Abstract
The DExD/H-box RNA-dependent ATPase Dbp5 plays an essential role in the nuclear export of mRNA. Dbp5 localizes to the nuclear pore complex, where its ATPase activity is stimulated by Gle1 and its coactivator inositol hexakisphosphate. Here, we present the crystal structure of the C-terminal domain of Dbp5, refined to 1.8 Å. The structure reveals a RecA-like fold that contains two defining characteristics not present in other structurally characterized DExD/H-box proteins: a C-terminal α-helix and a loop connecting β5 and α4, both of which are composed of conserved and unique elements in the Dbp5 primary sequence. Using structure-guided mutagenesis, we have identified several charged surface residues that, when mutated, weaken the binding of Gle1 and inhibit the ability of Gle1 to stimulate Dbp5's ATPase activity. In vivo analysis of the same mutations reveals that those mutants displaying the weakest ATPase stimulation in vitro are also unable to support yeast growth. Analysis of the correlation between the in vitro and in vivo data indicates that a threshold level of Dbp5 ATPase activity is required for cellular mRNA export that is not met by the unstimulated enzyme, suggesting a possible mechanism by which Dbp5's activity can be modulated to regulate mRNA export.
Keywords: crystal structure, DExD/H-box, nuclear pore complex
Nuclear mRNA export is an essential yet poorly understood step in the expression of all protein-encoding nuclear genes. mRNAs are synthesized in the nucleus, where they associate with several proteins into mRNA-protein complexes (mRNPs). mRNP composition changes during the process of mRNA maturation, which involves a number of modifications, including capping, splicing, and polyadenylation (1). All these processes seem to be interconnected to provide a mechanism by which only fully and correctly processed transcripts are competent for export to the cytoplasm and gain access to the translational machinery (2). mRNP export depends on the essential export receptor, a heterodimer of Mex67/Mtr2 (TAP/p15 in metazoans), which facilitates the passage of mRNAs through nuclear pore complexes (NPCs) by interacting with specific NPC components (3–5). Immunoelectron microscopy studies have revealed that mRNPs are remodeled during NPC engagement and export, with specific proteins dissociating from the mRNP at distinct stages of export (6). Proper remodeling during export is likely to be essential for providing directionality to the mRNA export process.
The conserved DExD/H-box protein Dbp5 is enriched at the cytoplasmic face of the NPC and has been suggested to play a critical role in the mRNP rearrangement observed on cytoplasmic entry (7–10). Members of the DExD/H-box protein family are required for all aspects of RNA processing and are thought to couple the energy of ATP hydrolysis to drive the remodeling of RNA molecules (11). DExD/H-box proteins possess several distinct biochemical activities in vitro, including double-stranded RNA duplex unwinding and the removal of RNA-associated proteins (12–15). However, the precise cellular functions of many DExD/H-box enzymes remain poorly understood. Mutations in DBP5 result in an accumulation of poly(A) RNA inside the nucleus, indicating a role for Dbp5 in mRNA export (7, 16). In vivo, Dbp5 has been shown to associate with mRNPs early in their biogenesis, and Dbp5 is present in both the nuclear and cytoplasmic compartments (9, 17). It is, however, enriched on the cytoplasmic face of the NPC via a direct interaction with the nucleoporin Nup159 (8, 9, 18). Additionally, the ATPase activity of Dbp5 is stimulated by another NPC component, Gle1 (19, 20). This activity is further stimulated by inositol hexakisphosphate (InsP6), which binds directly to Gle1 and modulates the Dbp5-Gle1 interaction (19, 20). A recent study has indicated that Dbp5 can function to remodel RNA-protein interactions in vitro because it is able to promote the dissociation of Nab2 from a Nab2-RNA complex (15). Nab2 associates with polyadenylated RNA but is not found associated with polysomes, indicating that it dissociates from poly(A) RNA before translation (21–23). Additionally, in vivo cross-linking studies have shown that reduced Dbp5 activity correlates with an increase in both Mex67 and Nab2 association with poly(A) RNA in vivo, consistent with the hypothesis that removal of these proteins from RNA is an in vivo function of Dbp5 (15, 24).
