Abstract
Hepatitis C virus (HCV) infection is a major cause of end-stage liver disease and a leading indication for liver transplantation. Current therapy fails in many instances and is associated with significant side effects. HCV encodes only a few proteins and depends heavily on host factors for propagation. Each of these host dependencies is a potential therapeutic target. To find host factors required by HCV, we completed a genome-wide small interfering RNA (siRNA) screen using an infectious HCV cell culture system. We applied a two-part screening protocol to allow identification of host factors involved in the complete viral lifecycle. The candidate genes found included known or previously identified factors, and also implicate many additional host cell proteins in HCV infection. To create a more comprehensive view of HCV and host cell interactions, we performed a bioinformatic meta-analysis that integrates our data with those of previous functional and proteomic studies. The identification of host factors participating in the complete HCV lifecycle will both advance our understanding of HCV pathogenesis and illuminate therapeutic targets.
Keywords: HCV, RNA interference, viral host factors, functional genomics, viral lifecycle
Hepatitus C virus (HCV) causes a chronic infection resulting in progressive liver damage. The virus enters hepatocytes via interactions between the viral envelope proteins, E1 and E2, and four known host receptors, CD81, Claudin-1, SBR-I, and occludin (1). Subsequent to entry, host ribosomes bind to the internal ribosomal entry site (IRES) of the HCV genome, and translate viral polyproteins on the rough endoplasmic reticulum (ER) (2). Host and viral proteases process the polyprotein into both structural (core, and envelope proteins, E1 and E2) and nonstructural proteins (p7, NS2–3, NS3, NS4A, NS4B, NS5A, and NS5B) (3). Oligomerization of NS4B distorts the host ER into membranous webs, which house HCV replication complexes (RCs), within which the RNA-dependent RNA polymerase, NS5B, transcribes viral genomic RNAs (2). Progeny viral genomes generated in the RCs are translocated to lipid droplet-containing organelles and assemble into virions, which then traffic to the cell surface for release (4). However, although progress has been made in the areas of entry and replication, the later stages of the viral lifecycle—assembly, budding, and maturation—remain less explored.
The in vitro study of HCV replication has benefited from the use of the replicon system, consisting of subgenomic or whole genome viral RNAs stably expressed in permissive tissue culture cells. Using the replicon system, several recent functional screens, using either partial or genome-wide siRNA libraries, have identified a number of host factors involved in HCV replication (5–7). However, the replicon system is unable to address the role of host factors in the full HCV lifecycle. The infection competent HCV cell culture system (HCVcc) recapitulates the complete lifecycle, thereby permitting a greater range of host-viral interactions to be studied (8–10). Two efforts have used the HCVcc system using limited candidate gene siRNA screens of either 65 or 140 curated targets (11, 12). Our previous functional genomic studies, and those of others, have revealed that viral dependencies on the host machinery are both intricate and extensive (13–15). Moreover, a great advantage may be gained in pursuing an unbiased genetic screening strategy in that unexpected connections and insights can be discovered. Given the complexity of the virus' relationship with the host cell, and the relatively unexplored mechanisms of HCV replication, we hypothesized that additional host factors might be found by completing an unbiased whole-genome siRNA screen with the HCVcc system.
Results
The siRNA Screen.
We used a two-part screen employing the infectious JFH-1 genotype 2a virus and the Huh 7.5.1 human hepatocellular cell line to find host proteins required by the HCVcc system. Part one of the screen involved transfecting the Huh 7.5.1 cells with siRNAs for 72 h, then challenging them with JFH-1 virus. After 48 h, the cells were stained and imaged for expression of the HCV core protein as a marker for productive viral infection (Fig. 1A). To find host factors involved in later stages of viral infection, we undertook part two of the screen, by exposing fresh Huh 7.5.1 cells to culture supernatant from part one for 48 h, then detecting core protein expression. This approach detects proteins needed for the complete viral lifecycle, from viral-host receptor binding to the completion of a second round of viral infection. In addition, genes functioning in either anti-viral responses or safeguarding the cells against the stress of infection, may be detected in this screen.
