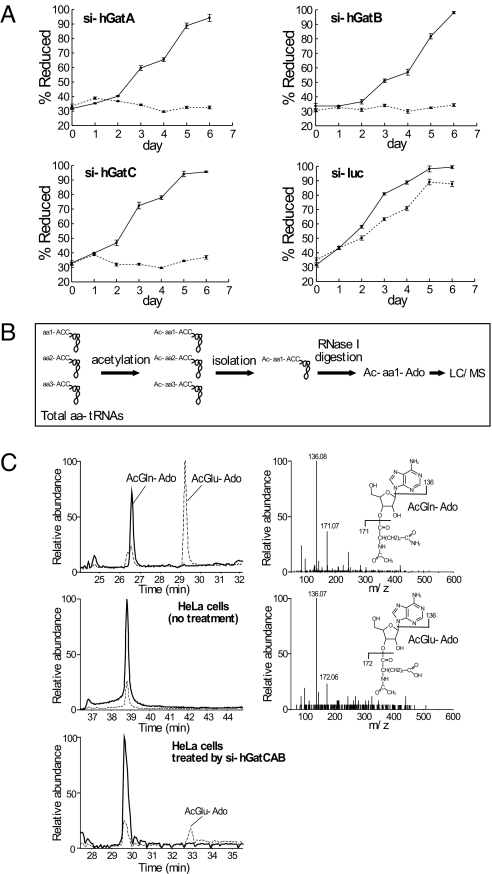
We then performed siRNA-mediated knockdowns of each subunit of the hGatCAB in HeLa cells and analyzed the growth properties of the siRNA-treated cells in glucose- or galactose-containing medium. Cells with defective mitochondrial function cannot proliferate in a non-fermentable growth medium, the carbon source of which is only consisted of galactose (22). The knockdown efficiency for each subunit was estimated by qRT-PCR and demonstrated that each mRNA in the siRNA-treated cells was decreased to less than 15% of the level in control cells. As shown in Fig. 2A, the growth of HeLa cells treated with siRNAs targeting each of the three subunits and cultured in galactose medium was significantly decreased as compared to siRNA-treated cells cultured in glucose medium. As a negative control, cells treated with a siRNA targeting luciferase were able to grow in galactose medium. These results clearly demonstrate that all of the subunits are required for mitochondrial function.
Fig. 2.
Growth of HeLa cells treated with siRNAs targeting hGatCAB and direct identification Glu-tRNAGln in the cells. (A) Growth curves for HeLa cells treated with siRNAs for hGatA (Upper Left), hGatB (Upper Right), hGatC (Lower Left), or luciferase (control, Lower Right). The siRNA-treated cells were cultured in glucose (solid line) or galactose (dotted line) containing medium. Each plot is the average of three independent cultures (bars, ± SD). (B) A schematic of the procedure for direct analysis of amino acids attached to an aa-tRNA is shown. Total aa-tRNAs obtained from the cells are acetylated and individual acetyl-aa-tRNAs are affinity-purified using the solid-phase DNA probe method, and are then digested by RNase I. The acetyl-aminoacyl-adenosine (Ac-aa-Ado) fragments originating from the 3′-terminus were analyzed by LC/MS. (C) Mass chromatograms detecting Ac-Gln-Ado (m/z 438, solid line) and Ac-Glu-Ado (m/z 439, dotted line); authentic Ac-Gln-Ado and Ac-Glu-Ado (Top Left) separately prepared from mouse liver, HeLa cells (no treatment, Middle Left) and HeLa cells treated by siRNAs targeting for all subunits of hGatCAB (Bottom Left). The small peak at m/z 439 (dotted line) that overlaps with the Ac-Gln-Ado (m/z 438) peak was assigned to the isotopic ion of Ac-Gln-Ado. Collision-induced dissociation (CID) spectra of Ac-Gln-Ado (Upper Right) and Ac-Glu-Ado (Lower Right) are shown. Product ions for the adenine base (BH2+, m/z 136), AcGln moiety (m/z 171), and AcGlu moiety (m/z 172) were clearly detected.
