Abstract
We used the muscle creatine kinase (MCK) conditional frataxin knockout mouse to elucidate how frataxin deficiency alters iron metabolism. This is of significance because frataxin deficiency leads to Friedreich's ataxia, a disease marked by neurologic and cardiologic degeneration. Using cardiac tissues, we demonstrate that frataxin deficiency leads to down-regulation of key molecules involved in 3 mitochondrial utilization pathways: iron-sulfur cluster (ISC) synthesis (iron-sulfur cluster scaffold protein1/2 and the cysteine desulferase Nfs1), mitochondrial iron storage (mitochondrial ferritin), and heme synthesis (5-aminolevulinate dehydratase, coproporphyrinogen oxidase, hydroxymethylbilane synthase, uroporphyrinogen III synthase, and ferrochelatase). This marked decrease in mitochondrial iron utilization and resultant reduced release of heme and ISC from the mitochondrion could contribute to the excessive mitochondrial iron observed. This effect is compounded by increased iron availability for mitochondrial uptake through (i) transferrin receptor1 up-regulation, increasing iron uptake from transferrin; (ii) decreased ferroportin1 expression, limiting iron export; (iii) increased expression of the heme catabolism enzyme heme oxygenase1 and down-regulation of ferritin-H and -L, both likely leading to increased “free iron” for mitochondrial uptake; and (iv) increased expression of the mammalian exocyst protein Sec15l1 and the mitochondrial iron importer mitoferrin-2 (Mfrn2), which facilitate cellular iron uptake and mitochondrial iron influx, respectively. Our results enable the construction of a model explaining the cytosolic iron deficiency and mitochondrial iron loading in the absence of frataxin, which is important for understanding the pathogenesis of Friedreich's ataxia.
Keywords: heme synthesis, iron, transferrin, frataxin
Many mechanisms involved in iron metabolism have been deciphered through the analysis of diseases or mutant animals. Understanding the mitochondrial iron-loading disease Friedreich's ataxia is of special interest, because it could aid in the clarification of mechanisms of mitochondrial iron transport (1).
Friedreich's ataxia is an inherited ataxia characterized by progressive neurologic and cardiologic degeneration and mitochondrial iron loading (1). This condition is caused by reduced expression of the mitochondrial protein frataxin (2). The precise function of frataxin remains unclear, but it has been implicated in mitochondrial iron metabolism, particularly iron-sulfur cluster (ISC) synthesis (3–5). Frataxin depletion results in mitochondrial DNA damage, mitochondrial iron accumulation, ISC deficiency, perturbed heme synthesis, and oxidative damage (6–9). In addition, recent studies have suggested cytosolic iron deficiency in Friedreich's ataxia models, as indicated by increased RNA-binding activity of iron regulatory protein 2, increased expression of transferrin receptor 1 (Tfr1), and decreased cytosolic ferritin (10, 11).
Three main mitochondrial iron utilization pathways are known: heme and ISC synthesis, and mitochondrial iron storage (1). The mitochondrion is the main site of ISC biosynthesis, providing ISCs for mitochondrial, cytosolic, and nuclear proteins (12). In humans, ISC assembly is facilitated by the product of the nitrogen fixation gene 1 (Nfs1), which provides sulfur (13) and the ISC scaffold proteins 1 and 2 (Iscu1/2) (1). Frataxin is involved in the formation of ISCs (14–17), and reduced activity of ISC-containing enzymes occurs early after frataxin depletion (7, 18).
The mitochondrion is the only site of heme synthesis. The final enzyme in this pathway is ferrochelatase (Fech) (1), an ISC-containing protein whose activity may be frataxin-dependent (19) and is down-regulated in the yeast frataxin homolog–depletion model (9). There is some evidence that frataxin plays a role in heme synthesis (9, 20, 21), although the precise mechanisms involved are unclear. Apart from the 2 iron-requiring synthetic pathways in the mitochondrion, mitochondrial ferritin (Ftmt) appears to sequester “free” mitochondrial iron (22, 23). The participation of frataxin in mitochondrial iron metabolism is evident from the frataxin deficiency observed in Friedreich's ataxia, which leads to mitochondrial iron loading. However, the exact function of frataxin in these pathways remains elusive and requires further study (1).
Because complications from cardiomyopathy are a frequent cause of death in Friedreich's ataxia, we used muscle creatine kinase (MCK) conditional frataxin knockout (mutant) mice, which lack frataxin in only the heart and skeletal muscle (7). In this model, the tissue-specific Cre transgene under the control of MCK promoter results in the conditional deletion of frataxin (7). Our aim was to explore the changes in iron metabolism due to frataxin deficiency. These mutants exhibit classical traits of the cardiomyopathy in Friedreich's ataxia, including cardiac hypertrophy, ISC enzyme deficiency, and marked mitochondrial iron accumulation (7, 11, 24). Thus, this model closely reflects the cardiac pathology in Friedreich's ataxia.
Here, we present results from cardiac tissues from MCK mutant and wild type (WT) mice that support a model describing the effects of frataxin deficiency on iron metabolism. Our findings demonstrate significant down-regulation of key molecules involved in all 3 pathways of mitochondrial iron utilization which contribute to mitochondrial iron loading. Further, we report alterations in gene and protein expression that allow increased cellular iron uptake, decreased cytosolic iron storage, and stimulation of mitochondrial iron uptake, leading to mitochondrial iron targeting and cytosolic iron deficiency.
