Abstract
Alternatively spliced brain-derived neurotrophic factor (BDNF) transcripts are targeted to distinct cellular compartments in neurons but the mechanisms underlying this sorting are unknown. Although only some BDNF isoforms are targeted to dendrites, we have found that the coding region common to all BDNF transcripts contains a constitutively active dendritic targeting signal and that this signal is suppressed in transcripts containing exons 1 or 4, which are restricted to the cell soma and proximal dendrites. This dendritic targeting signal is mediated by translin, an RNA-binding protein implicated in RNA trafficking, and is disrupted by the G196A mutation associated with memory deficits and psychiatric disorders. Molecular modeling and mutational studies indicate that the G196A mutation blocks dendritic targeting of BDNF mRNA by disrupting its interaction with translin. These findings implicate abnormal dendritic trafficking of BDNF mRNA in the pathophysiology of neuropsychiatric disorders linked to the G196A mutation.
Keywords: neuropsychiatric disorders, neurotrophins
Several lines of evidence indicate that targeting of BDNF mRNA to dendrites plays a key role in mediating synaptic plasticity (1–4). However, the molecular mechanisms regulating this process and the differential subcellular localization of alternatively spliced BDNF transcripts, remain to be clarified.
Multiple BDNF transcripts are generated by alternative splicing of one 5′ exon with a shared 3′ exon containing the entire BDNF coding region and either a short or long 3′ UTR sequence (5, 6). In recent studies, we have demonstrated that BDNF transcripts differ in their subcellular localization (7). Exon 1 and 4 transcripts are localized in the cell soma, while exon 2 and 6 transcripts show a somato-dendritic localization. Thus, splice variants appear to encode spatial localization signals used to preferentially regulate BDNF expression in different subcellular domains (2, 3). A recent study has suggested that the long 3′ UTR contains signals necessary for dendritic targeting of BDNF transcripts (4). However, it is unlikely that this mechanism can fully account for the differential dendritic targeting displayed by BDNF transcripts because more than one-third of exon 4 transcripts, which are retained in the soma, contain the long 3′ UTR. Conversely, more than one-half of exon 6 transcripts, an isoform that displays targeting to dendrites, contain the short 3′ UTR. To help define the mechanisms underlying differential localization of BDNF transcripts, we have tested the hypothesis that additional signals might be encoded by other BDNF mRNA regions.
Results
Differential Localization of BDNF Isoforms.
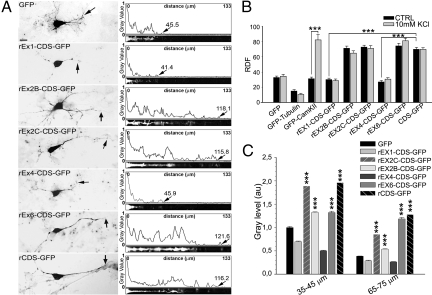
To test for the presence of dendritic targeting elements in the 5′ UTRs, we cloned the rat BDNF coding sequence (rCDS), upstream from and in-frame with the green fluorescent protein (GFP) reporter gene, either alone or preceded by one of the five most abundant rat BDNF 5′ UTR sequences (rEx1, rEx2B, rEx2C, rEx4, and rEx6) (5). Upon translation, all BDNF-GFP constructs had the expected molecular size of 54 kDa. The subcellular localization of GFP chimaeric constructs was detected by in situ hybridization with an antisense riboprobe highly specific for GFP (Fig. 1Aand Fig. S1). Control constructs had the expected localization and consisted of GFP alone (proximal dendritic localization), GFP-α-tubulin (somatic localization) (8) and GFP-α subunit of Ca2+/calmodulin-dependent protein kinase II (activity-dependent targeting to dendrites) (9). (Fig. S1). The dendritic distribution of BDNF transcripts was quantified in two ways. First, the relative dendritic filling index (RDF) was determined by the distance from the soma at which the in situ signal reached and remained below background levels for the remainder of the dendrite (MDDL = maximal distance of dendritic labeling) (10) divided by the length of the dendrites as determined by MAP2 immunostaining (Fig. S1). Second, we also carried out a densitometric analysis of the labeling intensity along the entire extent of the dendrite.
Fig. 1.
Dendritic mRNA localization of BDNF-GFP chimaeras. (A) In situ labeling of neurons transfected with GFP or BDNF-GFP transcripts. Arrows show the MDDL. (Scale bar, 10 μm.) Densitometric traces of linearized dendrites (ImageJ-straighten) are shown. (B) rEx2B-CDS-, rEx2C-CDS-, rEx6-CDS-, and rCDS-GFP show distal dendritic localization and comparable RDF. rEx1- and rEx4-CDS-GFP display proximal dendritic localization with lower RDF values than rCDS-GFP (***, P < 0.001). Depolarization had no effects, except for CaMKII-GFP (***, P < 0.001). (C) Densitometric analysis shows a rapid decline to background for rEx1-CDS-GFP and rEx4-CDS-GFP while the other chimaeric mRNAs remained elevated at 35–45 or 65–75 μm from the soma (***, P < 0.001 vs. rEx1-CDS-GFP). Error bars, SEM.