To obtain direct evidence that hGatCAB is a bona-fide Glu-AdT that can generate Gln-tRNAGln in mitochondria, we devised a method to directly analyze amino acids attached to individual tRNAs isolated from cells (23). The method is outlined in Fig. 2B. If the indirect pathway is involved in Gln-tRNAGln formation in human mitochondria, once the activity of hGatCAB is down-regulated by siRNAs targeting one of the subunits, Glu-tRNAGln (the misaminoacylated form) should accumulate in the cell. HeLa cells were harvested 72 h after transfection with three siRNAs targeting each of the hGatCAB subunits, and total RNA, including aa-tRNAs, was immediately extracted under acidic conditions at low temperature. To stabilize the aminoacyl-bond of each aa-tRNA, the α-amino group of the aminoacyl moiety was acetylated by acetic anhydride as described previously (23). The mt tRNAGln was isolated using a solid-phase DNA probe method (15, 23), and then completely digested by RNase I, yielding acetyl-aminoacyl-adenosines (Ac-aa-Ado) from the 3′-terminus of the tRNA. The Ac-aa-Ado species were determined by LC/MS analysis. Known control samples of Ac-Glu-Ado (m/z 439) and Ac-Gln-Ado (m/z 438) were clearly detected as singly-charged positive ions in separated peaks of a mass chromatogram (Fig. 2C). In mt tRNAGln isolated from untreated HeLa cells, only Ac-Gln-Ado was detected. In contrast, when hGatCAB was knocked down, Ac-Glu-Ado was clearly detected in the isolated mt tRNAGln samples (Fig. 2C). The chemical structure of Ac-Glu-Ado was confirmed by assignment of product ions produced by collision-induced dissociation (Fig. 2C, Right). These results provide the direct evidence that Gln-tRNAGln is generated from Glu-tRNAGln mediated by hGatCAB in human mitochondria.
In Vitro Reconstitution of Amidotransfer Reaction by hGatCAB.
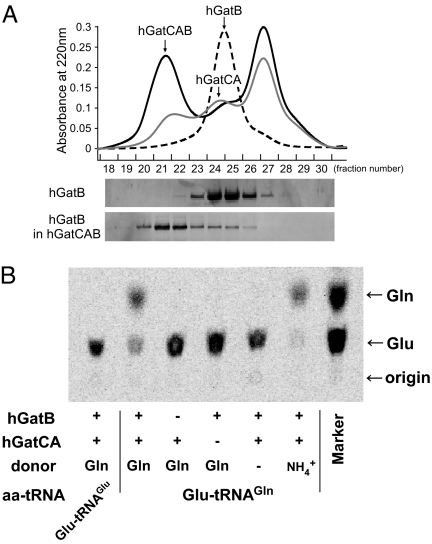
To reconstitute Gln-tRNAGln formation in vitro by hGatCAB, each subunit was recombinantly expressed in E. coli. According to the crystal structure of bacterial GatCAB (24), GatC interacts extensively with the hydrophobic core of GatA. Therefore, hGatC was coexpressed with hGatA to obtain an hGatCA complex in soluble form. To test the molecular interaction between recombinant hGatCA and hGatB, mixtures were analyzed by gel filtration chromatography (Fig. 3A). Each fraction was analyzed by SDS/PAGE and the His-tag of hGatB was detected by Western blotting (Fig. 3A). When hGatB alone was subjected to gel filtration chromatography, hGatB eluted at peak fractions 24 and 25. When hGatB was mixed with hGatCA, the hGatB signal was detected in higher molecular weight fractions (fractions 21 and 22). According to the retention times of molecular size markers, a heterotrimeric hGatCAB of approximately 132 kDa was formed.
Fig. 3.
