Abstract
Eukaryotes and bacteria regulate the activity of some proteins by localizing them to discrete subcellular structures, and eukaryotes localize some RNAs for the same purpose. To explore whether bacteria also spatially regulate RNAs, the localization of tmRNA was determined using fluorescence in situ hybridization. tmRNA is a small regulatory RNA that is ubiquitous in bacteria and that interacts with translating ribosomes in a reaction known as trans-translation. In Caulobacter crescentus, tmRNA was localized in a cell-cycle–dependent manner. In G1-phase cells, tmRNA was found in regularly spaced foci indicative of a helix-like structure. After initiation of DNA replication, most of the tmRNA was degraded, and the remaining molecules were spread throughout the cytoplasm. Immunofluorescence assays showed that SmpB, a protein that binds tightly to tmRNA, was colocalized with tmRNA in the helix-like pattern. RNase R, the nuclease that degrades tmRNA, was localized in a helix-like pattern that was separate from the SmpB-tmRNA complex. These results suggest a model in which tmRNA-SmpB is localized to sequester tmRNA from RNase R, and localization might also regulate tmRNA-SmpB interactions with ribosomes.
Keywords: RNase R, SmpB, tmRNA
Cells use spatial as well as temporal regulation of gene products to control normal physiological events and to respond to environmental challenges. Precise localization of proteins is used in eukaryotes and bacteria for processes such as cytoskeleton formation, cytokinesis, replication, and a wide array of signal transduction pathways (1). Eukaryotes also localize RNAs for regulatory control. For example, Xenopus and Drosophila localize mRNAs to regulate developmental timing (2, 3), and yeast localize the ash1 mRNA to the bud tip of dividing cells to control mating type switching (4). In contrast, there have been no reports of localization of endogenous bacterial RNAs. Here, we examine the localization of a small regulatory RNA, tmRNA, in the model bacterial species Caulobacter crescentus.
tmRNA is a highly abundant and widespread RNA that interacts with translating ribosomes in a reaction known as trans-translation (5–8). The termini of tmRNA fold into a structure that mimics the acceptor stem and TΨC arm of tRNAAla, and tmRNA is charged with alanine by alanyl-tRNA synthetase. Instead of an anticodon arm, tmRNA contains three to four pseudoknots and a specialized ORF. During trans-translation, tmRNA enters translating ribosomes that are near the 3′ end of an mRNA. tmRNA displaces the mRNA, and a peptide tag encoded in the tmRNA reading frame is added to the C terminus of the nascent polypeptide. The tmRNA peptide tag is recognized by several proteases, targeting the protein for rapid degradation (9). This reaction also releases the ribosome and promotes degradation of the mRNA that was being translated. A small protein, SmpB, binds to the tRNA-like domain of tmRNA and is required for tmRNA structure, stability, and activity (10). tmRNA and SmpB have been found in every bacterial species investigated. They are essential in some bacteria, and in others they are required for virulence (11, 12), antibiotic resistance (13), symbiosis (14), and cell-cycle control (15). In C. crescentus, tmRNA and SmpB are required for correct timing of DNA replication and differentiation (15).
C. crescentus is a model system for studies of bacterial differentiation and cell-cycle regulation because it is easy to obtain synchronized cultures of cells in G1 phase, called swarmer cells. Swarmer cells execute a developmental program that coordinates initiation of DNA replication with morphological differentiation into stalked cells. During differentiation, the polar flagellum is ejected and a stalk grows at the same site. Stalked cells complete S phase and divide, producing a stalked cell and a new swarmer cell. trans-Translation is required for correct timing of events during the developmental program. Cells lacking ssrA (the gene encoding tmRNA) or smpB do not initiate DNA replication at the correct time, and all subsequent cell-cycle–regulated events are delayed. Consistent with this phenotype, tmRNA and SmpB levels are temporally regulated as a function of the cell cycle (10, 15). tmRNA and SmpB accumulate during the G1–S phase transition, when both molecules are synthesized and stable. Proteolysis of SmpB in early S phase allows degradation of tmRNA by the exoribonuclease RNase R, dramatically decreasing the levels of tmRNA-SmpB complex in stalked cells (10). The experiments described here show that tmRNA, SmpB, and RNase R are localized to specific sites within the cytoplasm, and suggest that spatial as well as temporal mechanisms are used to regulate trans-translation.