Analysis of the chimaeric BDNF constructs by the RDF method indicated that they can be divided into two groups. rEx1-CDS-GFP and rEx4-CDS-GFP constructs were localized within the first 30% of the dendritic compartment (RDF = 30.12 ± 1.93, n = 122; RDF = 26.98 ± 1.57, n = 149, respectively, difference with GFP-tubulin P < 0.001; not statistically different from GFP; Fig. 1B). In contrast, labeling for rEx2B-CDS-GFP, rEx2C-CDS-GFP, and rEx6-CDS-GFP extended into distal dendrites (Ex2B = 71.36 ± 2.84, n = 196; Ex2C = 72.82 ± 2.64, n = 174; Ex6 = 74.56 ± 3.12, n = 175, respectively). Remarkably, the localization of these constructs parallels that of the corresponding endogenous transcripts in vivo even though they lack the BDNF 3′ UTR (7). Thus, these findings are consistent with the presence of dendritic targeting signals in exons 2 and 6. However, surprisingly, rCDS-GFP shows a comparable distal localization (RDF = 69.97 ± 2.38, n = 151; P < 0.001 vs. GFP, rEx1 and rEX4-rCDS-GFP). This unexpected finding indicates that, conversely, the CDS contains a dendritic targeting signal that is suppressed by exons 1 and 4. These results were corroborated by using the alternate quantification method, densitometric analysis at regular intervals along the dendrites. At 75 μm from the soma, the in situ signals for isoforms 2B, 2C, 6, and the CDS were significantly higher than those obtained for exons 1 and 4 (Fig. 1C). Of note, KCl did not affect the localization of any of the BDNF-GFP chimaeric mRNAs or GFP-tubulin (Fig. 1C). However, as expected, it did induce a robust increase in the RDF value of the control GFP-CaMKII construct from 30% (RDF = 31.27 ± 2.08 under basal conditions, n = 93) to 80% of the dendritic length after KCl treatment (RDF = 82.49 ± 6.68, n = 68; KCl vs. unstimulated P < 0.001). Bath application of NT3 or BDNF (50 ng/mL) for 3 h, known to enhance mRNA trafficking into dendrites (10–12), did not affect the dendritic localization of rCDS-GFP or GFP mRNAs (Fig. S2). In addition, using the MS2 RNA-tagging system, we confirmed that dendritic trafficking of rCDS labeled granules is not affected by KCl treatment (Fig. S3). Most MS2-rCDS granules (75%) had anterograde or retrograde movements with average velocity 0.12 μm/s ± 0.018 (max 0.26 μm/s; min. 0.07 μm/s), while the remaining 25% oscillated around a fixed position. In summary, the rat BDNF coding region appears to contain a constitutively active dendritic targeting signal that is not responsive to electrical activity, NT3 or BDNF and is suppressed by exons 1 or 4.
To rule out the possibility that the apparent differences in the extent of dendritic targeting of various BDNF constructs might be due to their ability to cause dendritic growth (13), we performed Sholl analysis on 8DIV hippocampal neurons 24 h after transfection with either GFP alone, BDNF CDS-GFP, small interfering (si)RNA against BDNF, or bath application of either BDNF (50 ng/mL) or the Trk signal transduction inhibitor K252α (10 nM). None of these treatments caused a statistically significant modification of the dendrogram up to 150 μm from the cell soma (Fig. S4). Also, densitometric analysis on cell somas of transfected neurons showed equal expression levels of these transcripts (Fig. S4).
Constitutive Dendritic Targeting Signal in the Coding Region.
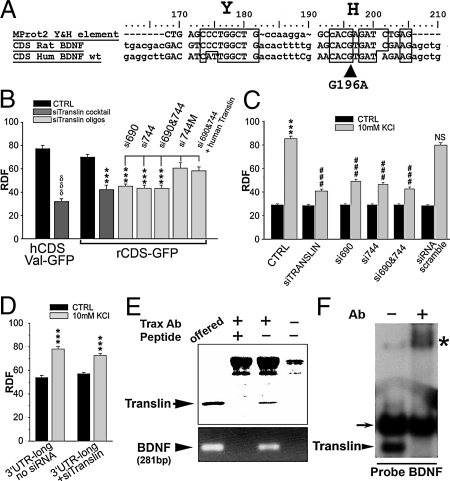
To help identify the dendritic targeting signal(s) present in the coding region, the rat and human sequences were scanned with BIOEDIT and ClustalW software to search for conserved recognition sequences for RNA binding proteins involved in dendritic RNA targeting. This sequence analysis revealed the presence of a strikingly conserved, bipartite recognition site (rBDNF CDS nucleotides 173–181 and 193–205; Fig. 2A), for the single stranded DNA/RNA binding protein translin, also known as testis brain-RNA binding protein (TB-RBP) (14, 15). These two elements are 70% and 80% identical to the rat Protamine-2 Y' and H' elements, respectively. Of note, the position of the H element within the human BDNF sequence overlaps the location of the G196A (Val66Met) mutation (16) (arrow in Fig. 2A), which has been shown to impair dendritic localization of BDNF and is associated with cognitive impairment and neuropsychiatric disorders (17). To determine whether this region is required for dendritic trafficking of the CDS-GFP chimaeric construct, we tested two deletion chimaeras: Δ (68–212)-rCDS-GFP and Δ (1–202)-rCDS-GFP. Both constructs had somatic expression levels that did not differ from that of GFP and displayed RDF values lower than that of the intact rCDS-GFP sequence (P < 0.001; Fig. S5), suggesting that these deletions disrupt the constitutive dendritic targetic signal present in the CDS.