Results
Microarray Analysis Shows Extensive Alteration in Gene Expression in the Hearts of 10-Week-Old MCK Mutants.
We examined gene expression via whole genome gene array using hearts from MCK mutants compared with WT littermates. We compared gene expression in the mutant and WT littermates at age 4 weeks, when no overt phenotype is evident, and at age 10 weeks, when cardiomyopathy is evident, including marked mitochondrial iron loading (7, 11).
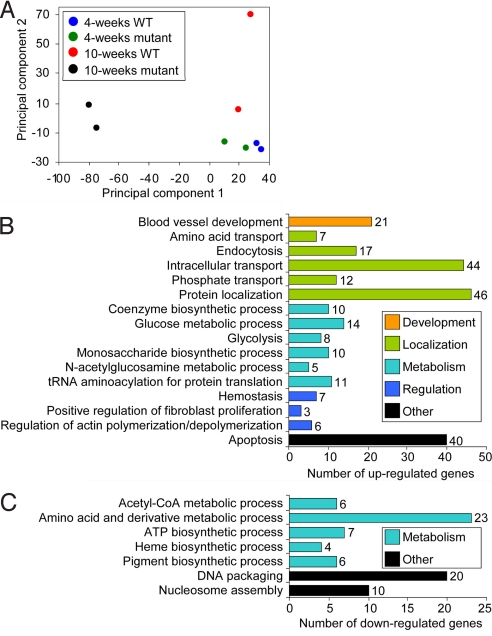
Examining these microarray data from the 4- and 10-week-old mutants and WT littermates using principal component analysis (25) revealed extensive deviation between the genome-wide expression profile of the 10-week-old mutants and their WT littermates, but little difference between the 4-week-old mutants and their WT littermates (Fig. 1A). Comparing the mutants and WTs at each age, this corresponds to 1,732 genes significantly altered at 10 weeks and only 5 genes significantly altered at 4 weeks (Table S1). Only 12 genes were significantly altered between the 4-week-old and 10-week-old WT mice, presumably reflecting developmental alterations. In marked contrast, 1,353 genes were altered between the 4-week-old and 10-week-old mutants (Table S1).
Fig. 1.
Microarray analysis of 4- and 10-week-old WT and mutant [frataxin (Fxn) knockout] mice. (A) Principal component plot showing the overall differential expression profile between WT and mutants of different ages. (B and C) Functional clustering of significantly (P < .05) altered genes in 10-week-old mutants showing various biological processes significantly (P < .05) up-regulated (B) or down-regulated (C).
To explore whether frataxin deletion affects any specific biological processes, we analyzed the list of significantly altered genes in 10-week-old mutants using the functional annotation software DAVID, according to the classification of biological processes assigned by the Gene Ontology Consortium (26). We found that various processes were significantly (P < .05) modulated, including development, molecular localization, metabolism, and regulation (Fig. 1 B and C). The top 3 up-regulated general gene groups function in protein localization, intracellular transport, and apoptosis. In contrast, the top 3 groups of genes down-regulated are involved in amino acid and derivative metabolic processes, DNA packaging, and nucleosome assembly. The importance of these alterations to the pathogenesis of Friedreich's ataxia is unclear, but the down-regulation of genes involved in heme synthesis (Fig. 1C) is of particular significance to this analysis.
Frataxin Deficiency Alters the Expression of Genes Involved in Iron Metabolism in the Heart.
The mechanism involved in mitochondrial iron accumulation in Friedreich's ataxia remains unclear (27), and deciphering it is imperative for understanding the function of frataxin in iron homeostasis and the pathogenesis of Friedreich's ataxia. To investigate whether decreased frataxin causes disturbance in the expression of genes involved in iron metabolism, we examined a list of genes exhibiting significant (P < .05) differential expression, leading to the identification of 18 genes associated with iron metabolism (Table S2). These were not the only genes identified from the microarray that were significantly differentially expressed; the remainder play roles in cardiac hypertrophy and other functions. Considering the role of frataxin in mitochondrial iron metabolism (1), we focused on the alteration of gene expression in these pathways.
Altered Cardiac Iron Metabolism Indicates Cytosolic Iron Deficiency and Mitochondrial-Targeted Iron Transport.