Results
tmRNA Is Localized in C. crescentus.
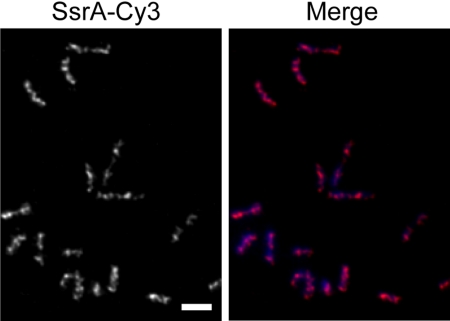
Fluorescence in situ hybridization (FISH) has been used to observe the subcellular localization of DNA and RNA in eukaryotic cells, and several studies in bacteria have used FISH to study the localization of DNA segments (16, 17). We adapted FISH to observe RNA localization in C. crescentus. To probe for tmRNA, a DNA oligonucleotide probe complementary to the tmRNA reading frame sequence was synthesized and conjugated to the fluorophore Cy3 (SsrA-Cy3). Fixed C. crescentus cells were probed with SsrA-Cy3 to determine whether tmRNA was localized within the bacterial cytoplasm. A localized fluorescence signal was observed in 68% of cells (n = 122) (Fig. 1). These cells contained a pattern of four to six regularly spaced foci that in some cases were connected with bands or extended filaments of fluorescence signal. Of the cells, 32% had very little fluorescence signal. To ensure that the fluorescence pattern was a result of specific hybridization of SsrA-Cy3 with tmRNA, FISH experiments were repeated using cells deleted for ssrA, the gene encoding tmRNA. The fluorescence signal after probing ΔssrA cells with SsrA-Cy3 was indistinguishable from mock FISH experiments using wild-type or ΔssrA cells in which no SsrA-Cy3 was added, indicating that SsrA-Cy3 specifically recognized tmRNA. These results suggest that when tmRNA is expressed in C. crescentus, it is localized in a discrete pattern in the cytoplasm.
Fig. 1.
Localization of tmRNA. C. crescentus cells were probed for tmRNA localization using FISH. Fluorescence signal from SsrA-Cy3 (Left) and merged image of DAPI (blue) and SsrA-Cy3 (magenta) signals (Right) are shown. Scale bar, 1 μm.
The pattern of tmRNA localization is similar to that observed for proteins that form helical structures in C. crescentus and other bacteria. For example, MreB in C. crescentus (18), Escherichia coli (19), and Bacillus subtilis (20), Mbl in B. subtilis (21), FrzS in Myxococcus xanthus (22), and RNase E in E. coli (23) are each localized to helical structures in the cell, and immunofluorescence images of these proteins produce patterns of regularly spaced foci and bands. Therefore, we will refer to the pattern of regularly spaced foci and bands as a helix-like pattern. Because FISH requires fixed cells, it is not possible to obtain true three-dimensional information. However, the FISH results for tmRNA are consistent with localization of this RNA to a helical structure.
tmRNA Localization Changes During the Cell Cycle.
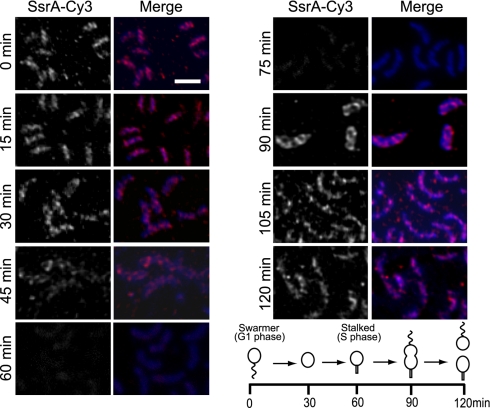
To determine whether the localization of tmRNA changes during the cell cycle, the FISH experiments were repeated using synchronized C. crescentus cultures (Fig. 2). In swarmer (G1 phase) cells, tmRNA was localized in a helix-like pattern. This pattern was retained during the G1-S phase transition. In stalked cells, after DNA replication had been initiated, the fluorescence signal was reduced, consistent with degradation of most tmRNA during this portion of the cell cycle. The residual fluorescence signal in these stalked cells was homogeneously distributed throughout the cytoplasm. As S phase progressed, the fluorescence signal increased, forming a helix-like pattern over the entire length of the predivisional cells. These results indicate that in swarmer and predivisional cells, when tmRNA is abundant, it is localized in a helix-like pattern, but that degradation of tmRNA in stalked cells coincides with delocalization of the remaining tmRNA.