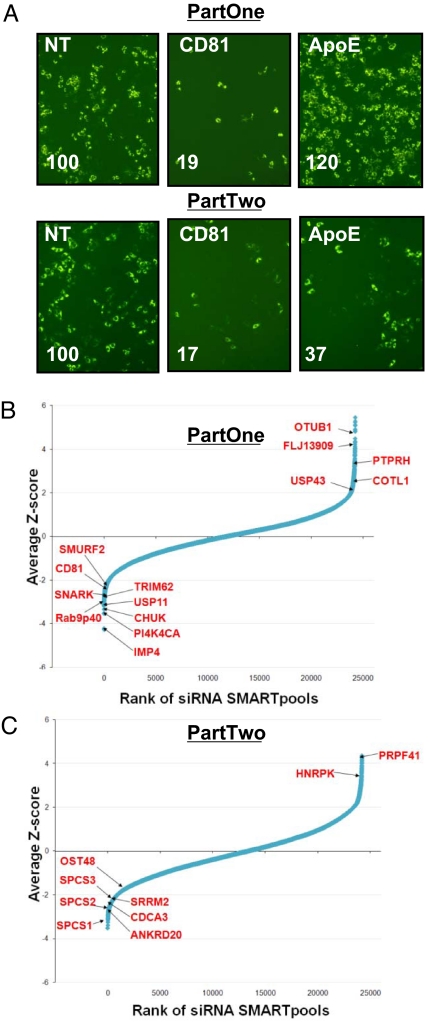
Fig. 1.
The siRNA screen. (A) siRNAs against CD81 or ApoE inhibit HCV infection (green: HCV core). Percent infected (lower left corner) is relative to control throughout (Fig. S1A and B). NT, Nontargeting siRNA #2. Magnification, 20×. (B and C) The results of the HCVcc siRNA screen part one (B) and part two (C) are shown with the siRNA SMARTpools ranked in order of average Z-score, from lowest (decreased infection) to highest (increased infection). The position of known HCV-host factors and several newly identified genes that scored in the screen are indicated.
The screen was optimized using siRNAs against the host proteins, CD81 (part one) and ApoE (part two). ApoE, a host protein involved in lipoprotein biosynthesis, has been shown to be required for infectious particle formation (16). siRNAs against CD81 resulted in inhibition of part one infection (5–6-fold, Fig. 1A and Fig. S1A and B). siRNAs targeting ApoE, however, did not affect infection in part one, but did inhibit HCV propagation in part two by 2–3 fold, in keeping with its previously assigned role in infectious particle formation (Fig. 1A and Fig. S1A and B). The levels of infection were verified using quantitative RT-PCR and found to be proportional to the levels determined by imaging (Fig. S1C and D). This platform was then used for a genetic screen with a whole-genome siRNA library (Dharmacon siGenome, 19,470 pools of four siRNAs per gene). Pools were classified as hits (decreased infection) if the percentage of infected cells was <50% of the plate mean in either part one and two, or in part two alone, and did not decrease cell numbers to <50% of the plate mean in part one. Using these criteria, we selected 407 pools for further validation (2% of the total genes screened, Fig. 1B). siRNA pools whose transfection increased core protein expression by >150% of the plate mean in either part one or two, were also selected (increased infection) for validation (114 pools, 0.6%, Fig. 1C). In the validation round, we tested the four siRNAs from each pool separately, and found that 262 out of the 521 total pools selected (50%) were confirmed with at least two siRNAs, decreasing the likelihood of off-target effects [Dataset S1, Materials and Methods, the list of all genes scoring in the primary round is shown in Dataset S2 (17)]. Forty-four genes decreased viral propagation with multiple siRNAs predominantly in part two, implicating them in late-stage viral infection (Dataset S1).
siRNA Screen Bioinformatics and Meta-analysis.