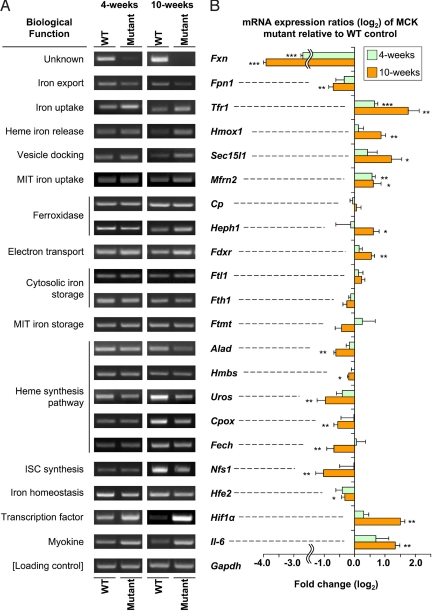
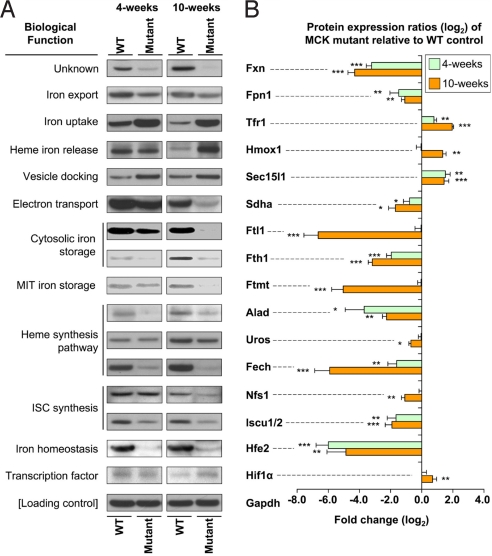
The functions of the iron metabolism genes (Table S2) with significantly altered expression can be grouped into the categories: cellular iron metabolism, heme biosynthesis, ISC biosynthesis, iron-sensing proteins, cytokines, and energy metabolism. These genes include ferroportin 1 (Fpn1), Tfr1, heme oxygenase 1 (Hmox1), ferritin light chain (Ftl1), exocyst complex component 6 (Sec15l1), mitoferrin 2 (Mfrn2), ceruloplasmin (Cp), and hephaestin (Heph). Significant changes in expression were confirmed for most of the genes at the mRNA level by RT-PCR (Fig. 2) and at the protein level by Western blot analysis (Fig. 3). The exceptions were Cp and Ftl1, which exhibited no change in mRNA level. Expression of Heph and Mfrn2 could not be confirmed at the protein level, due to the unavailability of antibodies.
Fig. 2.
RT-PCR analysis of iron metabolism–related genes selected from Affymetrix GeneChips that were significantly (P < .05) differentially expressed between 4- and 10-week-old WT and mutant (Fxn knockout) mice. (A) RT-PCR confirming significant differential expression of genes observed in Affymetrix GeneChips. (B) Densitometry of RT-PCR analysis. *, P < .05; **, P < .01; ***, P < .001. Results shown in (A) are representative of 3–6 experiments, and those in (B) are the mean ± SD of 3–6 experiments.
Fig. 3.
Western blot analysis of significantly (P < .05) differentially expressed iron metabolism–related genes and densitometric analysis from 4- and 10-week-old WT and mutant (Fxn knockout) mice. (A) Western blot confirming the differential gene expression from the RT-PCR analysis shown in Fig. 2. (B) Densitometric analysis. *, P < .05; **, P < .01; ***, P < .001. Results shown in (A) are representative of 3–6 experiments, and those in (B) are the mean ± SD of 3–6 experiments.
mRNA and protein expression of the iron exporter Fpn1 were decreased in 4-week-old mutants, but more markedly and significantly (P < .01) decreased in 10-week-old mutants (Figs. 2 and 3). Tfr1, the primary source of iron uptake (28), was significantly (P < .01) increased at the mRNA and protein levels in the 4-week-old mutants and especially in the 10-week-old mutants. Both up-regulation of Tfr1 and down-regulation of Fpn1 are consistent with our previous findings using MCK mutants (11). These observations suggest increased iron import and decreased iron export, respectively, and are indicative of a response to cytosolic iron deficiency. The significant (P < .05) up-regulation of Hmox1 mRNA and protein levels in the 10-week-old mutants indicates liberation of “free iron” through heme catabolism. The marked and significant (P < .001) decrease in ferritin-H (Fth1) and -L (Ftl1) chain protein in the 10-week old mutants (Fig. 3) also confirmed the cytosolic iron deficiency.
Furthermore, mRNA and protein expression of Sec15l1 (Figs. 2 and 3) and mRNA expression of mitochondrial iron importer Mfrn2 (Fig. 2) were significantly (P < .05) up-regulated. The exocyst complex component Sec15l1 has been implicated in assisting transferrin (Tf) Tfr1-containing endosome uptake (29, 30). Thus, the alterations in these 2 molecules indicate increased iron uptake and mitochondrial iron import. This increase in mitochondrial iron trafficking would exacerbate the cytosolic iron deficiency and facilitate mitochondrial iron overload. This could lead to a constant iron flux being trafficked to the mitochondrion, where iron utilization pathways are down-regulated (see below), leading to mitochondrial iron accumulation (7, 11).
Frataxin Deficiency in the Heart Leads to Suppression of Proteins in Mitochondrial Iron Utilization Pathways.
It was of interest to find that in the microarray, the expression of key molecules in heme and ISC biosynthesis, as well as iron storage, were significantly decreased in the mutant. In terms of heme synthesis, there was significant (P < .05) down-regulation of coproporphyrinogen oxidase (Cpox), hydroxymethylbilane synthase (Hmbs), 5-aminolevulinate dehydratase (Alad), uroporphyrinogen III synthase (Uros), and ferrochelatase (Fech) (Table S2). In the ISC pathway, there was significant down-regulation of Nfs1 (1, 13) (Table S2).