In vitro reconstitution of Gln-tRNAGln formation by hGatCAB. (A) Gel filtration chromatography (Superdex 200) of hGatCA (gray line), hGatB (dotted line), and a mixture of hGatCA and hGatB (black line) detected by UV absorption at 220 nm. Elution profiles of hGatB alone (Upper) and hGatB in the hGatCAB complex (Lower) were analyzed by Western blotting using an anti-His-tag antibody. (B) In vitro reconstitution of Gln-tRNAGln formation by the recombinant hGatCAB. Phosphor-image of the TLC analysis of [14C]-labeled Gln and Glu deacylated from aa-tRNAs in transamidation experiments. The assays were performed in the presence (+) or absence (−) of hGatB or hGatCA. Gln or NH4+ was used as an amide donor. [14C]-labeled Glu-tRNAGlu or Glu-tRNAGln was used as a substrate, as indicated below the panel. Positions of Gln and Glu on TLC were determined using [14C]Gln and [14C]Glu as markers (right lane).
We next examined the in vitro amidotransferase activity of the reconstituted hGatCAB (Fig. 3B). When Glu-tRNAGln was used as a substrate, Gln-tRNAGln was clearly formed in the presence of hGatCAB and an amide donor, Gln or NH4+. This reaction did not occur when Glu-tRNAGlu was used as a substrate, in the absence of hGatB or hGatCA, or in the absence of an amide donor (Gln or NH4+). We were able to successfully reconstitute Gln-tRNAGln formation in vitro using the recombinant mtGluRS and hGatCAB.
Exclusion of the Mischarged Intermediate of tRNAGln by Mitochondrial EF-Tu Surveillance.
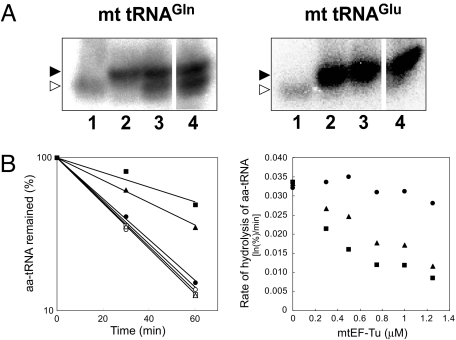
The indirect pathway involves a mischarged intermediate of tRNAGln. If Glu-tRNAGln were to participate in translation, incorporation of the incorrect amino acid would reduce the fidelity of mitochondrial protein synthesis. A previous report has demonstrated that chloroplast EF-Tu does not recognize Glu-tRNAGln, generated as a mischarged intermediate in the biogenesis of Gln-tRNAGln, in the organelle (25). Thus, we hypothesized that a similar mechanism may maintain the fidelity of translation in human mitochondria. If mtEF-Tu only weakly binds Glu-tRNAGln, Glu-tRNAGln should be rapidly deacylated in mitochondria. To examine this possibility, we separated tRNAs by acid-urea polyacrylamide gel electrophoresis and examined the state of the aa-tRNAs in human mitochondria by northern blotting (26). In the control cells treated by siRNA targeting luciferase, both mt tRNAs for Gln and Glu were fully aminoacylated (Fig. 4A, lane 2). When hGatA was knocked down by siRNA, the deacylated form of mt tRNAGln appeared (Fig. 4A, lane 3), while no change in the status of aminoacylation of mt tRNAGlu was detected. Moreover, when the three subunits of hGatCAB were inactivated simultaneously, the deacylated form of mt tRNAGln increased, while mt tRNAGlu was still fully aminoacylated (Fig. 4A, lane 4). These results suggest that Glu-tRNAGln generated by siRNA-mediated knock down of hGatCAB tends to be rapidly deacylated in mitochondria, probably due to the low affinity of Glu-tRNAGln for mtEF-Tu.
Fig. 4.