Two recent studies have also implicated a role for Dbp5 and Gle1 in translation termination (25, 26). Mutations in DBP5 and GLE1 result in both increased sensitivity to translation inhibitors and inefficient translation termination, suggesting a mechanistic coupling between the processes of mRNA export and translation (25, 26).
In this study, we present the crystal structure of the C-terminal domain (CTD) of Dbp5 from Saccharomyces cerevisiae, demonstrating that Dbp5, like other structurally characterized DExD/H-box proteins, adopts a RecA-like fold. Using structure-directed mutagenesis, we demonstrate that one surface of the CTD is a critical point of contact between Dbp5 and its activator Gle1. Point mutations that weaken the Gle1-Dbp5 interaction also lead to a reduced ability of Gle1 to activate Dbp5's ATPase activity in vitro. In vivo, these mutations display growth defects and accumulate poly(A) RNA in the nucleus, indicating a defect in mRNA export. Our data refine the model of Dbp5's ATPase activity's involvement in mRNA export, suggesting that a threshold of Dbp5 activation by Gle1 and InsP6 is required for mRNA export. One intriguing implication of these data is the possibility that bulk mRNA export is affected in vivo by modulating Dbp5's ATPase activity, possibly under conditions of cellular stress.
Results
Crystal Structure of the CTD of Dbp5.
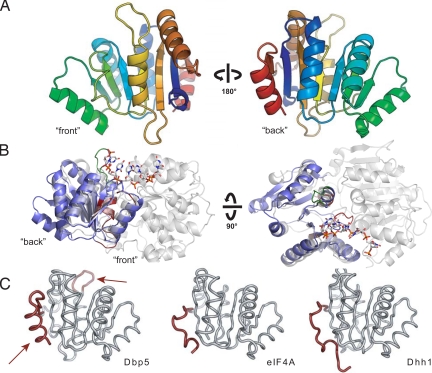
To understand how Gle1 stimulates the activity of the DExD/H-box ATPase Dbp5, it is critical to identify the surface of Dbp5 required for the interaction with Gle1. The N-terminal domain of Dbp5 was previously shown to target Dbp5 to the NPC via its interaction with the nucleoporin Nup159, but no protein interaction partners for the CTD of Dbp5 have been characterized (18). Studies on the eIF4A–eIF4G DEAD-box protein/activator complex revealed that the CTD of eIF4A contains the predominant eIF4G binding surface (27, 28). Additionally, dominant mutations in Dbp5 that enhance Gle1 stimulation of Dbp5's ATPase activity are located in the CTD (19). These observations led us to test whether Gle1 interacts with the CTD of Dbp5. Toward this goal, we determined and analyzed the crystal structure of the Dbp5 CTD at a resolution of 1.8 Å [supporting information (SI) Table S1]. The CTD adopts a RecA-like αβα fold similar to those of other DEAD-box proteins (Fig. 1A). This domain contains three conserved motifs: motifs IV and V, which are involved in substrate recognition, and motif VI, which participates in nucleotide binding. These motifs face the interdomain cleft when the CTD is modeled on the crystal structure of the full-length DEAD-box protein Vasa (Fig. 1B). Vasa is a DEAD-box protein from Drosophila involved in germline development and was one of the earliest members of this family for which a crystal structure was obtained in the presence of an RNA substrate (29). When compared with other DEAD-box proteins, the CTD of Dbp5 possesses two unique features (Fig. 1C). One feature is a 6-residue insertion in the loop connecting β5 and α4. This Dbp5-specific insertion is required for export of heat shock mRNAs at 42 °C and bulk mRNA export at 42 °C following preincubation at 37 °C and has been proposed to function in the regulation of mRNA export under heat stress conditions (32). The other feature is an α-helix at the C-terminus of the protein, representing a sequence element that is conserved in all Dbp5 homologs but is not present in other structurally characterized DEAD-box proteins.
Fig. 1.