Fig. 2.
tmRNA localization is cell-cycle regulated. Cells were sampled from synchronized cultures and tmRNA localization was determined using FISH. Fluorescence signal from SsrA-Cy3 (Left) and merged image of DAPI (blue) and SsrA-Cy3 (magenta) signals (Right) are shown. Scale bar, 1 μm. The time that each sample was taken is indicated, and the corresponding stage of the cell cycle is indicated in the schematic diagram.
SmpB Is Colocalized with tmRNA.
Biochemical data indicate tmRNA and SmpB form a tightly associated 1:1 complex (Kd = 1–2 nM), and that SmpB and tmRNA are present at approximately equimolar amounts in vivo; so it is likely that almost all SmpB is bound to tmRNA in C. crescentus cells (10). To determine whether SmpB is localized in the same pattern as tmRNA, cells were observed using immunofluorescence microscopy (IFM) with an antibody raised against SmpB. SmpB signal was localized in 72% of cells observed (n = 94), with a pattern of four to six regularly spaced foci, similar to the pattern observed for tmRNA (Fig. 2). IFM experiments using cells deleted for smpB showed no detectable fluorescence and were indistinguishable from mock IFM experiments in which no anti-SmpB antibody was added. To further confirm the SmpB localization pattern, IFM experiments were repeated using cells expressing a SmpB variant with an M2 epitope at the C terminus (SmpB-M2) probed with anti-M2 antibody conjugated to Cy3. These images showed fluorescence with a pattern identical to experiments performed with the anti-SmpB antibody [supporting information (SI) Fig. S1]. Therefore, the localization pattern observed using IFM was caused by localized SmpB protein.
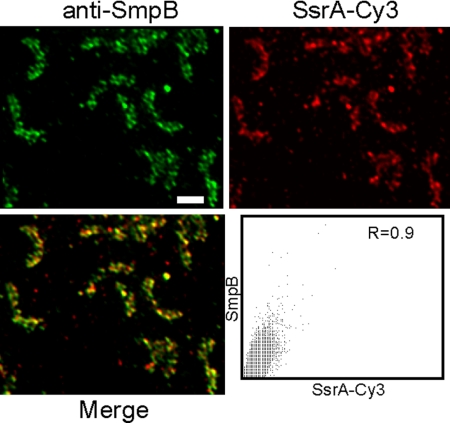
To test whether tmRNA and SmpB were colocalized, fixed C. crescentus cells were probed for both tmRNA, using SsrA-Cy3, and SmpB, using anti-SmpB antibody (Fig. 3). A merged image of the tmRNA signal, shown in red, and the SmpB signal, shown in green, produced a predominantly yellow pattern, indicating that the tmRNA and SmpB foci were colocalized. The degree of colocalization was quantified by plotting the red and green intensity for each pixel and measuring the overlap coefficient (R) (24). Using this technique, images of signals that are colocalized will result in R = 1.0, and images of signals that are not related will have R = 0.5. The tmRNA and SmpB images had R = 0.9, indicating a high degree of colocalization.
Fig. 3.
SmpB is colocalized with tmRNA. Localization patterns of SmpB and tmRNA were detected in the same cells using FISH (red, Upper Right) and IFM (green, Upper Left) and FISH (red, Upper Right). Merged image (Lower Left) shows regions of overlap (yellow). For each pixel, the intensity from the SmpB channel was plotted versus the intensity from the tmRNA channel (Lower Right), and the overlap coefficient (R) was calculated. Scale bar, 1 μm.
SmpB Is Required for tmRNA Localization.