Fig. 2.
Role of translin in BDNF mRNA targeting. (A) Conserved translin-binding Y and H elements in human and rat BDNF and mouse Protamine2. (B) Inhibition of the rat and human CDS-GFP RDF values with an siRNA translin mixture (rat ***, P < 0.001; human δδδ, P < 0.001 vs. control), or two siRNA oligos used either alone or in combination (si690, si744), mutated si744 (si744M) had no effect. Inhibition of CDS-GFP dendritic targeting by si690 and si744 was rescued by co-transfection with siRNA-insensitive human translin. (C) Endogenous BDNF mRNA dendritic targeting up-regulation by KCl (***, P < 0.001 Crtl+KCl vs. Crtl), is inhibited by translin siRNA (###, P < 0.001 si-translin+KCl vs. Crtl+KCl), or si690 and si744, but not by scrambled siRNA (NS, not significant). (D) GFP-3′ UTRlong displays an activity-dependent targeting to dendrites in response to 3 h 10 mM KCl that is not affected by si-translin. (E) (Upper) trax antibody immunoprecipitates Translin (arrowhead). Coprecipitation of translin is blocked by omission of Trax antibody or preincubation with its antigen peptide. (Lower) RT-PCR analysis of the immunoprecipitated RNA:protein complexes. BDNF mRNA was detected only in the “offered” sample and the IP pellet containing translin. (F) A radiolabelled segment of the human CDS region containing the Y and H elements (nucleotides 176–211) forms two gel-shift bands after incubation with rat brain extracts. The lower band (arrowhead “Translin”) is “supershifted” with Trax antibody (marked with *). Upper band (small arrow) reflects binding of this probe with an unrelated complex.
Based on these encouraging observations, we proceeded to assess the involvement of translin in dendritic targeting of BDNF mRNA by using an siRNA “cocktail” against translin (si-translin), generated by digestion of a 300-bp fragment of translin (Fig. 2B). Si-translin produced 72% down-regulation of endogenous translin mRNA and 73% reduction of translin protein in hippocampal cultures without interfering with control GAPDH RNA or tubulin protein expression (Fig. S6). Dendritic targeting of both rat and human BDNF CDS-GFP constructs was significantly reduced by si-translin (P < 0.001; Fig. 2B) leading to RDF values close to that of GFP alone (GFP RDF = 32.06 ± 3.12, n = 58). Similar effects on dendritic targeting of transfected BDNF rCDS-GFP, were obtained by silencing translin with two defined siRNA oligo sequences used either alone or in combination (si690, si744; Fig. 2B), that we had found silenced endogenous translin in rat cortical cultures (Fig. S6). In contrast, a scrambled siRNA oligo generated by inserting multiple mismatches into si744 (si744M) had no effect on BDNF rCDS-GFP dendritic targeting (Fig. 2B). As human translin contains multiple mismatches with both si690 and si744, we assessed whether it would reverse their inhibitory effects. As expected, cotransfection of human translin with si690 and si744 rescued the constitutive dendritic targeting of the BDNF rCDS-GFP construct (Fig. 2B), confirming that the inhibitory effect of these siRNA oligos is due to silencing endogenous translin. Taken together, these studies demonstrate that translin mediates the constitutive dendritic targeting signal located in the CDS and this targeting mechanism is conserved in rat and human BDNF mRNAs.
As the CDS targeting signal was identified with an artificial, truncated BDNF construct, its physiological relevance was still unclear. Accordingly, we examined translin's role in dendritic targeting of endogenous, full-length BDNF mRNA. To this end, primary hippocampal cell cultures were transfected with si-translin or a control, scrambled, siRNA and then depolarized with 10 mM KCl for 3 h. In accordance with previous studies (4), depolarization induced a strong RDF increase for endogenous full-length BDNF mRNA (from CTRL = 29.18 ± 0.74 =, to 85.37 ± 2.18 = 10 mM KCl; P < 0.001). However, in the presence of si-translin and KCl, the RDF only increased to 40.85 ± 1.6 (P < 0.001 with respect to KCl alone), while a scrambled siRNA had no significant effect (Fig. 2C). Similarly, activity-induced dendritic targeting of endogenous BDNF mRNA was abolished by transfecting neurons with si690 and si744 either alone or in combination (Fig. 2C) Of note, si-translin did not block the ability of KCl to induce dendritic trafficking of the long 3′ UTR fragment of BDNF mRNA that encodes an electrical activity-dependent targeting signal (Fig. 2D). Taken together, these results suggest that the constitutive dendritic targeting signal located in the CDS, which depends on translin, also plays a critical role in mediating the inducible targeting of full-length BDNF mRNA into distal dendrites.