Among the host factors whose depletion decreased HCV infection in the primary screen, we recovered both positive controls, CD81 and ApoE, and over thirty proteins previously implicated by functional genomic studies to be needed for the replication of HCV or West Nile virus (WNV), a related flavivirus (Dataset S1 and Dataset S2). The screen recovered six of the 26 host proteins determined by Randall et al. to decrease JFH-1 levels by >3-fold after intensive siRNA targeting (Dataset S2) (11). Among these shared candidates are the kinases MAPK1 and Raf1/MAPK3, the RNA-binding protein, ELAVL1 and the RNA-helicase, DDX3X. Proteomic studies, including the HCV infection mapping project (I-MAP) and experiments by Randall et al., have reported a direct physical interaction between Raf1/MAPK3, Stat3 and DDX3X and one or more of the HCV-encoded proteins (11, 20).
This screen detected 15 of 96 factors recently reported by Tai et al. in a whole-genome siRNA screen using an HCV subgenomic replicon (Dataset S2) (5); the common genes include the kinases PI4KA and NUAK/SNARK, and the enzymes HAS1, CTSF, and IDS. NUAK/SNARK, an AMPK-related kinase functioning in the cellular stress response, was one of nine host factors found by Ng et al. to be necessary for HCV replicon function in a siRNA screen targeting approximately 4,000 genes (7). A critical role for PI4KA was also apparent in an HCVcc-based screen using a preselected library of genes implicated in endocytic trafficking (12). In that study, 7 of the 140 genes tested were needed by HCV, including the actin-regulating kinase, ROCK2, which we also recovered in our screen (12). Consistent with Tai et al., we observed that multiple subunits (ARCN1, COPA, and COPB1) of the Golgi-associated retrograde transport complex, coatomer 1 (COP1), were needed for JFH-1 infection (Dataset S2) (5). The requirement for two COP1 subunits (COPA and COPB2) for HCV replication, was also observed in the trafficking-focused screen (12).
Interestingly, our study detected 14 genes in common with those previously found in an RNAi screen for human proteins required for WNV infection, among them the enzymes BCKDHA, RPS6KL1, and USP11 (Dataset S2) (14). The WNV screen detected host factors involved in viral entry through replication, and thus would not find genes detected in part two of our study. However, the small degree of overlap (4.9%), between the host genes needed by these related viruses could arise in part from viral-specific host factor requirements, as well as differences in experimental methodologies. For example, none of the HCV siRNA screening efforts identified the ER-associated-degradation pathway (ERAD) as important, even though multiple components were required for WNV replication (14), indicating differential dependencies.
Analysis for functionally enriched categories demonstrates a significant number of HCV host factors reside in the nucleus (82, P = 0.002). This includes ribonucleoprotein complex components (e.g., RBM22, SRRM2, UBA52), and transcription factors (e.g., BRF1, E2F2, ERCC5, FOXE3, MLXIPL, SMAD5, SMAD6, TRRAP, WWTR1). Classification of the genes into higher level molecular function and biological process categories showed significant enrichment for kinases (14 in total, P = 0.01), factors involved in protein metabolism and modification (P = 0.03), oncogenesis (P = 0.006), nucleic acid binding (P = 0.01) and nucleic acid metabolism (P = 0.03, Fig. S1E and F and Dataset S3).
We encountered several pathways and macromolecular complexes consistent with known phases of the viral lifecycle. HCV entry depends on four host cell receptors, and our screen recovered two, CD81 and claudin 1. Our results confirm that cyclophilin A (CypA) is the peptidyl-prolyl cis-trans isomerase required for HCV infection (18). Therefore, CypA is the likely target of the anti-HCV cyclosporin analogue, Debio-025, thus demonstrating that RNAi can find high-yield leads for host-directed anti-viral therapies (HDAVs, 19). Depletion of three of five signal peptidase components (SPCS1, 2 and 3), which process the HCV envelope proteins E1 and E2 in the rER (3), inhibited infection in part two, validating their role in pathogenesis (P = 6.14×10−4) and the design of this portion of the screen. Glycosylation of the E1 and E2 viral envelope proteins is needed for HCV entry (21), suggesting an explanation for the role of OST48/DDOST, a component of the rER oligosaccharyltransferase complex, which we detected as a late-acting HCV host factor. Our efforts to find HIV-dependency factors (HDFs) also found that OST48 was required for late-stage viral replication, consistent with its playing a role in glycosylation of the viral envelope spike (13).