Structure of the Dbp5 CTD. (A) “Front” and “back” views of the secondary structure features of Dbp5. (B) Position of the Dbp5 CTD in the context of a full-length DExD/H-box ATPase. Side and top views of Dbp5 CTD (blue) superposed on the Vasa helicase from Drosophila (gray) (29) show the orientation of the CTD in the context of an active RNA helicase assembly. The front of the domain faces the active site cleft and contains the conserved DExD/H sequence motifs (shown in red), whereas the back is distal to the interdomain interaction surface. The Dbp5-specific six amino-acid insertion is highlighted in green. (C) Comparison of Dbp5 CTD with the CTDs of eIF4A (30) and Dhh1 (31). Dbp5 CTD possesses two elements shown in red: an extended loop element after strand β5 (Top, arrow) and a conserved α-helix (Bottom, arrow) at its C-terminus that are not present in other structurally characterized DExD/H-box proteins.
Characterization of the Gle1 Interaction Surface on Dbp5.
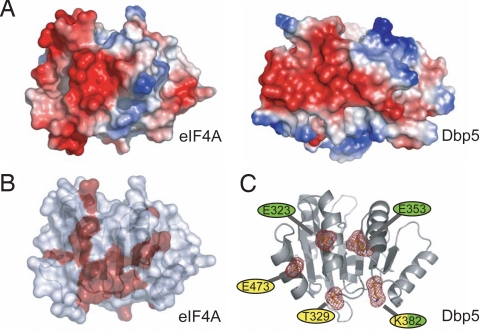
Comparison of the surface electrostatic potential of the CTDs of Dbp5 and eIF4A reveals some similarity in the charge distribution on the back surfaces of these proteins (Fig. 2A). A crystal structure of the eIF4A–eIF4G complex, together with an NMR-based map of the eIF4A–eIF4G interaction, revealed that this region of eIF4A is the predominant eIF4G binding surface (Fig. 2B) (27, 28). The equivalent surface in Dbp5 contains several conserved or invariant residues. We used site-directed mutagenesis to disrupt five residues on this surface and tested whether they play a role in the Gle1-mediated activation of Dbp5 (Fig. 2C). Three conserved acidic residues were individually mutated to lysine or alanine; two are at positions equivalent or nearly equivalent to acidic residues that participate in the eIF4A–eIF4G interaction (E323 and E353), and the third is an invariant residue in the conserved C-terminal helix of Dbp5 (E473). Two additional highly conserved residues were also targeted. K382, which is in a position equivalent to a basic residue (R312) involved with the eIF4A–eIF4G association, was mutated to glutamic acid, and T329, a nearly invariant residue on this surface, was mutated to alanine (Fig. S1). Full-length Dbp5 mutant variants were expressed, purified, and tested for in vitro ATPase activity. With the exception of Dbp5K382E, whose activity is about 64% that of WT, all other variants retained approximately WT levels of basal ATPase activity (Fig. 3A and Table S2). Additionally, the Michaelis constant (Km) for ATP for all the variants remains largely unchanged from WT, with the exception of the E353K variant, which is roughly 2-fold higher (Fig. S2). Importantly, all variants retained the ability to be stimulated by RNA to at least a WT extent, demonstrating that the mutant proteins are functional for RNA binding and ATP hydrolysis and that the surface mutations do not affect the overall folding of Dbp5 (Fig. 3B).
Fig. 2.
Selection of surface residues for mutagenesis. (A) eIF4A and Dbp5 have similar electrostatic surface potentials on the “back” sides of their CTDs. Areas of negative charge are indicated in red, and areas of positive charge are indicated in blue. (B) Surface residues on eIF4A involved in interactions with eIF4G are indicated in red (28). (C) Residues targeted for site-directed mutagenesis. Invariant residues (yellow) and residues corresponding to eIF4A/4G interaction hotspots (green) were chosen for in vitro and in vivo characterization.