RT-PCR confirmed the significant (P < .05) down-regulation observed in the microarray for these latter genes at age 10 weeks (Fig. 2). Western blot analysis also was performed (Fig. 3), but due to the limited antibody availability, only a subset of molecules was analyzed: Alad, Uros, Fech, Nfs1, and Iscu1/2. Although Iscu1/2 was not differentially expressed in the microarray, it was included for Western blot analysis, because Iscu and Nfs1 play crucial roles in ISC synthesis (1), and assessing this was vital.
Western blot analysis (Fig. 3) confirmed the significant (P < .01) down-regulation of all molecules in the heme and ISC synthetic pathways that were identified in 10-week-old mutants by the microarray (Table S2) and RT-PCR (Fig. 2). Although altered expression of the mitochondrial iron storage molecule, Ftmt, was not identified in the gene array, significant (P < .001) down-regulation of Ftmt protein expression was found (11) (Fig. 3). Together with our finding of down-regulation of heme and ISC biosynthesis, this demonstrates suppression of mitochondrial iron utilization.
Another gene up-regulated in the mutant was ferredoxin reductase (Fdxr) (Table S2), and this was confirmed by RT-PCR to be significant (P < .01) in 10-week-old mutants compared with WT mice (Fig. 2). The importance of this molecule in iron metabolism is highlighted by the fact that depletion of yeast Fdxr is known to cause dysregulation of iron metabolism (31). Ferredoxin facilitates electron transfer to enzymes, including those involved in heme synthesis (32). In this study, Fdxr up-regulation may be related to heme a synthesis, which is required for oxidative phosphorylation. Our proteomic studies have shown that frataxin depletion leads to energy metabolism deficiencies in MCK mutants, leading to metabolic compensation in an attempt to overcome the deficit (24). This may explain the Fdxr up-regulation.
MCK Mutants Exhibit Decreased Cardiac Heme Content.
Considering our observations of decreased expression of key enzymes in heme biosynthesis and increased Hmox1 expression, we explored whether frataxin deficiency leads to reduced total cardiac heme in mutants. Analysis of heart heme revealed a significant (P < .001) reduction in the 10-week-old mutants to almost half that of the WT mice (7.3 ± 0.6 vs. 12.5 ± 1.5 nmol heme/mg protein). There was no significant difference in heme levels between the 4-week-old mutants and WT mice.
Frataxin Deficiency in the Heart Affects Genes That Modulate Hepcidin Expression.
We also identified a number of other significantly differentially expressed genes that regulate the important hormone hepcidin, which modulates systemic iron levels (28, 33) (Table S2). Confirmation of alterations in mRNA expression by RT-PCR (Fig. 2) in 10-week-old mutants revealed significant (P < .01) up-regulation of hypoxia inducible factor-1α (Hif1α), significant (P < .01) up-regulation of interleukin-6 (Il6), and significant (P < .05) down-regulation of hemochromatosis type 2 (Hfe2). Western blot analysis of Hif1α and Hfe2 protein confirmed the RT-PCR results, demonstrating significant (P < .01) up-regulation of Hif1α and significant (P < .01) down-regulation of Hfe2 in the 4-week-old and 10-week-old mutants (Fig. 3). In contrast, Il6 protein was difficult to detect, probably because of its rapid secretion from myocytes.
Systemic Dysregulation of Iron Metabolism Occurs in MCK Mutants.
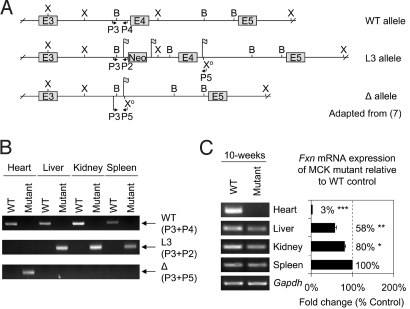
Considering that our data illustrate alterations in expression of molecules that play crucial roles in regulating systemic iron metabolism via their effects on hepcidin (e.g., Il6) (28, 33), it was of interest to examine the iron content of organs other than the heart in the mutants. As shown previously (7), the MCK promoter leads to deletion of frataxin in striated muscle, but not in other tissues, as confirmed in this study by genomic PCR (Fig. 4 A and B). Hence, not only the heart was significantly (P < .001) iron-loaded in 10-week-old MCK mutants, but also the liver, kidney, and spleen (Table S3). This indicated that deletion of frataxin in only the muscle of MCK mutants resulted in changes in systemic iron metabolism.
Fig. 4.
Alterations in frataxin mRNA expression in extracardiac tissues in the MCK mutant (Fxn knockout) mice relative to the WT mice are not due to Fxn deletion. (A) Conditional deletion of mouse frataxin exon 4 as reported previously (7). From top to bottom: WT allele, loxP-flanked Fxn exon 4 allele (L3), and Cre-mediated exon 4 deleted allele (Δ). The Δ allele was derived from the exon 4 deletion of the L3 allele via a cross with a CMV-Cre line (7). Flag, loxP site; B, BamHI; E, exon; P, primer; X, XbaI; Xo, modified XbaI. (B) The L3 allele was identified in the liver, kidney, and spleen, but not the heart, of the mutants, demonstrating the muscle-specific deletion of exon 4. This results in the Δ allele in the mutant heart only. PCR conditions were as described previously (7, 11). (C) RT-PCR showing pronounced down-regulation of frataxin mRNA in the heart and less marked reductions in the liver and kidney of 10-week-old mutants relative to WT mice of the same age. Densitometry data are reported as the mean ± SD of 3 experiments. *, P < .05; **, P < .01; ***, P < .001.