Translin Binds to BDNF mRNA.
As these results suggest that endogenous BDNF mRNA binds to translin in vivo, we proceeded to test this prediction in immunoprecipitation experiments. Because translin forms a heteromeric complex with its partner protein, trax (translin-associated factor x), we used trax antibodies to immunoprecipitate the translin/trax complex (15) from rat forebrain homogenates and detected BDNF mRNA by RT-PCR in the immunoprecipitates (Fig. 2E). Omission of the antibody or its preincubation with the antigen peptide blocked immunoprecipitation of both translin and BDNF mRNA. Thus, these results indicate that BDNF mRNA is associated with the translin/trax complex in vivo. To check whether translin binds to the segment of the CDS containing adjacent Y- and H-like elements, we performed gel-shift studies with forebrain homogenates using this segment as probe (Fig. 2F). These studies confirmed that endogenous translin/trax complex binds to this sequence.
Translin Binding to BDNF mRNA: Role of G196.
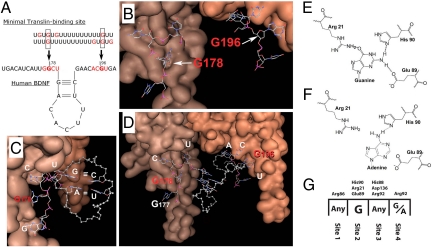
Previous studies demonstrated that guanine residues play a critical role in conferring high-affinity binding of RNA oligos to the translin/trax complex (18) (Fig. 3A). To understand how translin binds to BDNF mRNA, we modeled the interaction between the 3D structure of human translin, obtained from the Protein Data Bank (PDB entry 1j1j), and the segment of human BDNF mRNA containing the putative translin binding site and the G196 residue. The interaction of a human translin dimer with this RNA sequence was simulated by incremental construction of the RNA in the binding site of the protein. Accordingly, we first searched for potential binding sites on a translin monomer using the GRID algorithm (see SI Methods). This analysis revealed that the most relevant binding clefts are delimited by the charged amino acid residues 21, 86, 88, 90, 92, and 136, which is in striking agreement with the residues previously identified as critical for binding in mutational studies of translin (19). The second step involved calculating the 3D structure of the complex formed between the RNA nitrogen bases and the translin binding clefts identified in the previous step. The GRID docking algorithm GLUE identified four adjacent binding sites for individual ribonucleotide bases on each translin monomer: the first one is mainly formed by Arg-86, the second one is delimited by His-90, Arg-21, and Glu-89, and the third and fourth sites are less distinct, as they are globally delimited by His-88, Asp 136, and Arg 92 (Fig. 3 B–D and G). While the first and third sites do not show a strong preference among the four standard nucleotides (<0.2 kcal/mol, as calculated by the force field; see SI Methods for details), the second binding site is extremely selective for guanine. This finding fits well with previous binding studies that identified G residues as being critical for ligand recognition (18) (Fig. 3 B–D). In fact, when we simulated the binding of a G residue in site 2, a rigid network of three hydrogen bonds was formed (Fig. 3 E–G). In contrast, the hydrogen bond network formed by Arg-21, His-90, and Glu-89 in translin is almost completely lost when adenine is substituted for guanine, causing an energy penalty of 6.8 kcal/mol. This value is consistent with the loss of two strong hydrogen bonds. The fourth binding site shows a preference for either purine base since the energy difference calculated for the binding of guanine or adenine is modest (0.7 kcal/mol). Binding of pyrimidines is still possible, although the energy penalty is around 2 kcal/mol. Next, we modeled the interaction between a translin dimer and a 48-nt-long sequence spanning the H' and Y' elements of human BDNF mRNA (see Fig. 2A) After attempting to dock both 5′-3′ and 3′-5′ orientations in the binding site, we found that only the orientation shown in Fig. 3 B–D is compatible with continuity of the RNA oligos. In this conformation, the RNA strand forms an “M”-like configuration. The central part of the M is likely to form a stem loop structure consistent with the prediction of the mfold program (Fig. 3 C and D), whereas each of the external bars of the M (ascending and descending, respectively) interacts with the four-nucleotide binding site present on each translin monomer. A striking feature of this model is that it places G196, which is located in one of these external bars, in the highly selective site 2 of one of the translin monomers and, therefore, raises the possibility that the G196A SNP would impair binding of this BDNF oligo to translin. Another noteworthy aspect of the model is that the stem structure contains two stable Watson and Crick pairings A-U and G-C (Fig. 3 C and D). See Fig. S7 for other views of the translin-RNA complex.
Fig. 3.