Status of Gln-tRNAGln in cells treated with siRNAs targeting hGatCAB and measuring the binding affinity of mtEF-Tu with Glu-tRNAGln. (A) Total aa-tRNAs were separated by acid-urea PAGE, and mt tRNAGln (Left) and mt tRNAGlu (Right) from HeLa cells treated with the siRNAs targeting luciferase (lane 2), hGatA (lane 3) or all three subunits of hGatCAB (lane 4) were detected by northern blotting. The lane 1 signal in each panel represents the deacylated tRNAs prepared by alkaline-treatment of aa-tRNAs. Positions of the aa-tRNA and deacyl-tRNA are shown by the closed and open arrowheads, respectively. (B) (Left) Time course experiments of the hydrolysis protection assay for Glu-tRNAGlu (square), Glu-tRNAGln (circle), and Gln-tRNAGln (triangle) in the presence (closed symbols) or absence (open symbols) of bovine mtEF-Tu. The remaining aa-tRNAs were measured and plotted. (Right) The rates of hydrolysis of aa-tRNAs measured in the presence of different concentrations of mtEF-Tu.
To investigate whether mtEF-Tu plays a role in the exclusion of Glu-tRNAGln from mitochondrial protein synthesis, we conducted hydrolysis protection assays to estimate the binding affinity of mtEF-Tu for Glu-tRNAGln. The aminoacyl moiety of an aa-tRNA is spontaneously hydrolyzed with a characteristic half-life. In the presence of EF-Tu/GTP, the tRNA is protected from deacylation by the specific binding of EF-Tu/GTP to the aminoacyl moiety of the aa-tRNA. We compared the rate of deacylation for Glu-tRNAGln, Gln-tRNAGln, and Glu-tRNAGlu in the absence or presence of mtEF-Tu/GTP (Fig. 4B). Without mtEF-Tu/GTP, the three aa-tRNAs were rapidly deacylated at similar rates with a half-life of ≈20 min. In the presence of mtEF-Tu/GTP, the deacylation rate of both Gln-tRNAGln and Glu-tRNAGlu clearly decreased, indicating that mtEF-Tu/GTP efficiently binds to aa-tRNAs charged with their cognate amino acid and protects them from deacylation. In contrast, the rate of deacylation of the misaminoacylated Glu-tRNAGln was unchanged in the presence of mtEF-Tu/GTP (Fig. 4B, Left). To confirm these results, we plotted the rate of hydrolysis by titrating mtEF-Tu. The rate of deacylation for both cognate aa-tRNAs decreased with increasing concentrations of mtEF-Tu (Fig. 4B, Right). In contrast, the rapid deacylation of Glu-tRNAGln was maintained even in the presence of high concentrations of mtEF-Tu (Fig. 4B, Right). These results demonstrate that Glu-tRNAGln has a low binding affinity for mtEF-Tu, suggesting a mechanism by which mischarged aa-tRNAs are rejected by the mitochondrial translation system. After Glu-tRNAGln is converted to Gln-tRNAGln by hGatCAB, it recovers a strong binding affinity to mtEF-Tu and is available for mitochondrial protein synthesis.
Cytoplasmic tRNAGln Is a Potential Substrate for mtGluRS and hGatCAB.
It was reported that cytoplasmic (cyto) tRNAGln is imported into human mitochondria (13), indicating cyto tRNAGln might be charged with Gln, and participate in mitochondrial translation, although we have revealed in this study that mt tRNAGln is a major substrate in the indirect pathway in human mitochondria. To examine cyto tRNAGln to be a substrate for mtGluRS and hGatCAB, we isolated cyto tRNAGln from human placenta using the chaplet column chromatography (15), and carried out glutaminylation of cyto tRNAGln by recombinant mtGluRS and hGatCAB. In fact, cyto tRNAGln was slightly glutamylated by mtGluRS (Fig. S4A) and converted to Gln-cyto tRNAGln (Fig. S4B), but with low activity compared with mt tRNAs for Glu and Gln, implying that the imported cyto tRNAGln can be slightly glutaminylated in human mitochondria.