We then analyzed frataxin mRNA expression in the heart, liver, kidney, and spleen of 10-week-old mutant and WT mice (Fig. 4C). Despite the fact that the frataxin gene was not deleted in the liver, kidney, or spleen (Fig. 4B), frataxin mRNA expression in the mutant was significantly (P < .05) decreased in the liver and kidney compared with the WT mice (Fig. 4C). These alterations in frataxin expression potentially could be mediated by systemic factors involved in modulating iron metabolism. However, the extent of frataxin down-regulation (58–80% of the WT) in the liver, kidney, and spleen was not as pronounced as the marked and significant (P < .001) decrease of expression in the mutant heart (3% of the WT; Fig. 4C). Ablation of frataxin expression in the heart was not complete, because it was a total organ preparation containing other cells besides myocytes.
Discussion
Elucidation of mitochondrial iron trafficking pathways is important for understanding heme and ISC synthesis and the pathogenesis of diseases due to disturbances in these pathways (1). Because frataxin is localized in the mitochondrion and plays a role in iron metabolism, the MCK mutant, which demonstrates clear mitochondrial iron loading (7), provides a unique model for examining the role of frataxin in these pathways.
Myocardial Iron Metabolism is Altered by Frataxin Deficiency.
Our study enables construction of a model of the defective iron metabolism in the mutants (Fig. 5). There was significant down-regulation of molecules that play key roles in the 3 main pathways of mitochondrial iron utilization: heme and ISC biosynthesis, and mitochondrial iron storage (Figs. 2 and 3). Thus, mitochondrial iron would be insufficiently used for heme and ISC synthesis, both of which are normally effectively transported out of the mitochondrion for cytosolic use (1). Direct evidence of these metabolic defects was provided by the decreased cardiac heme in the mutants, diminished activity and expression of ISC enzymes [e.g., succinate dehydrogenase (Sdha); see Fig. 3 and refs. 7, 11, and 24], and increased mitochondrial iron loading in the mutants (7, 11).
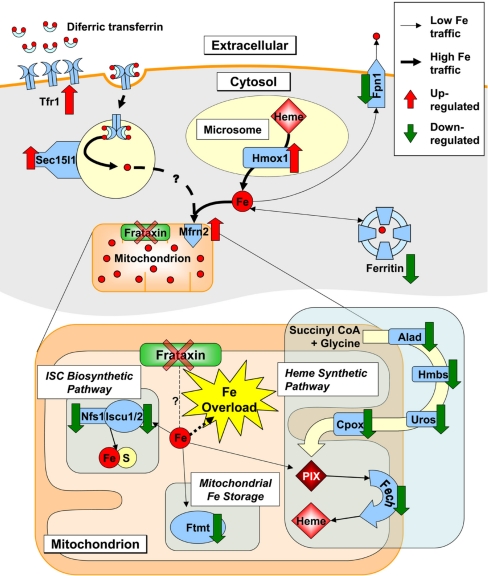
Fig. 5.
Model of altered iron metabolism due to frataxin deficiency in MCK mutant hearts. Frataxin deficiency leads to increased mitochondrial-targeted iron uptake and cytosolic iron deficiency, facilitated by (i) Tfr1 up-regulation, increasing transferrin iron uptake; (ii) Fpn1 down-regulation, preventing iron release; (iii) Hmox1 up-regulation, increasing cytosolic heme catabolism; and (iv) Sec15l1 up-regulation, potentially aiding iron uptake. Iron is then taken up more avidly by the mitochondrion via increased Mfrn2. Moreover, there is down-regulation of 3 major pathways of mitochondrial iron utilization—ISC synthesis, heme synthesis from iron incorporation into protoporphyrin IX (PIX), and mitochondrial iron storage (Ftmt). The decreased iron utilization in these pathways reduces iron export from the mitochondrion as heme and ISCs. This suppression, together with increased iron uptake, decreased iron release, and iron targeting to the mitochondrion, leads to the marked mitochondrial iron loading (1).
Previous studies have shown that mitochondrial iron uptake and utilization for heme synthesis are not well coupled, with no obvious negative feedback mechanism apparent (34, 35). This is evident when heme synthesis is prevented using succinylacetone, a specific inhibitor of Alad (34, 35). Inhibition of this or other enzymes in this pathway (1) results in pronounced mitochondrial iron loading in reticulocytes (34, 35). Under these conditions, the inhibition of heme synthesis leads to increased iron uptake from Tf and decreased ferritin iron uptake (34, 35). In fact, iron continues to enter the mitochondrion despite the absence of heme synthesis to use and then export it from the mitochondrion as heme, leading to mitochondrial iron loading (34, 35). Hence, these observations when heme synthesis is inhibited in erythroid cells are very similar to our findings in the mutant hearts.