Three-dimensional modeling of translin dimer binding to human BDNF mRNA. (A) Minimal consensus RNA sequence for binding to the translin complex with two critical G residues separated by a poly U spacer of variable length. In human BDNF, spacing of the relevant G residues (G178 and G196, in bold) is constrained by formation of a stem-loop. (B and C) Interaction of G178 and G196 and position of the stem-loop on BDNF mRNA with a translin dimer (monomers in two different brown shades). (D) Modeling of the eight BDNF mRNA nucleotides interacting with a translin dimer. (E) Translin amino acids His-90, Arg-21, Glu-89 form three hydrogen bonds with G196 (normal BDNF) but only one with A196 (mutated BDNF, in F). (G) Summary of the nucleotide:amino acid selectivity for each of the four binding sites on translin.
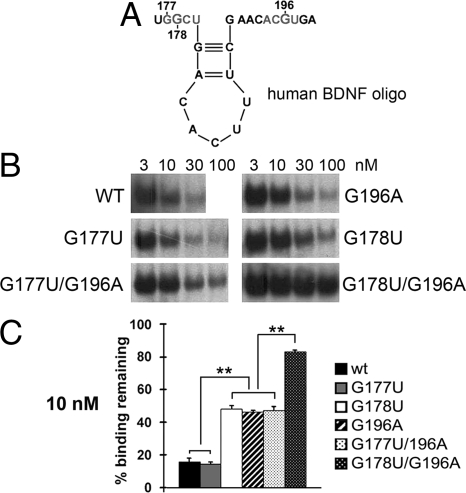
To test the prediction made by in silico 3D modeling that the G196A SNP impairs binding of the BDNF oligo to the translin/trax complex, we performed gel-shift competition assays with a wild-type human BDNF oligo spanning nucleotides 176–199 and the G196A version of the same segment. Since the docking simulation also predicts that G178 binds to the highly selective site 2 on the other translin monomer, we also examined the impact of mutating this G residue, as well as its neighbor G177 (Fig. 4A). While 10 nM of wild type or G177U BDNF RNA was sufficient to displace about 85% of a labeled rat protamine-2 oligo probe containing the translin binding site, the mutated G196A or G178U oligos were able to displace only 50% of the protamine-2 probe, as would be expected if these oligos have lower affinity for the complex (Fig. 4 B and C). In agreement with the model, mutating both these key G residues, G178 and G196, almost completely abolished the ability of this oligo to inhibit binding of the protamine-2 probe. In contrast, mutating G177 and G196 did not decrease its inhibitory activity below of that of the single G196A mutation (Fig. 4 B and C). Taken together, these competition data strongly support the in silico modeling that predicts G178 and G196 to be the crucial residues for binding of BDNF mRNA to translin.
Fig. 4.
G178 and G196 mediate binding to translin. (A) Stem-loop structure and the nucleotides (in gray) interacting with translin in the oligo used for these experiments. (B) In competition gel-shift assays, significant loss in inhibitory potency is produced by the G178U and G196A substitutions. The double mutation G178U/G196A markedly decreases its ability to compete with the WT Prm-2 probe. The G177U mutant does not differ from WT, and the inhibitory activity of the G177U/G196A double mutant equals that of G196A mutant. (C) Quantification of the gel-shift competition assay. Bars, means ± SEM. **, P < 0.01 (paired t test).
G196A Mutation Impairs Dendritic Targeting of BDNF mRNA.
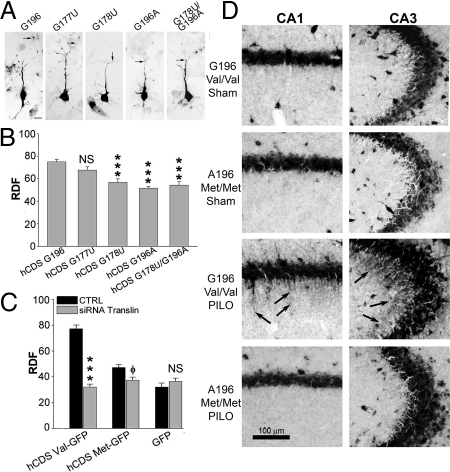
Because translin mediates the CDS dendritic targeting signal and the G196A SNP impairs translin binding to this segment, these findings imply that the G196A SNP will disrupt dendritic targeting of the CDS. To test this prediction, primary rat hippocampal neurons were transfected, with the human BDNF-GFP chimeras containing either the normal G196 allele (Val) or the mutated G196A (Met) version. In addition, we tested whether mutations at G177 and G178 residues would exert similar effects on BDNF mRNA targeting as they do on binding to the translin complex. As predicted, we found that dendritic localization of the G196A form of human BDNF transcripts (hCDS Met-GFP) was markedly reduced compared with the normal allele (P < 0.001, Fig. 5B). Consistent with the competitive binding experiments, G178U, as well as combined mutation of both G178 and G196, also decreased CDS dendritic localization. Of note, G196A, G178U, and G196A/G178U showed decreases of similar magnitude (≈30% decrease), suggesting that changing either of these key G residues is sufficient to achieve the maximal possible impairment of the translin-mediated dendritic targeting of the CDS-GFP construct (Fig. 5 A and B; P < 0.001). In contrast, G177U, which does not impair binding to translin, also did not affect targeting of the human BDNF CDS (Fig. 5B). Of note, si-translin has little further inhibitory effect on targeting of the G196A construct (hCDS Met-GFP), even though it strongly inhibits either rat or human wild-type CDS. (Fig. 5C). The marked difference in dendritic targeting noted between G196 (Val) and G196A (Met) BDNF constructs was not due to their effects on the degree of arborization of transfected neurons, because we did not find any difference in the total length of processes of the in situ positive neurons 24 h posttransfection with mutant or normal human BDNF constructs (hCDS Val-GFP = 186.35 ± 2.05 μm; hCDS Met-GFP = 184 ± 6.65 μm).