Discussion
Approximately two decades ago, Söll's group reported that no GlnRS activity was detected in the S100 fraction of mouse liver mitochondria (5), strongly suggesting that the indirect pathway consisted of ND-GluRS and Glu-AdT is involved in Gln-tRNAGln formation in mammalian mitochondria. We have shown here that the recombinant mtGluRS was indeed an ND-GluRS that efficiently glutamylated mt tRNAs for both Glu and Gln. In the crystal structure of the GluRS-tRNAGlu complex (27), Arg-358 in discriminating type GluRS (D-GluRS) from T. thermophilus specifically recognizes C36 of tRNAGlu. The D-GluRS Arg-358 is supposed to sterically interfere with the G36 position in the tRNAGln, resulting in a lack of recognition of tRNAGln. Arg-358 is considered to be a signature residue of D-GluRS. In the ND-GluRS of T. elongatus (21), this position is replaced by Gly so that it accommodates the G36 of tRNAGln, resulting in recognition of the tRNAs for both Glu and Gln. The residue corresponding to Arg-358 in mammalian mtGluRS is His (Fig. S2), which is expected to recognize both C36 and G36 of mt tRNAsGlu/Gln as an ND-GluRS.
Homologs of bacterial Glu-AdT subunits are found in the nuclear genomes of eukaryotes. In Saccharomyces cerevisiae, pet112 encodes a mitochondrial protein that is homologous to gatB (8). Pet112 is essential for mitochondrial translation, and loss of Pet112 can be complemented by the Bacillus gatB gene (7). YMR293c/HER2 is a gatA homolog of S. cerevisiae (28), and its gene product is localized in mitochondria (29). A null mutant of YMR293c exhibited abnormal mitochondrial morphology (6) and respiratory defects (28). Although an apparent homolog of the bacterial gatC is not encoded in the S. cerevisiae genome, a fungi-specific subunit gatF was recently discovered, and a new heterotrimeric GatFAB has been found as Glu-AdT in yeast mitochondria (9). However, yeast mtGluRS is a D-GluRS and unable to aminoacylate mt tRNAGln in vitro (9, 10). This discrepancy was solved by the finding that cyto GluRS is imported in mitochondria and glutamylates mt tRNAGln, which is then subjected to Gln-tRNAGln formation by GatFAB (9). In plants, mitochondrial Gln-tRNAGln formation is achieved by the indirect pathway mediated by ND-GluRS and a GatCAB-type Glu-AdT, and both of these enzymes are also imported into chloroplasts (11). Therefore, the dual-targeted ND-GluRS and Glu-AdT ensure both mitochondrial and chloroplastic Gln-tRNAGln formation in plants. In Drosophila, the mutant benedict (bene) was shown to disrupt the Drosophila homolog of the gatA gene (30). Null mutants of the gatA homolog grew slowly and never achieved wild-type size, and gatA null larvae died before pupariation (31). Thus, gatA is required for mitotic and endoreplicating tissues in the larval stages of Drosophila. Here, we have identified the human homologs of the bacterial gatA, gatB, and gatC. These homologs were localized in the mitochondria and are required for respiratory function. In addition, we directly observed Glu-tRNAGln as a misaminoacylated form of Gln-tRNAGln in cells in which hGatCAB was inactivated by RNAi. These results demonstrate that Gln-tRNAGln formation in human mitochondria is mediated by ND-GluRS and hGatCAB, which acts as a Glu-AdT. We successfully reconstituted a heterotrimeric complex of hGatCAB and demonstrated its transamidation activity in vitro.
It was reported that mammalian cyto tRNAGln is imported into mitochondria (13). To investigate the fate of the imported cyto tRNAGln, we examined cyto tRNAGln to be converted to Gln-tRNAGln in vitro using the recombinant mtGluRS and hGatCAB. It was shown that cyto tRNAGln could be charged with Glu and converted to Gln-tRNAGln, although its activity is not as efficiently as that of mt tRNAGln. Thus, the imported cyto tRNAGln potentially might have a supportive role to be used in mitochondrial translation. Use of the cyto tRNA in mitochondria may be required for adaptation of the cell to some environmental stresses (32). Further studies are needed to clarify this issue.