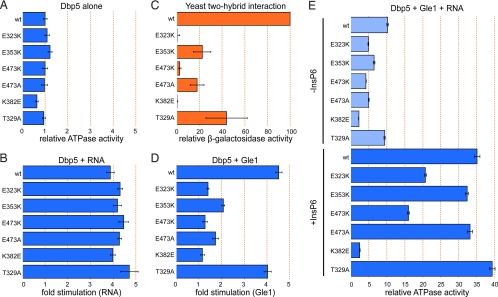
Fig. 3.
Dbp5 surface mutations affect Gle1 binding and ATPase activation. (A) Intrinsic ATPase activity of Dbp5 variants alone. Steady-state ATPase assays were performed using 1 μM Dbp5 and 2 mM ATP. Values are normalized to WT Dbp5. (B) Fold stimulation by RNA. ATPase assays were performed as in A, except poly(A) RNA was added to a final concentration of 25 μg/mL. Values are fold stimulation over each variant's intrinsic activity. (C) Dbp5-Gle1 interaction as measured by the yeast two-hybrid assay. Binding drives expression of β-galactosidase. Activity is represented as the percentage of the WT Dbp5-Gle1 interaction. (D) Fold stimulation by Gle1. ATPase assays were performed as in A, except Gle1 was added to a final concentration of 2 μM. Values are fold stimulation over each variant's intrinsic activity. (E) ATPase activity in the presence of Gle1 and RNA without (Upper, light blue) or with (Lower, dark blue) InsP6. ATPase assays were performed as in A, except poly(A) RNA was added to 25 μg/mL, Gle1 to 2 μM, and InsP6 (when present) to 1 μM. Values are normalized as described in A (WT Dbp5 without any stimulators is set to 1). In all graphs, error bars represent the standard error of at least three independent experiments.
We then tested the ability of the Dbp5 variants to bind to Gle1. The interaction between Dbp5 and Gle1 in vitro is weak but can be robustly detected by the 2-hybrid assay (19). We performed 2-hybrid analysis between full-length Dbp5 constructs and a C-terminal Gle1 fragment (257–538) driving the expression of a β-galactosidase reporter. None of the Dbp5 variants interacted with Gle1 as tightly as the WT protein (Fig. 3C). Three variants in particular, K382E, E473K, and E323K, show a drastically reduced interaction by this method, suggesting that these residues are important for the binding of Dbp5 to Gle1.
To assess whether the reduced interactions observed by the 2-hybrid assay correlate with a reduction in the stimulation of Dbp5 by Gle1, we repeated the in vitro ATPase assay in the presence of Gle1. In accordance with the 2-hybrid analysis, none of the variant proteins are stimulated by Gle1 to the extent of the WT protein (Fig. 3D and Fig. S3). The three variants that showed the weakest interaction by 2-hybrid assay are stimulated only minimally by Gle1. This reduction in stimulation may be attributable to a weaker interaction between Gle1 and Dbp5, or the interaction may be altered in such a way that Gle1 is unable to stimulate Dbp5 fully, although the 2-hybrid assay results suggest the former. In the presence of Gle1, RNA, and InsP6, the differences in ATPase activity between the variants becomes even more pronounced, with the K382E variant showing the most drastic defect: this variant displays only a 4-fold increase over its basal activity, whereas WT Dbp5 shows a 35-fold increase (Fig. 3E, Lower). Together, these results demonstrate that the C-terminal surface of Dbp5 defined by residues K382, E473, and E323 plays an important role in binding to Gle1, and is therefore critical for Gle1's ability to stimulate Dbp5's ATPase activity.
The Interaction Between Dbp5 and Gle1 Is Important for Cell Growth and mRNA Export.