Fig. 5.
Impact of human G196A mutation on BDNF mRNA targeting. (A) In situ hybridization on hippocampal neurons transfected with human BDNF CDS-GFP with mutations at G177,G178 and G196. Arrows show the MDDL. (Scale bar, 10 μm.) (B) hCDS-G178U-, hCDS-G196A-, and hCDS-G178U/G196A-GFP mRNAs show lower RDF values than hCDS-G196-GFP (***, P < 0.001), hCDS-G177U had no effect (NS). (C) Translin silencing impairs dendritic targeting of human BDNF CDS mRNA (***, P < 0.001) but has a smaller effect on hCDS Met-GFP (phi symbol, P < 0.05 vs. hCDS Met-GFP without siRNA). Error bars, SEM. (D) In situ hybridization for BDNF mRNA on hippocampal sections from wt (Val/Val) and Met/Met knock-in mice. In saline-treated mice, BDNF mRNA is confined to the cell body layers of CA1 (Left) and CA3 (Right) in both wt and Met/Met mice (Upper). Upon pilocarpine treatment for 3 h (PILO), BDNF mRNA labeling is apparent in dendritic regions (stratum radiatum) of CA1 and CA3 neurons in Val/Val mice, but not in Met/Met mutant mice (Lower). (Scale bar, 100 μm.)
In the next set of experiments, we used G196A knock-in mice (20) to investigate whether this mutation also affects BDNF mRNA dendritic targeting in vivo. In adult G196 (Val/Val) mice, BDNF mRNA is targeted to distal dendrites of hippocampal neurons upon stimulation with the proepileptic drug pilocarpine (Fig. 5D), in agreement with previous studies done in rats (21). Although Val/Val and Met/Met mice showed comparable seizure activity in response to pilocarpine, dendritic targeting of BDNF mRNA in hippocampal neurons was abolished in G196A (Met/Met) mice (Fig. 5D). Thus, these findings indicate that the inhibitory effect of G196A on dendritic targeting is not limited to the truncated CDS transcript but also applies to endogenous, full-length BDNF mRNA.
Discussion
This study provides compelling evidence that the translin/trax complex mediates dendritic targeting of BDNF mRNA. We found that the BDNF coding region common to all BDNF splice variants is sufficient to direct constitutive targeting of BDNF mRNAs to the distal dendritic compartment. Characterization of this signal has demonstrated that it: (i) can be overridden by the 5′ UTRs of BDNF mRNA isoforms (exon 1 and 4) that are retained in the cell body and proximal dendrites, (ii) is mediated by translin, and (iii) is blocked by the G196A mutation. Furthermore, several lines of evidence indicate that the G196A SNP blocks dendritic targeting of BDNF mRNA by disrupting its interaction with the translin/trax complex. First, an RNA oligo spanning the G196 position binds with high affinity to the translin/trax complex and insertion of the G196A SNP inhibits their interaction. Second, computer modeling of the binding of this oligo to a translin dimer identified a second G residue (G178) as a critical nucleotide for binding to the complex, while the adjacent G residue (G177) is not. Mutation of G178, but not G177, inhibits binding to the translin complex and also markedly impairs dendritic targeting of the CDS-GFP construct. The similar effects of G178A and G196A mutations on translin binding affinity and RNA transport imply that the G196A substitution impairs dendritic trafficking of BDNF mRNA through the disruption of its interaction with translin.
Dendritic trafficking of BDNF transcripts displays two key features: activity dependence and transcript selectivity. Although we showed that 3′ UTR plays a critical role in mediating activity-dependent trafficking of BDNF transcripts, this segment is unlikely to mediate transcript selectivity because both the short and long forms of the 3′ UTR are contained in BDNF transcripts that are retained in the soma/proximal dendrites, as well as those targeted to distal dendrites. Our results indicate that transcript selectivity is mediated by a constitutively active dendritic targeting element (DTE) located in the CDS that can be overridden by signals located in the 5′ UTR of transcripts that are retained in the soma/proximal dendrites, but not by those targeted to distal dendrites (Fig. 6). The ability of si-translin to block activity-dependent targeting of both the truncated CDS-GFP construct and endogenous, full-length transcripts, but not the isolated 3′ UTR, indicates that both a translin-dependent DTE in the CDS and a translin-independent DTE in the 3′ UTR are required to mediate activity-dependent trafficking of full-length BDNF transcripts. Thus, according to this scenario, even though the isolated 3′ UTR segment undergoes activity-dependent dendritic targeting, the 3′ UTR DTE is not sufficient to drive activity-dependent targeting of the full-length transcript. Further confirmation of this model is provided by our finding that the G196A SNP, which by definition acts selectively on the CDS DTE, blocks pilocarpine-induced dendritic trafficking of BDNF mRNA in vivo.