Previously unappreciated HCV dependencies were also detected. Components of the nucleolus were well represented in the nuclear-associated factors enriched for by the screen (TOP1, TCOF1, PNO1, NOP2, NOL6, NOP5/NOP58, NOC4L, HEATR1, and IMP4). TCOF1 physically associates with nucleolar proteins, and is the gene responsible for Treacher Collins Syndrome, a severe congenital craniofacial developmental disease (22). Mice deficient in TCOF1 display ribosome biogenesis deficiencies suggesting that cells depleted in TCOF1 may be unable to meet the increased demand for protein production required by HCV (22). Four of nine components of the Ccr4-Not complex (CNOT1, CNOT2, CNOT3, and CNOT6L) were required for HCV infection. The Ccr4-Not complex is a global regulator of gene expression that functions in transcription and polyadenylation (23). Mutation of each of the components of the orthologous yeast Ccr4-Not complex revealed that each subunit regulates the expression of a largely unique set of genes (24). Therefore, the components identified in this screen may control distinct sets of HCV host factors.
Integration of HCV-Host Factor Interactions.
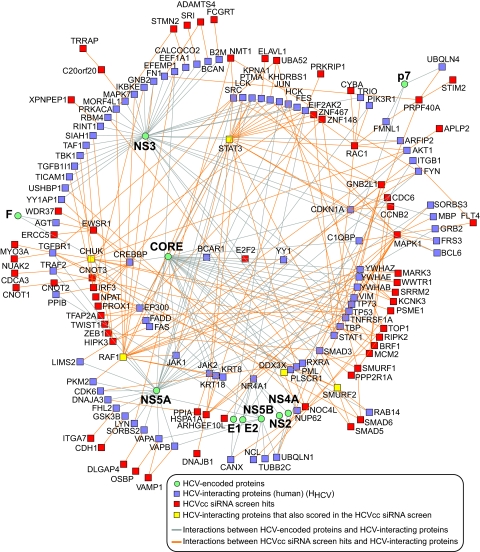
To provide a more comprehensive view of HCV-host interactions, we integrated the functional results of our study with the proteomic and literature mining data of the HCV infection mapping project (I-MAP), undertaken by Chassey et al. (Fig. 2) (20). The I-MAP used parallel yeast two-hybrid screens employing HCV proteins as prey and two human cDNA libraries as bait. The resulting protein-protein interaction (PPI) network was then extended using literature mining and a large human PPI database. In addition to providing functional evidence for the importance of nine proteins found by the I-MAP (CHUK, CTGF, DDX3X, PI4KA, RAF1, STAT3, FBLN5, RNF31, SMURF2), our network extension analysis revealed numerous first-order (direct) interactions between 63 HCV host factors found in this screen and those found in the I-MAP (P = 0.0001, Fig. 2). This places the candidates detected using RNAi into the interaction neighborhood of the HCV proteome (Fig. 3).
Fig. 2.
Network showing connections between HCV proteins, HCV-interacting human proteins and candidate proteins from the HCVcc siRNA screen. Network extension analysis of HCV-host protein interactions derived from the infection mapping project (I-MAP, 20) revealed extensive first-order (direct) interactions between host factors implicated in the I-MAP and those found in the HCVcc siRNA screen (enrichment P = 0.0001).
Fig. 3.
Network clusters derived from Fig. 2 showing interactions between HCV-host factors from the siRNA screen and host proteins interacting with HCV core, E1, E2, F, p7 or NS components (from the infection mapping project (I-MAP, 20) thereby placing the siRNA screen hits in the network neighborhood of the respective HCV proteins. For clarity, interactions between HCV proteins and their human interacting partners are not displayed, but are shown in Fig. 2.