Considering the foregoing, it is notable that Alad and 4 other molecules in the heme synthesis pathway are down-regulated by frataxin deficiency in the mutant. This, together with defective ISC biosynthesis and decreased Ftmt, would lead to decreased mitochondrial iron utilization and excess “free” iron that cannot be exported from the mitochondrion as heme or ISCs. This would contribute to the mitochondrial iron loading observed (Fig. 5). Supporting our findings, a direct interaction between purified frataxin and Fech has been reported, suggesting a role for frataxin in heme synthesis (9, 20). Indeed, we have suggested that frataxin acts as a metabolic switch to regulate mitochondrial iron utilization between heme and ISC biosynthesis (8). Moreover, an investigation in frataxin knockout mice showed decreased Isu1, Cpox, and Fech mRNA transcripts (not protein levels) (21), which is validated by our findings.
The decrease in mitochondrial iron utilization appears to be only part of the defect caused by frataxin deficiency that leads to mitochondrial iron accumulation. Coupled with defective mitochondrial iron utilization in the mutant, our findings indicate at least 4 mechanisms responsible for the cytosolic iron deficiency and increased mitochondrial iron influx that contribute to mitochondrial iron overload (Fig. 5). First, there was marked Tfr1 up-regulation in the mutants (Figs. 2 and 3), leading to increased 59Fe uptake from 59Fe-Tf in vivo in the hearts of the mutants (11). Second, there was down-regulation of Fpn1, decreasing cellular iron release. Third, there was increased Hmox1, which catabolizes cytosolic heme, concurrent with decreased protein expression of Ftl1 and Fth1 (Fig. 3), leading to reduced cytosolic iron incorporation and storage (Fig. 5). We have directly shown this effect in MCK mutants in vivo using 59Fe-Tf uptake studies (11). These alterations in protein expression explain our studies in vivo demonstrating lower amounts of 59Fe incorporated from 59Fe-Tf into the cytosol of mutant relative to WT hearts (11). Fourth, the increased expression of Sec15l1 (Figs. 2 and 3), a proposed mammalian exocyst protein (29, 30), and Mfrn2, a mitochondrial iron importer (28), in the mutant could facilitate mitochondrial iron influx, which is enhanced in this animal model (11). This iron is obtained via increased iron uptake from Tf by Tfr1, increased liberation of cytoplasmic iron from heme via Hmox1, and down-regulation of iron storage in cytosolic ferritin. Considering this in terms of enhanced mitochondrial iron uptake, Sec15l1 plays a role in the Tf cycle via its association with Rab11, a GTPase involved in vesicular trafficking (29, 30). Inactivation of Sec15l1 alters recycling of Tf-containing endosomes, increasing Tfr1 exocytic vesicular release and reducing cellular iron uptake (29, 30). Therefore, increased Sec15l1 in the mutant could contribute to increased iron uptake and mitochondrial iron loading, particularly in association with increased Mfrn2 expression, exacerbating the effect of the latter (Fig. 5).
Collectively, the reduction in mitochondrial iron utilization, coupled with the augmented iron uptake and decreased storage and utilization of iron in the cytosol, coalesce to lead to a mechanism that targets iron to the mitochondrion in the mutant (Fig. 5). These alterations in cellular and mitochondrial iron trafficking cannot be considered a primary or secondary event of frataxin deficiency. Nonetheless, these processes do lead to mitochondrial iron overload, which is considered detrimental (1, 11).
Systemic Iron Metabolism is Altered in Frataxin Deficiency in Heart and Muscle.
Our findings also provide evidence of alterations in systemic iron metabolism in the mutants, resulting in increased spleen, liver, and kidney iron (Table S3). In fact, we found modified Hif1α, Il6, and Hfe2 expression in mutant hearts (Table S2; Fig. 2). These molecules are involved in regulating hepcidin, which plays a key role in systemic iron homeostasis (28).
Of note, Il6 has been identified as a myokine (i.e., produced by myocytes) and can modulate metabolism in the absence of inflammation (36). Thus, it could be a regulator in the MCK mutants, where the deletion of frataxin occurs only in the heart and skeletal muscle (Fig. 4A; ref. 3). Il6 acting systemically via hepcidin (28, 33) could result in Fpn1 down-regulation in the heart (Fig. 3) and other tissues and thereby contribute to the increased iron levels (Table S3). Moreover, Il6 can lead to iron loading of the liver and brain in other animal models (37, 38).
In conclusion, we have identified alterations in gene and protein expression that occur in the MCK mutants that explain the cytosolic iron deficiency and mitochondrial iron overload occurring in frataxin deficiency (Fig. 5). Frataxin deficiency leads to down-regulation of 3 key mitochondrial iron utilization pathways (ISC synthesis, heme synthesis, and mitochondrial iron storage). There is also increased iron availability for mitochondrial iron uptake mediated by increased Tfr1, decreased iron release by reduced Fpn1, increased heme catabolism, down-regulation of cytosolic ferritin, and increased expression of molecules that could enhance cell and mitochondrial iron uptake (Sec15l1 and Mfrn2, respectively). This study is important for deciphering the role of iron in the pathogenesis of Friedreich's ataxia.