According to biochemical and structural studies (24), the bacterial GatCAB strictly recognizes the first U1–72A base pair and a specific number of bases in the D-loop of tRNAGln as identity elements. The first U1–72A base pair is conserved in mt tRNAGln (Fig. 1A), while the corresponding base pair of mt tRNAGlu is G1–72U, indicating that the first base pair also acts as a positive determinant recognized by hGatCAB. According to the crystal structure of the archaeal GatDE complexed with tRNAGln (33), the tail domain of GatE closely contacts the D-loop/T-loop assembly region, in particular the G18-C56 base pair, which indicates that the interaction between the D- and T-loop is critical for the correct and efficient recognition of tRNAGln by Glu-AdT. In most tRNAs from the three domains of life, G18G19 in the D-loop and T54Ψ55C56 in the T-loop are well conserved (34, 35). In contrast, most mammalian mt tRNAs have variations in the numbers of bases with no consensus sequences in either the D- or T-loop (36). In fact, mammalian mt tRNAs frequently lack the conserved D- and T-loop interactions (37). However, unlike other mt tRNAs, mammalian mt tRNAGln species commonly have the canonical D- and T-loops, containing a G18G19 D-loop and U54U55C56 T-loop sequence (Fig. S5) (38). Although the complex structure of GatCAB-tRNAGln is not available, considering that GatCAB strictly recognizes the number of bases in the D-loop of tRNAGln (24), we suggest that hGatCAB recognizes the canonical D-loop/T-loop interaction of mt tRNAGln. Although there has been a high mutation pressure in the D- and T-loops of mt tRNAs during the evolution of the mitochondrial translation system, the canonical D-loop/T-loop sequence of the mt tRNAGln may be preserved since it functions as an identity element recognized by hGatCAB.
In 1994, Sprinzl and coworkers reported that chloroplast EF-Tu did not efficiently bind Glu-tRNAGln (25). This was the first reported instance of translational fidelity monitored by EF-Tu. Similarly, we demonstrated here that Glu-tRNAGln was not recognized by mtEF-Tu and, after conversion to Gln-tRNAGln, mtEF-Tu binding affinity was recovered. These observations suggest that the rejection of misaminoacylated tRNAs appears to be a common feature of EF-Tu. Molecular recognition of the amino acid moiety of an aa-tRNA by EF-Tu has been studied by Uhlenbeck and coworkers (39, 40). tRNAGlu binds more tightly to EF-Tu than tRNAGln does. In addition, EF-Tu exhibits the strongest binding affinity to the Gln moiety among 20 species of aminoacyl moieties. Our data, here, suggest that mtEF-Tu may have a similar preference for the amino acid moieties attached to their cognate tRNAs.
Materials and Methods
Identification of the Amino Acid Attached to mt tRNAGln in the Cell.
The isolation of individual species of aa-tRNA and analysis of the attached amino acids were conducted according to the method of Suzuki et al. (23). Briefly, HeLa cells (7.5 × 105 cells) were cultured and transfected with a mixture of three siRNAs targeting all subunits of hGatCAB (final concentration, 3.3 nM for each). At 72 h post-transfection, total RNA was extracted from the cells, as described in SI Text. The amino groups of the amino acid moieties of the aa-tRNAs in the total RNA fraction were acetylated with acetic anhydride to stabilize the aminoacyl bond (23). The acetylaminoacyl-tRNAs were then isolated from the total RNA by a solid-phase DNA probe method using a 3′-biotinylated oligonucleotide with a sequence identical to that used for northern blotting, which was performed by immobilizing the oligonucleotide on streptavidin-agarose (GE Healthcare). For further purification, the isolated aa-tRNAs were electrophoresed on a 12% polyacrylamide gel containing 7 M urea. The purified acetylaminoacyl-tRNA was digested with RNase I in 50 mM NaOAc at 37 °C for 1 h and subjected to an LC/MS analysis as described (16, 17). Other methods are decribed in SI Text.
Supplementary Material
Acknowledgments.
We thank the Suzuki laboratory members, especially to Drs. Yohei Kirino and Narumi Shigi-Hino, for technical advice and many fruitful discussions. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (to T.S.); the Global COE Program for Chemistry Innovation (to A.N.); a JSPS Fellowship for Japanese Junior Scientists (to Takeo S.); and a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T. S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907602106/DCSupplemental.