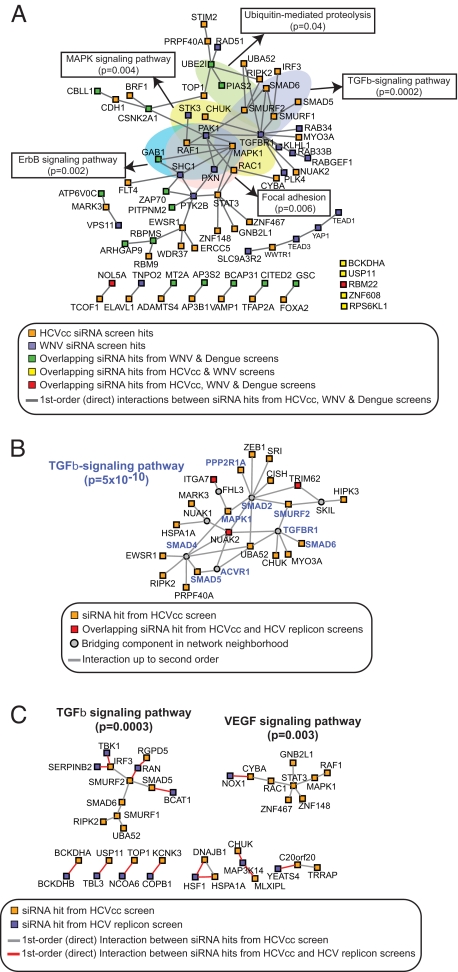
HCV and WNV belong to the Flaviviridae family, whose members include the human pathogens dengue virus and Japanese encephalitis virus (25). We explored the possibility of uncovering common features presented by host-Flaviviridae interactions. Among the overlapping hits and first-order interactions between host factors required for HCV, WNV and dengue virus siRNA screens (14), a striking enrichment of pathway components and common network modules involved in TGF-β signaling (P = 0.0002), ErbB signaling (P = 0.002), MAPK signaling (P = 0.004), focal adhesion (P = 0.006), and ubiquitin-mediated proteolysis (P = 0.04) was observed (Fig. 4A). These may represent important shared functional modules used by Flaviviridae. A comparison of this study to the genome-wide HCV replicon screen detected common network modules associated with TGF-β signaling, perhaps delineating the portion of the viral lifecycle that relies on this pathway (P < 0.0005, Fig. 4 B and C) (5).
Fig. 4.
Common features of host-Flaviviridae interactions. (A) Common network modules for HCV-dependency factors identified in siRNA screens for Flaviviridae. The network was generated using first-order (direct) interactions between siRNA hits from HCV, WNV, and dengue virus screens to identify common functional modules potentially used by Flaviviridae. Network clusters obtained were examined for enrichment (P < 0.05) in KEGG signaling pathways. (B and C) Common network modules for HCV host factors identified in the HCVcc and HCV replicon siRNA screens. (B) Network cluster generated using second order interactions for overlapping siRNA hits reveals a significant enrichment for the TGF-β-signaling pathway (P = 5 × 10−10). (C) First order interactions identify connections between HCV replicon screen hits and a number of TGF-β- and VEGF pathway-associated siRNA hits from the HCVcc screen. Other binary or triple-member connections include subunits (BCKDHA and BCKDHB) of the branched chain keto acid dehydrogenase E1 enzyme complex, and components (TRRAP and YEATS4) of the NuA4 histone acetyltransferase complex.
An estimated 25–30% of HIV-infected individuals are chronically coinfected with HCV (26). Defining host factors and pathways upon which both HIV and HCV depend could suggest a common treatment strategy. Comparing the host factors recovered from the HCVcc and HIV siRNA screens reveals ten genes, DDX3X, DNAJB1, ETF1, HEATR1, MAP4, NMT1, OST48, SPCS3, SUV420H1, and Rap9p40/RABEPK that are needed for replication of both viruses (13). Rab9p40 functions in the movement of late endosomes to the trans Golgi network (TGN), and along with its interaction partners, Rab9 and PIKfyve, is required for the particle assembly of multiple viruses (HIV, Marburg, Ebola and measles virueses), although the mechanism is not understood (27, 28). This study, together with that of Tai et al., adds HCV to the list of Rab9p40-dependent viruses.