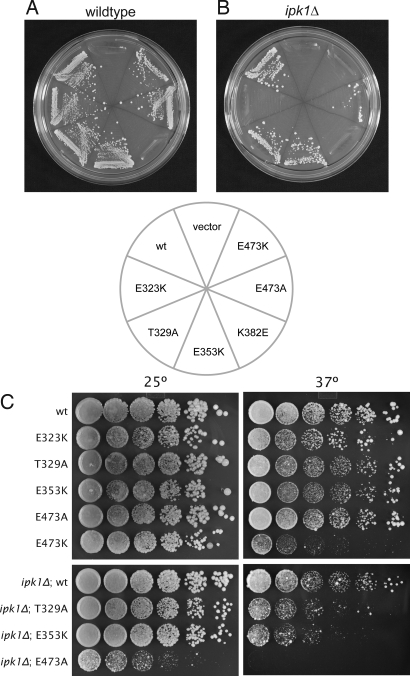
We next tested the ability of the genes encoding these Dbp5 variants to support yeast growth by testing for their ability to complement a dbp5 null mutation in vivo. In an otherwise WT setting, dbp5K382E was unable to support yeast growth, whereas all other variants were able to complement the null mutation (Fig. 4A). Because the interaction between Dbp5 and Gle1 is positively regulated by InsP6, we also tested for complementation in an ipk1Δ background (19). IPK1 encodes the enzyme inositol 1,3,4,5,6-pentakisphosphate 2-kinase, which produces InsP6 and has been shown to be synthetic lethal with alleles of GLE1, NUP159, and DBP5 (18, 19, 33, 34). In ipk1Δ cells, which lack the InsP6 coactivator, the E323K and E473K variants are unable to support growth, as is the K382E variant (Fig. 4B). Of note, these three Dbp5 alleles display the weakest interaction with Gle1 by the two-hybrid assay, the lowest activation by Gle1, and lowest overall ATPase activity in the in vitro ATPase assay (Fig. 3), demonstrating a very tight correlation between our in vitro and in vivo results. Furthermore, these data suggest that minimal Dbp5 ATPase activity is required for yeast cells to survive. In the presence of InsP6, this threshold is not met in cells expressing the dbp5K382E allele. In the absence of InsP6, the E323K and E473K variants also fail to reach the threshold and are unable to support growth.
Fig. 4.
Dbp5 surface mutations affect cell viability. (A) Plasmid shuffle in WT background. Cells harboring a dbp5Δ mutation on the chromosome and a pURA3-DBP5 plasmid were transformed with LEU2-marked plasmids expressing the indicated dbp5 variants, grown in URA-positive medium to allow for loss of the URA3-marked plasmid, and tested for their ability to grow on 5-fluoro-orotic acid. The cells containing the dbp5K382E variant do not grow, demonstrating that this variant is unable to complement the null dbp5Δ mutation. (B) As in A, except in the presence of the ipk1Δ mutation, which prevents InsP6 production. In this sensitized background, the E323K and E473K mutations, in addition to the K382E mutation, are unable to support growth. (C) Viable strains from A and B were tested for their ability to grow at different temperatures. Five-fold serial dilutions of yeast from overnight cultures were prepared and plated on rich media and incubated for 5 days at the indicated temperatures. In an ipk1Δ background, the dbp5E473A mutation confers a temperature-sensitive phenotype: it supports growth at 25 °C but not at 37 °C. Additionally, the dbp5E473K allele in an otherwise WT background is sick at 37 °C.
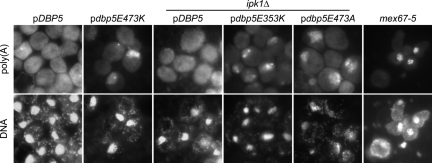
In addition to these three nonconditional mutations, we identified a fourth variant, E473A, which displayed both temperature-sensitive and cold-sensitive phenotypes: dbp5E473A was able to complement the dbp5Δ mutation at room temperature but did not support growth at 37 °C or 16 °C (Fig. 4C and Fig. S4). Additionally, the E473K variant is sick at both elevated and lowered temperatures (Fig. 4C and Fig. S4). We performed in situ hybridization experiments against poly(A) RNA in all the viable strains for cells grown at 30 °C or after a 4-h shift to either 16 °C or 37 °C. At 37 °C, the E3473K variant, as well as the E353K and E473A variants in an ipk1Δ background, shows an accumulation of the poly(A) signal inside the nucleus, suggesting that the viability of these cells is compromised because of a defect in mRNA export (Fig. 5), although the possibility that these mutations also affect Dbp5's role in translation has not been excluded (25, 26). Additionally, the E353K and E473A variants in an ipk1Δ background at 30 °C show a weak mRNA export defect in some cells (Fig. S5). No mRNA export defect could be observed for all the other variants and temperatures tested.