Materials and Methods
Animals.
Mutants homozygous for deletion of Frda exon 4 (Fig. 4 A and B) and WT mice were used and genotyped as described previously (7). All animal work was approved by the University of Sydney's Animal Ethics Committee.
Microarray Processing.
Total RNA was isolated from hearts of 8 female littermates: two 4-week-old WTs, two 4-week-old mutants, two 10-week-old WTs, and two 10-week-old mutants. Total RNA was isolated using TRIzol (Invitrogen). First-strand cDNA synthesis and biotin-labeled cRNA were performed and hybridized to the mouse Affymetrix GeneChip 430 2.0 (39).
Microarray Data Analysis.
A 2-phase strategy was used to identify differentially expressed genes (39). First, genome-wide screening was performed using Affymetrix GeneChips. Then, low-level analysis was performed with Affymetrix GeneChip Operating Software 1.3.0, followed by the GC robust multiarray average (GCRMA) method for background correction and quantile–quantile normalization of expression. Tukey's method for multiple pairwise comparisons was applied to acquire fold-change estimations. Tests for significance were calculated and adjusted for multiple comparisons by controlling the false discovery rate at 5% (40).
Definitive evidence of differential expression was obtained from RT-PCR assessment of samples used for the microarray analysis and at least 3 other independent samples. Principal component analysis was performed by standard methods (25). Functional annotation of genes was assigned via Gene Ontology (http://www.geneontology.org) and classifications obtained through DAVID (http://david.abcc.ncifcrf.gov).
RT-PCR and Western Blot Analysis.
RT-PCR was performed (11) using the primers listed in Table S4. Western blot analysis was performed (11) using antibodies against frataxin (US Biological); Tfr1 (Invitrogen); Fpn1 (D. Haile, University of Texas Health Science Center); Hmox1 (AssayDesigns); Sdha, Gapdh, and Iscu1/2 (Santa Cruz Biotechnology); Fech (H. Dailey, University of Georgia, Biomedical and Health Sciences Institute); Hfe2 (S. Parkkila, University of Tampere, Institute of Medical Technology); Nfs1, Uros, and Alad (Abnova); Sec15l1 (N.C. Andrews, Duke University); Ftl1, Fth1, Ftmt (S. Levi, San Raffaele Institute); and Hif1α (BD Biosciences).
Heme Assay.
Hearts were exhaustively perfused and washed with PBS (0.2% heparin at 37 °C) to remove blood. After homogenization, heme was quantified using the QuantiChrom Heme Assay (BioAssay Systems).
Tissue Iron Stores.
Tissue iron was measured via inductively coupled plasma atomic emission spectrometry (39).
Statistical Analysis.
Data were compared using the Student t-test. Data were considered statistically significant when P < .05. Results are expressed as mean ± SD or mean ± SEM.
Supplementary Material
Acknowledgments.
We thank H. Puccio and M. Koenig (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for providing the MCK mice. This work was supported by grants from the National Health and Medical Research Council of Australia, Muscular Dystrophy Association USA, Friedreich's Ataxia Research Association USA, Friedreich's Ataxia Research Association Australia, and the Canadian Institutes of Health Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906784106/DCSupplemental.
References
- 1.Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondrion: Novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 2.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAATTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 3.Babcock M, et al. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 4.Rotig A, et al. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, et al. Mrs3p, Mrs4p, and frataxin provide iron for Fe-S cluster synthesis in mitochondria. J Biol Chem. 2006;281:22493–22502. doi: 10.1074/jbc.M604246200. [DOI] [PubMed] [Google Scholar]
- 6.Wilson RB, Roof DM. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat Genet. 1997;16:352–357. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 7.Puccio H, et al. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 8.Becker EM, Greer JM, Ponka P, Richardson DR. Erythroid differentiation and protoporphyrin IX down-regulate frataxin expression in Friend cells: Characterization of frataxin expression compared to molecules involved in iron metabolism and hemoglobinization. Blood. 2002;99:3813–3822. doi: 10.1182/blood.v99.10.3813. [DOI] [PubMed] [Google Scholar]
- 9.Lesuisse E, et al. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1) Hum Mol Genet. 2003;12:879–889. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 10.Li K, Besse EK, Ha D, Kovtunovych G, Rouault TA. Iron-dependent regulation of frataxin expression: Implications for treatment of Friedreich ataxia. Hum Mol Genet. 2008;17:2265–2273. doi: 10.1093/hmg/ddn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitnall M, et al. The MCK mouse heart model of Friedreich's ataxia: Alterations in iron-regulated proteins and cardiac hypertrophy are limited by iron chelation. Proc Natl Acad Sci USA. 2008;105:9757–9762. doi: 10.1073/pnas.0804261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber J, Lill R. Biogenesis of iron-sulfur proteins in eukaryotes: Components, mechanism and pathology. Mitochondrion. 2002;2:71–86. doi: 10.1016/s1567-7249(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 13.Land T, Rouault TA. Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol Cell. 1998;2:807–815. doi: 10.1016/s1097-2765(00)80295-6. [DOI] [PubMed] [Google Scholar]