Discussion
Previous efforts have discovered important steps in HCV replication, yet many events remain uncharacterized (2, 3, 25). Using RNAi and the HCVcc system, we have completed a genome-wide screen to find host factors required by HCV. The screen was validated by the “rediscovery” of >30 host factors previously implicated in HCV pathogenesis, including two host coreceptors, CD81 and claudin 1. We then performed a bioinformatic metaanalysis to synthesize the information from our own work and earlier functional and proteomic screens for both HCV and related Flaviviridae host factors. This analysis revealed a strong statistical enrichment for several host cell pathways and complexes, as well as multiple direct interactions between the functionally defined data and one from a comprehensive proteomic study (20). Many interesting connections have emerged from the integration of these efforts that will serve as a resource for future investigations.
We also identified ten host genes that are needed by HCV and HIV. These genes may represent common dependencies that could be exploited in instances of coinfection. The observation that Rab9p40 is needed by the HCV replicon, demonstrates that it acts before viral particle formation, perhaps influencing the generation of the membranous web, given the juxtaposition of this structure and the TGN. Rab9p40 contains a predicted β-propeller structure homologous to known phosphatidyl inositol (PI)-binding domains (28), which might provide a functional link to PI (4)-phosphate generated in the ER by PI4KA (29).
TGF-β levels are elevated in patients living with HCV, with even higher levels being present in HIV-HCV coinfected individuals (30). Elevated TGF-β levels enhance HCV replication in vitro and correlate with accelerated liver fibrosis in vivo (31, 32). Gene expression profiling of Huh 7.5.1 liver cells infected with the JFH-1 virus discovered increased transcription of TGF-β pathway genes, including Smad7 and 9 (33). SMAD7 dampens the pathway's activity via a TGF-β-induced negative feedback loop (33, 34). Our data provide further evidence of the importance of this pathway for HCV replication and lend functional relevance to the above-mentioned gene expression profiling studies, in that both positive (SMAD5) and negative regulators (SMAD6, SMURF1, and SMURF2) of the TGF-β pathway were detected. Together, these results suggest that intact TGF-β pathway homeostasis may benefit the virus.
Whereas RNAi technology represents a revolution in mammalian genetics, it also has caveats and must be interpreted with significant caution. Major concerns include false positives, arising from off-target effects (OTEs), where the promiscuous actions of the siRNA results in the observed phenotype being because of the depletion of a second unintended target, as well as false negatives, stemming in part from inefficient targeting or nonspecific toxicity (17). We attempted to minimize false positives by testing our primary candidates with multiple individual siRNAs (Dataset S2) (17). We also integrated several HCV host factor data sets, and revealed statistically enriched complexes and pathways. To preserve new connections and minimize bias, we performed our bioinformatics analysis after the selection of our validated gene set. Thus it appears prudent to view the gene candidates confirmed with two or more siRNAs, or in two or more studies, either proteomically or functionally, as being the highest confidence host genes involved in HCV infection. However, given the variation in screening methodologies, and the current levels of false negatives, it is likely that many genes uniquely indicated in these studies will ultimately be shown to partake in the viral lifecycle.
The genes identified by these efforts represent a starting point for defining a complete set of HCV-host interactions. HCV host factors are potentially valuable targets for therapies to combat the more recalcitrant HCV strains. An example of an HDAV against HCV is Debio 025, which inhibits HCV infection by interfering with the host CypA protein (35). Our confirmation of HCV's reliance on CypA illustrates that forward genetics can find host proteins that can be targeted to block infection.
In light of the heterogeneous course of chronic HCV infection, the variable response of patients to therapy, and the estimated 20% of individuals that are exposed to HCV but clear the infection, a reasonable assumption is that polymorphisms in HCV host factors may in part underlie these observations. There is mounting evidence from genome wide association studies that rare polymorphisms may collectively account for a sizable percentage of the variation observed in heritable traits, including the course of chronic infections (36). Functional genomic and proteomic studies of HCV infection, and the meta-analyses of these combined efforts, may therefore provide targets upon which to focus the search for causal genetic variations that affect HCV infection.