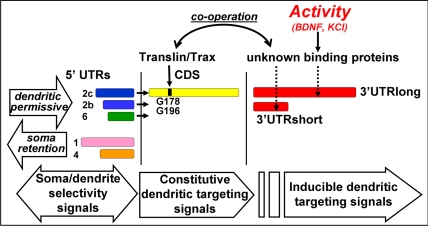
Fig. 6.
Proposed model of BDNF mRNAs sorting mechanisms. Multiple 5′ UTR sequences with either inhibitory or permissive targeting properties determine transcript-selective sorting. Translocation to dendrites is mediated by constitutive dendritic targeting signals in the CDS that require binding of the translin/trax complex at the G178-G196 residues. The 3′ UTR region(s) contain inducible dendritic targeting signals that respond to KCl and BDNF and cooperate with the constitutive signals in the CDS mediated by the translin/trax complex.
Our finding that the G196A SNP blocks dendritic trafficking of BDNF mRNA implies that phenotypic changes induced by this mutation, such as reduced hippocampal dendritic complexity and volume, as well as memory deficits (22, 23), and susceptibility to mood disorders (24) may be due to this effect. This evidence links defects in dendritic trafficking with susceptibility to eating and mood disorders, and reduced memory performance. From this perspective, it is also noteworthy that translin deletion in mice impairs performance in several behavioural paradigms used to assess learning, memory, and anxiety (25). However, it is important to emphasize that this deficit in subcellular BDNF mRNA sorting may not account completely for the biological deficits produced by this mutation. Recent studies have suggested that this SNP may also impair BDNF protein sorting by disrupting its interaction with sortilin, a vesicular membrane protein implicated in protein trafficking (26). Thus, conceivably, the G196A SNP interferes with BDNF processing by disrupting trafficking of both BDNF mRNA and protein through distinct mechanisms.
Methods
Chimaeric GFP Constructs.
Total RNA was extracted from whole rat brain using TriZol Reagent (Invitrogen). Total RNA was reverse-transcribed into cDNA and amplified with exon-specific forward and a common reverse primer pair (details in SI Methods). PCR fragments were cloned into pEGFP-N1 vector (Clontech).
MS2-Time-Lapse BDNF mRNA Trafficking.
The plasmid GFP-MS2-NLS encoding an MS2 phage coat protein in fusion with GFP and a nuclear localization sequence (NLS) was cotransfected with the CDS of rat BDNF linked with 6× multimerized MS2 binding site (6×BS) (27). The GFP-MS2-NLS fusion protein bound to the 6×BS insert was visualized in video time-lapse experiments in living neurons for 20 min at room temperature using a Sensicam cooled video camera (TPO) on a Nikon-S300 inverted microscope. Image stacks were analyzed with National Institutes of Health (NIH) ImageJ.
RNAi Interference.
RNAi “cocktails” against translin and CPEB1 were generated by RNAseIII (Ambion) cleavage of relatively long (≈300 nt) double-stranded RNA generated from T7-promoter containing PCR products (details in SI Methods). siRNA oligo sequences are described in SI Methods.
Cell Cultures.
Primary hippocampal neurons were prepared from P2 rats as described (10). SK-N-BE neuroblastoma cells were cultured in D-MEM supplemented with 10% FBS and antibiotics (Euroclone). Cells were transfected at 7 DIV with 1 μg plasmids or 19 nM siRNA mixture and 2 μL Lipofectamine 2000 (Invitrogen).
Animal Treatment.
Animals were treated according to the institutional guidelines in compliance with the European Council Directive 86/609 and NIH Guide for the Care and Use of Laboratory Animals. Two-month-old val/val or met/met mice (20) were pretreated with methylscopolamine (1 mg/kg, i.p.) 30 min before i.p. injection of 250 mg/kg pilocarpine or saline.
In Situ Hybridization, Immunocytochemistry, and Western Blotting.
In situ hybridization on brain sections or cultures was performed as described (10, 21). See SI Methods for details. For immunocytochemistry, cells were incubated with rabbit anti-MAP2 antibody (1:1,000, Roche) and anti-rabbit-FITC (1:100, Dako). For Western blots, hippocampal culture lysates were incubated with rabbit anti-translin (1:2,000) antibody or mouse monoclonal anti α-tubulin (1:20,000,Sigma).
Quantitative Imaging Analysis and Statistics.