Fig. 5.
Dbp5 alleles confer a defect in mRNA export. Cells were grown to midlog phase at 30 °C and shifted to 37 °C for 30 min (mex67–5) or 4 h (all others) before fixation. In situ hybridization was performed using a fluorescein-labeled oligo-(dT)40 probe. The mex67–5 is shown as a positive control because it displays a strong and rapid accumulation of poly(A) inside the nucleus on temperature shift. DNA was stained with DAPI.
Discussion
In an effort to dissect the molecular details of Dbp5 activation by Gle1, we solved the crystal structure of the CTD of Dbp5. This structure allowed us to identify a surface on Dbp5 that is critical for the interaction with Gle1. Using structure-directed mutagenesis, we identified 3 charged residues that, when individually mutated, led to decreased ATPase activation by Gle1 and a concomitant reduction in viability. Interestingly, 2 previously identified dominant Dbp5 mutations also map to the identified Gle1 interaction surface (19). The crystal structure allows us to investigate the environment surrounding these residues. The change from threonine to proline at position 322 is likely to have a helix-destabilizing effect, and the leucine-to-valine substitution at position 327 may affect the packing of the central helix with the Dbp5-specific C-terminal helix (Fig. S6). A plausible explanation is that rearrangements of these Dbp5 helices occur on Gle1 binding and that this is facilitated by the specific amino-acid substitutions of the dominant mutations. The Dbp5-Gle1 interaction surface is in a similar position to the predominant interaction surface between eIF4A and eIF4G, suggesting a conserved mechanism by which these two DExD/H-box proteins, and potentially other members of the DExD/H-box family, interact with and are stimulated by their cellular activators (27, 28). Interestingly, in addition to an interaction with the C-terminus, eIF4G can weakly interact with the N-terminal RecA domain of eIF4A (27). It will be important to test if similar weak interactions also exist between the N-terminal domain of Dbp5 and Gle1. As in the case of eIF4A and eIF4G, such interactions could help to close the conformation between the two RecA domains of Dbp5 and could contribute to the observed increased affinity for ATP and the increased ATPase activity in the presence of Gle1 (19, 20). It should also be pointed out that N-terminal domains in Gle1, which are not present in the recombinant protein used for our in vitro ATPase experiments, could influence the Gle1-Dbp5 interaction. Secondary structure prediction indicates that the C-terminus of Gle1 adopts a purely α-helical fold and falls into a similar structural category as eIF4G, further indicating that these proteins may interact with members of the DExD/H-box family in a similar manner. Further studies, including structure determination of Gle1 or a Gle1/Dbp5 complex, will be needed to investigate these ideas.
Our structure reveals two interesting differences between Dbp5 and other DExD/H-box protein structures: an α-helix, formed by a C-terminal extension beyond the predicted RecA-like fold, and a loop near the predicted RNA binding site, formed by a 6-residue insertion between motifs V and VI. The sequences that form these structural variations are conserved among Dbp5 orthologs but are not found in other DExD/H-box proteins. The insertion loop is close to the putative RNA binding surface, suggesting that it might affect or regulate the interaction of Dbp5 with RNA substrates. For example, it could play a role in the dissociation of specific protein-RNA complexes, either by forming a wedge that drives between protein and RNA or by kinking the RNA to a point where the RNA-protein interaction is no longer favored. However, it should be noted that these residues are not essential for viability or for mRNA export at temperatures up to 37 °C (32). Interestingly, they have been shown to play a role in the export of heat-shock mRNA at 42 °C; however, at present, it remains unclear how this loop would specifically affect the export of heat-shock mRNAs at elevated temperatures (32).