Experimental Methods
See SI Text for additional experimental details.
siRNA Screen: Part 1.
siRNAs were transfected into the Huh 7.5.1 cells at a 50 nM final concentration, using Oligofectamine (Invitrogen) in a 384-well format (Corning 3712). After 72 h, the cells were infected with the JFH-1 genotype 2a virus. After 48 h, the media was replica plated onto a plate with Huh 7.5.1 cells. The part 1 cells were then fixed and stained with anti-HCV core 6G7 monoclonal antibody provided by Drs. Harry Greenberg and Xiaosong He, (Stanford University) using an Alexa Fluor 488 antibody (A11001, Invitrogen). Cells were imaged with an Image Express Micro (Molecular Devices) and analyzed with Metamorph Cell Scoring software (Molecular Devices Inc.).
Part 2.
Forty-eight hours after the transfer of the part one supernatant, the part two cells were stained for HCV core expression and imaged as in part 1.
Cell Lines.
Huh 7.5.1 cells were grown in DMEM 10% FBS.
Virus.
JFH-1 was propagated and infectivity was titrated as described (8, 37).
Supplementary Material
Acknowledgments.
We thank the ICCB-Longwood: C. Shamu, S. Chang, S. Rudnicki, S. Johnston, K. Rudnicki, D. Wrobel, and Z. Cooper. Funding support: A.L.B. (Harvard Center for AIDS Research), A.N. (Crohns and Colitis Foundation of America), R.J.X. (Center for the Study of Inflammatory Bowel Disease Genetics and Genomics Core and National Institutes of Health Grant DK043351). The ICCB-Longwood is in part supported by a grant to T. Mitchison (National Cancer Institute). This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, and the New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (National Institutes of Health Grant U54 AI057159 to Dennis Kasper). S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907439106/DCSupplemental.
References
- 1.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 4.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 5.Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Supekova L, et al. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J Biol Chem. 2008;283:29–36. doi: 10.1074/jbc.M703988200. [DOI] [PubMed] [Google Scholar]
- 7.Ng TI, et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 8.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 11.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger KLC, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, et al. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flisiak R, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 20.de Chassey B, et al. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goffard A, Dubuisson J. Glycosylation of hepatitis C virus envelope proteins. Biochimie. 2003;85:295–301. doi: 10.1016/s0300-9084(03)00004-x. [DOI] [PubMed] [Google Scholar]
- 22.Sakai D, Trainor PA. Treacher Collins syndrome: Unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41:1229–1232. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 24.Azzouz N, et al. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 26.Andersson K, Chung RT. Hepatitis C Virus in the HIV-infected patient. Clin Liver Dis. 2006;10:303–320. doi: 10.1016/j.cld.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Murray JL, et al. Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J Virol. 2005;79:11742–11751. doi: 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shisheva A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol Int. 2008;32:591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balla A, Balla T. Phosphatidylinositol 4-kinases: Old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Blackard JT, et al. Intrahepatic cytokine expression is downregulated during HCV/HIV co-infection. J Med Virol. 2006;78:202–207. doi: 10.1002/jmv.20528. [DOI] [PubMed] [Google Scholar]
- 31.Lin W, et al. HIV increases HCV replication in a TGF-beta1-dependent manner. Gastroenterology. 2008;134:803–811. doi: 10.1053/j.gastro.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Wilson LE, et al. Progression of liver fibrosis among injection drug users with chronic hepatitis C. Hepatology. 2006;43:788–795. doi: 10.1002/hep.21091. [DOI] [PubMed] [Google Scholar]
- 33.Walters KA, et al. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99:2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flisiak R, et al. The cyclophilin inhibitor Debio 025 combined with PEG IFNalpha2a significantly reduces viral load in treatment-naïve hepatitis C patients. Hepatology. 2009;49:1460–1468. doi: 10.1002/hep.22835. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 37.Kato T, et al. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol. 2007;81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.