Images of labeled cultures were acquired with a CCD camera (Nikon ADX-1200) on a Nikon E800 microscope and analyzed with Image-ProPlus (Media Cybernetics). The MDDL was determined as described (10), each MDDL measure was divided by the mean dendritic length to obtain the RDF (%RDF). Statistical analysis of the RDF data were performed using ANOVA on ranks, with Bonferroni post-hoc test.
Gel-Shift Competition Assay.
Gel-shift competitions were performed as described (18). For competition studies, 5 μg protein extract was incubated with unlabeled competitors. See SI Methods for details.
Immunoprecipitation and RT-PCR.
Immunoprecipitation experiments were performed as described (28) using anti-trax antibody. RNA was extracted from IP samples with TRIzol (Invitrogen), RT-PCR was performed as described above and BDNF was detected with specific primer pairs (5′-CCATGAAAGAAGCAAACGT-3′,5′-CTCCAGCAGAAAGAGCAGA-3′).
Secondary Structure Prediction and Modeling Methods.
The 1J1J structure of human translin was downloaded from the PDB repository (29). A single monomer was analyzed with the GRID program, using eight different probes (DRY, C3, OH, O, N, N:, N2, NH). Computer prediction of RNA secondary folding structures was performed using the Web MFOLD server. See SI Methods for details.
Supplementary Material
Acknowledgments.
We thank Dr. B. Lu for providing human val and met BDNF-GFP (National Institute of Child Health and Human Development, Bethesda), Dr. Y. Mori (Osaka University, Osaka) for CaMKII-GFP, Prof. S. Kindler (University of Hamburg, Hamburg, Germany) for tubulin-GFP, R. Fiorelli and F. Rapino for technical help, and Dr. D. Ginty for helpful discussions. This work was supported by PRIN (E.T.), National Alliance for Research on Schizophrenia and Depression (Z.L.), and National Institute on Drug Abuse (J.M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902833106/DCSupplemental.
References
- 1.Tongiorgi E. Activity-dependent expression of brain-derived neurotrophic factor in dendrites: Facts and open questions. Neurosci Res. 2008;61:335–346. doi: 10.1016/j.neures.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Horch HW. Local effects of BDNF on dendritic growth. Rev Neurosci. 2004;15:117–129. doi: 10.1515/revneuro.2004.15.2.117. [DOI] [PubMed] [Google Scholar]
- 3.Soule J, Messaoudi E, Bramham CR. Brain-derived neurotrophic factor and control of synaptic consolidation in the adult brain. Biochem Soc Trans. 2006;34:600–604. doi: 10.1042/BST0340600. [DOI] [PubMed] [Google Scholar]
- 4.An JJ, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Blichenberg A, et al. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- 9.Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- 10.Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Righi M, Tongiorgi E, Cattaneo A. Brain-derived neurotrophic factor (BDNF) induces dendritic targeting of BDNF and tyrosine kinase B mRNAs in hippocampal neurons through a phosphatidylinositol-3 kinase-dependent pathway. J Neurosci. 2000;20:3165–3174. doi: 10.1523/JNEUROSCI.20-09-03165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowles RB, Kosik KS. Neurotrophin-3 signals redistribute RNA in neurons. Proc Natl Acad Sci USA. 1997;94:14804–14808. doi: 10.1073/pnas.94.26.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 14.Kwon Y, Hecht N. Binding of a phosphoprotein to the 39 untranslated region of the mouse protamine 2 mRNA temporally represses its translation. Mol Cell Biol. 1993;13:6547–6557. doi: 10.1128/mcb.13.10.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Wu Y, Baraban JM. The Translin/Trax RNA binding complex: Clues to function in the nervous system. Biochim Biophys Acta. 2008;1779:479–485. doi: 10.1016/j.bbagrm.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freudenberg-Hua Y, et al. Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population. Genome Res. 2003;13:2271–2276. doi: 10.1101/gr.1299703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Baraban J. High affinity binding of the Translin/Trax complex to RNA does not require the presence of Y or H elements. Brain Res Mol Brain Res. 2004;120:123–129. doi: 10.1016/j.molbrainres.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Aoki K, Suzuki K, Ishida R, Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–366. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 20.Chen ZY, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tongiorgi E, et al. BDNF mRNA and protein are targeted to discrete dendritic laminae by event that trigger epileptogenesis. J Neurosci. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bath KG, Lee FS. Variant BDNF (Val66Met) impact on brain structure and function. Cogn Affect Behav Neurosci. 2006;6:79–85. doi: 10.3758/cabn.6.1.79. [DOI] [PubMed] [Google Scholar]
- 23.Pezawas L, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Stein JM, et al. Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J Neurosci. 2006;26:2184–2196. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZY, et al. Sortilin controls intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J Neurosci. 2005;25:6156–6166. doi: 10.1523/JNEUROSCI.1017-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 28.Finkenstadt PM, et al. Somatodendritic localization of Translin, a component of the Translin/Trax RNA binding complex. J Neurochem. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura I, et al. Structure of human translin at 2.2 A° resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:674–679. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.