- 14.Foury F. Low iron concentration and aconitase deficiency in a yeast frataxin homologue–deficient strain. FEBS Lett. 1999;456:281–284. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- 15.Gerber J, Muhlenhoff U, Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutz T, Westermann B, Neupert W, Herrmann JM. The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol. 2001;307:815–825. doi: 10.1006/jmbi.2001.4527. [DOI] [PubMed] [Google Scholar]
- 17.Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet. 2002;11:2025–2036. doi: 10.1093/hmg/11.17.2025. [DOI] [PubMed] [Google Scholar]
- 18.Stehling O, Elsasser HP, Bruckel B, Muhlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: Evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 19.Bencze KZ, et al. Human frataxin: Iron and ferrochelatase binding surface. Chem Commun. 2007;14:1798–1800. doi: 10.1039/b703195e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Lyver ER, Knight SA, Lesuisse E, Dancis A. Frataxin and mitochondrial carrier proteins, Mrs3p and Mrs4p, cooperate in providing iron for heme synthesis. J Biol Chem. 2005;280:19794–19807. doi: 10.1074/jbc.M500397200. [DOI] [PubMed] [Google Scholar]
- 21.Schoenfeld RA, et al. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum Mol Genet. 2005;14:3787–3799. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- 22.Levi S, et al. A human mitochondrial ferritin encoded by an intronless gene. J Biol Chem. 2001;276:24437–24440. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 23.Nie GJ, Sheftel AD, Kim SF, Ponka P. Overexpression of mitochondrial ferritin causes cytosolic iron depletion and changes cellular iron homeostasis. Blood. 2005;105:2161–2167. doi: 10.1182/blood-2004-07-2722. [DOI] [PubMed] [Google Scholar]
- 24.Sutak R, et al. Proteomic analysis of hearts from frataxin knockout mice: Marked rearrangement of energy metabolism, a response to cellular stress and altered expression of proteins involved in cell structure, motility and metabolism. Proteomics. 2008;8:1731–1741. doi: 10.1002/pmic.200701049. [DOI] [PubMed] [Google Scholar]
- 25.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, et al. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandolfo M, Pastore A. The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J Neurol. 2009;256:9–17. doi: 10.1007/s00415-009-1003-2. [DOI] [PubMed] [Google Scholar]
- 28.Dunn LL, Suryo-Rahmanto Y, Richardson DR. Iron uptake and metabolism in the new millennium. Trends Cell Biol. 2007;17:93–100. doi: 10.1016/j.tcb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Lim JE, et al. A mutation in Sec15l1 causes anemia in hemoglobin-deficit (hbd) mice. Nat Genet. 2005;37:1270–1273. doi: 10.1038/ng1659. [DOI] [PubMed] [Google Scholar]
- 30.Zhang AS, Sheftel AD, Ponka P. The anemia of “haemoglobin-deficit” (hbd/hbd) mice is caused by a defect in transferrin cycling. Exp Hematol. 2006;34:593–598. doi: 10.1016/j.exphem.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Saxena S, Pain D, Dancis A. Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J Biol Chem. 2001;276:1503–1509. doi: 10.1074/jbc.M007198200. [DOI] [PubMed] [Google Scholar]
- 32.Barros MH, Carlson CG, Glerum DM, Tzagoloff A. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 2001;492:133–138. doi: 10.1016/s0014-5793(01)02249-9. [DOI] [PubMed] [Google Scholar]
- 33.Ganz T. Hepcidin and its role in regulating systemic iron metabolism. Hematol Am Soc Hematol Educ Program. 2006:29–35. doi: 10.1182/asheducation-2006.1.29. [DOI] [PubMed] [Google Scholar]
- 34.Adams ML, Ostapiuk I, Grasso JA. The effects of inhibition of heme synthesis on the intracellular localization of iron in rat reticulocytes. Biochim Biophys Acta. 1989;1012:243–253. doi: 10.1016/0167-4889(89)90104-3. [DOI] [PubMed] [Google Scholar]
- 35.Ponka P, Wilczynska A, Schulman HM. Iron utilization in rabbit reticulocytes: A study using succinylacetone as an inhibitor or heme synthesis. Biochim Biophys Acta. 1982;720:96–105. doi: 10.1016/0167-4889(82)90043-x. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen BK, Febbraio MA. Muscle as an endocrine organ: Focus on muscle-derived interleukin-6. Physiol Rev. 2008;88:1379–1406. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- 37.Kobune M, Kohgo Y, Kato J, Miyazaki E, Niitsu Y. Interleukin-6 enhances hepatic transferrin uptake and ferritin expression in rats. Hepatology. 1994;19:1468–1475. [PubMed] [Google Scholar]
- 38.Castelnau PA, et al. Abnormal iron deposition associated with lipid peroxidation in transgenic mice expressing interleukin-6 in the brain. J Neuropathol Exp Neurol. 1998;57:268–282. doi: 10.1097/00005072-199803000-00008. [DOI] [PubMed] [Google Scholar]
- 39.Dunn LL, Sekyere EO, Rahmanto YS, Richardson DR. The function of melanotransferrin: A role in melanoma cell proliferation and tumorigenesis. Carcinogenesis. 2006;27:2157–2169. doi: 10.1093/carcin/bgl045. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.