References
- 1.Tumbula DL, Becker HD, Chang WZ, Soll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Soll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curnow AW, et al. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frechin M, Duchene AM, Becker HD. Translating organellar glutamine codons: A case by case scenario? RNA Biol. 2009;6:31–34. doi: 10.4161/rna.6.1.7564. [DOI] [PubMed] [Google Scholar]
- 5.Schon A, Kannangara CG, Gough S, Soll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 6.Dimmer KS, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SI, Stange-Thomann N, Martins O, Hong KW, Soll D, Fox TD. A nuclear genetic lesion affecting Saccharomyces cerevisiae mitochondrial translation is complemented by a homologous Bacillus gene. J Bacteriol. 1997;179:5625–5627. doi: 10.1128/jb.179.17.5625-5627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulero JJ, Rosenthal JK, Fox TD. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Curr Genet. 1994;25:299–304. doi: 10.1007/BF00351481. [DOI] [PubMed] [Google Scholar]
- 9.Frechin M, Senger B, Braye M, Kern D, Martin RP, Becker HD. Yeast mitochondrial Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009;23:1119–1130. doi: 10.1101/gad.518109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinehart J, Krett B, Rubio MA, Alfonzo JD, Soll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pujol C, Bailly M, Kern D, Marechal-Drouard L, Becker H, Duchene AM. Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants. Proc Natl Acad Sci USA. 2008;105:6481–6485. doi: 10.1073/pnas.0712299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnefond L, Fender A, Rudinger-Thirion J, Giege R, Florentz C, Sissler M. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry. 2005;44:4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
- 13.Rubio MA, et al. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci USA. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sylvers LA, Rogers KC, Shimizu M, Ohtsuka E, Soll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993;32:3836–3841. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Suzuki T. Chaplet column chromatography: Isolation of a large set of individual RNAs in a single step. Methods Enzymol. 2007;425:231–239. doi: 10.1016/S0076-6879(07)25010-4. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 17.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, Suzuki T, Wada T, Saigo K, Watanabe K. Taurine as a constituent of mitochondrial tRNAs: New insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002;21:6581–6589. doi: 10.1093/emboj/cdf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirino Y, et al. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc Natl Acad Sci USA. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 21.Schulze JO, et al. Crystal structure of a non-discriminating glutamyl-tRNA synthetase. J Mol Biol. 2006;361:888–897. doi: 10.1016/j.jmb.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T, Ueda T, Watanabe K. A new method for identifying the amino acid attached to a particular RNA in the cell. FEBS Lett. 1996;381:195–198. doi: 10.1016/0014-5793(96)00107-x. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 25.Stanzel M, Schon A, Sprinzl M. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur J Biochem. 1994;219:435–439. doi: 10.1111/j.1432-1033.1994.tb19956.x. [DOI] [PubMed] [Google Scholar]
- 26.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 27.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat Struct Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 28.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 29.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 30.Morris JZ, Navarro C, Lehmann R. Identification and analysis of mutations in bob, Doa and eight new genes required for oocyte specification and development in Drosophila melanogaster. Genetics. 2003;164:1435–1446. doi: 10.1093/genetics/164.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris JZ, et al. Mutations in the Drosophila mitochondrial tRNA amidotransferase, bene/gatA, cause growth defects in mitotic and endoreplicating tissues. Genetics. 2008;178:979–987. doi: 10.1534/genetics.107.084376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamenski P, et al. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Oshikane H, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 34.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abe T, et al. tRNADB-CE: tRNA gene database curated manually by experts. Nucleic Acids Res. 2009;37:D163–168. doi: 10.1093/nar/gkn692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakita K, et al. Higher-order structure of bovine mitochondrial tRNA(Phe) lacking the 'conserved' GG and T psi CG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res. 1994;22:347–353. doi: 10.1093/nar/22.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Putz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA. 2007;13:1184–1190. doi: 10.1261/rna.588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.