While this manuscript was in preparation, several additional Dbp5 structures were published (35–38). Two of these studies showed the N-terminal domain of Ddx19, the human ortholog of Dbp5, in complex with Nup214 (Nup159 in yeast) (35, 36). One of these studies also showed Ddx19 in complex with RNA and demonstrated that Nup214 and RNA interact with Ddx19 through overlapping binding sites, suggesting that their binding is mutually exclusive (36). Consistent with this, it was shown that increasing amounts of Nup214 can inhibit the RNA stimulation of Ddx19's ATPase activity (36). Another structure of Ddx19 revealed that the extreme N-terminal extension forms a helix that fits between the two RecA folds and negatively regulates its ATPase activity (37). Finally, solution structures of the N-terminal domain and CTD of S. cerevisiae Dbp5, as well as a crystal structure of Saccharomyces pombe Dbp5, were reported (38). Interestingly, the C-terminal helix in our structure is observed in the solution structure of S. cerevisae Dbp5-CTD but is not present in any of the full-length structures (from human or S. pombe). Although this may suggest that this is a feature unique to the S. cerevisiae protein, it is intriguing that the conserved residue E473, which is important for the interaction between Dbp5 and Gle1, is located in this helix.
Some of the enzymatic parameters that we report here differ from our previously published results (19). Many of these differences can be explained by changes in specific activity of the protein preparations attributable to differences in the purification procedures and by changes in the reaction conditions for the ATPase assays. However, the relatively large increase in the stimulatory effect of RNA that we observe here cannot be easily explained by subtle changes in the specific activity of Dbp5. Interestingly, it has recently been reported that the N-terminal region of the human ortholog of Dbp5, Ddx19, forms an α-helical “switch” that negatively regulates ATPase activity (37). Constructs that lack this region are more active than the full-length Ddx19 and are not stimulated by the addition of RNA (37). We have found that the N-terminal region of Dbp5 is slowly degraded over time as the purified protein is stored. It is therefore possible that some of the differences observed in this work compared with our previous results, particularly in RNA stimulation, are attributable to differing amounts of degradation at the N-terminus. In this work, we were therefore careful to use preparations of Dbp5 that were as fresh as possible and to purify all Dbp5 variants in parallel, enabling us to compare the relative rates of ATPase activity between the different Dbp5 variants accurately.
Our data show a tight correlation between in vitro ATPase activity of Dbp5 and in vivo viability. These data are consistent with previous work indicating that activation of Dbp5's ATPase activity is an essential function of Gle1 (19). Our data suggest further that there exists a threshold level of Dbp5 ATPase activity required for yeast viability and mRNA export that is not met by the Dbp5 enzyme alone. This is demonstrated by the ATPase assays containing Dbp5, Gle1, and RNA, either in the presence or absence of InsP6 (Fig. 3E). These assays are the closest in vitro approximations to the in vivo setting of active Dbp5 in either a WT cell (with InsP6) or an ipk1Δ cell (without InsP6) (Fig. 4 A and B). In these assays, all conditions that gave less than ≈5-fold WT Dbp5 activity represent in vivo settings in which the Dbp5 variant failed to support growth, whereas conditions that yielded between 6.5- and 40-fold WT Dbp5 activity represent those in vivo settings in which yeast cells were viable. The in vitro conditions that represent the temperature-sensitive dbp5E473A; ipk1Δ strain show ATPase activity at ≈5-fold WT, which is very close to the identified threshold for viability.
Although it is not necessarily surprising that there exists a threshold level of Dbp5 ATPase activity that is required for mRNA export, it is interesting that this threshold is only met in the presence of its in vivo activators Gle1, InsP6, and RNA. This opens the possibility that, in vivo, the ATPase activity of Dbp5 could be modulated to regulate mRNA export. For example, it has been reported that during heat shock, bulk mRNA export is blocked and polyadenylated RNA accumulates inside the nucleus (39). Whether this is the result of decreased Dbp5 ATPase activity is unknown, but potential regulation of Dbp5 could occur through the modulation of InsP6 levels or availability. It would be therefore interesting to analyze if cellular InsP6 levels fluctuate under various growth conditions. Further studies will be required to investigate if and when Dbp5 activity is modulated in vivo and if mRNA export is regulated in response to stresses or the activation of cellular signaling pathways.
Methods
Yeast Strains and Plasmids.
Yeast strains used in this study are listed in Table S3. Plasmids used in this study are listed in Table S4. For information on plasmid construction, see SI Text.
Protein Purification and In Vitro Assays.
Protein purification and steady-state ATPase assays were performed essentially as previously described (19), with some exceptions. Briefly, His6-Dbp5 variants were purified in two steps by Ni-affinity and cation exchange. His6-MBP-Gle1 (244–538) was purified in one step by Ni-affinity. For details, see SI Text.
Two-Hybrid Analysis.
β-Galactosidase assays were performed as previously described (19). Briefly, cells were grown to an A600 of 0.4–0.6 in media containing 2% (wt/vol) galactose, collected, and disrupted using chloroform and SDS. β-Galactosidase activity was detected by using ortho-nitrophenyl-β-galactoside as a substrate and measuring an increase in A420.
Microscopy.
In situ hybridization against poly(A) RNA was done as previously described (19). Briefly, cells were grown in yeast extract/peptone/dextrose medium at 30 °C to an A600 of 0.2 to 0.6 and, when indicated, were transferred to 37 °C for 30 min (mex67–5) or 4 h (all others) before fixation. Poly(A) RNA was detected using a fluorescein-labeled oligo (dT)40 probe, and DNA was visualized using DAPI.
Crystallization and Structure Determination.
Selenomethionine-labeled Dbp5-CTD (residues 296–482) was purified as previously described (18). For details, see SI Text.
Supplementary Material
Acknowledgments.
We thank Christiane Brune for excellent technical assistance with the in situ hybridizations and Ben Montpetit for providing protein for additional ATPase assays. We also thank Sarah Munchel and Johanna Carroll for critically reading the manuscript, members of the Weis and Berger laboratories for helpful discussions, and members of the Kuriyan and Marqusee laboratories for access to equipment and workspace. This work was supported by a National Science Foundation Graduate Research Fellowship (to Z.Y.D.) and by National Institutes of Health Research Grant GM58065 (to K.W.). J.M.B. acknowledges support from the G. Harold and Leila Y. Mathers Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition footnote: The atomic coordinates and structure factors have been deposited with the Research Collaboratory for Structural Bioinformatics Database (ID code 3GFP).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902251106/DCSupplemental.
References
- 1.Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 2008;582:1987–1996. doi: 10.1016/j.febslet.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 2.Stutz F, Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 3.Izaurralde E. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur J Cell Biol. 2002;81:577–584. doi: 10.1078/0171-9335-00273. [DOI] [PubMed] [Google Scholar]
- 4.Segref A, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katahira J, et al. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng SS, et al. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahm Y, et al. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein ymr 255p. EMBO J. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocak S, Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 12.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 13.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 14.Fairman ME, et al. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 15.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Jin SB, Bjorkroth B, Wieslander L, Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002;21:1177–1187. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Weirich CS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 20.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 21.Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: A yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green DM, et al. Nab2p is required for poly(A) RNA export in saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 23.Windgassen M, et al. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Gross T, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 26.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schutz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 30.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rollenhagen C, Hodge CA, Cole CN. The nuclear pore complex and the DEAD box protein Rat8p/Dbp5p have nonessential features which appear to facilitate mRNA export following heat shock. Mol Cell Biol. 2004;24:4869–4879. doi: 10.1128/MCB.24.11.4869-4879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 34.Miller AL, et al. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J Biol Chem. 2004;279:51022–51032. doi: 10.1074/jbc.M409394200. [DOI] [PubMed] [Google Scholar]
- 35.Napetschnig J, et al. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci USA. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 37.Collins R, et al. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan JS, et al. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol. 2009;388:1–10. doi: 10.1016/j.jmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN. Regulation of mRNA export in response to stress in saccharomyces cerevisiae. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.