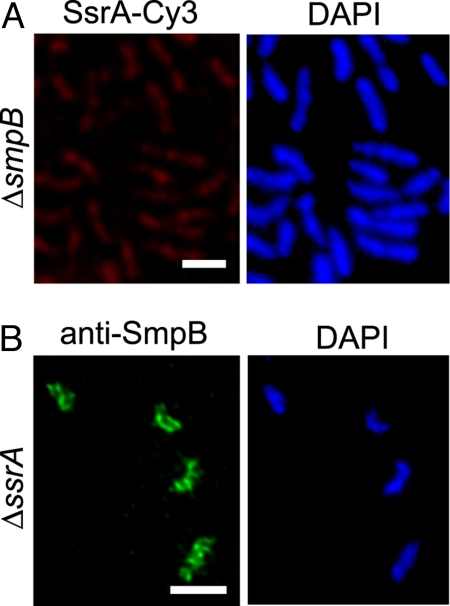
To determine whether tmRNA localization requires SmpB, FISH experiments with SsrA-Cy3 were repeated using ΔsmpB cells. The tmRNA signal was decreased in the absence of SmpB, consistent with previous data showing tmRNA levels are decreased by 90% in ΔsmpB cells (10). The remaining SsrA-Cy3 signal was homogeneously distributed throughout ΔsmpB cells (Fig. 4A), suggesting that tmRNA is not localized in the absence of SmpB. To exclude the possibility that tmRNA delocalization in ΔsmpB cells was caused by a low amount of tmRNA, tmRNA localization was examined in cells that lack RNase R (Fig. S2). RNase R degrades tmRNA in stalked cells, and tmRNA levels are constitutively high in cells deleted for vacB, the gene encoding RNase R (10). tmRNA was delocalized in ΔvacB stalked cells, even though tmRNA remained abundant (Fig. S2). Therefore, SmpB is required for tmRNA localization.
Fig. 4.
SmpB is required for tmRNA localization. (A) Localization of tmRNA in a ΔsmpB strain using FISH (Left) and the same cells stained with DAPI (Right) are shown. (B) Localization of SmpB in a ΔssrA strain using IFM with anti-SmpB antibody (Left) and the same cells stained with DAPI (Right) are shown. Scale bars, 1 μm.
To determine whether tmRNA is required for SmpB localization, localization of SmpB was examined in ΔssrA cells. The SmpB localization pattern in ΔssrA cells was indistinguishable from that observed in wild-type cells (Fig. 4B), indicating that SmpB is localized in the absence of tmRNA.
tmRNA Activity Does Not Affect Localization.
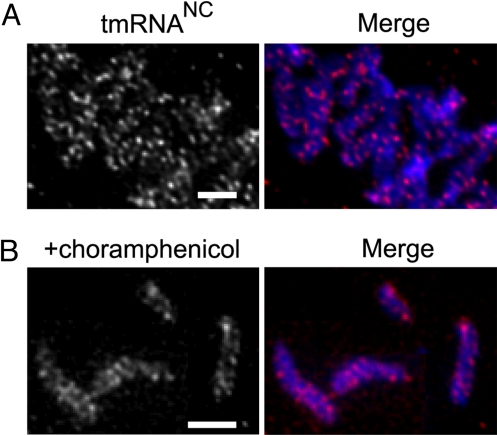
Because SmpB has no trans-translation activity in ΔssrA cells (25), and studies in E. coli have shown that SmpB does not interact with ribosomes in the absence of tmRNA (26), the localization of SmpB in ΔssrA cells suggests that trans-translation activity is not required for SmpB localization. To further test this hypothesis, localization of tmRNANC, an inactive variant of tmRNA, was probed. tmRNANC has a point mutation changing the G:U base pair in the acceptor stem to a G:C base pair. Similar mutations in other species result in a tmRNA that is not charged with alanine in vivo and is therefore inactive for trans-translation (27, 28). Northern blots showed that C. crescentus tmRNANC was not charged with alanine in vivo, and tmRNANC did not complement the ssrA phenotype in a ΔssrA strain, confirming that this variant is not active. FISH assays using cells containing tmRNANC but no wild-type tmRNA showed a fluorescence pattern identical to that of wild-type cells (Fig. 5A). Thus, trans-translation activity is not required for localization of either tmRNA or SmpB.
Fig. 5.
tmRNA localization does not require activity. (A) Localization of the inactive variant tmRNANC expressed in ΔssrA cells was determined using FISH. (B) Cells were treated with chloroamphenicol to inhibit translation, and localization of tmRNA was determined using FISH. Fluorescence images from SsrA-Cy3 (Left Panels) and a merged image of SsrA-Cy3 (magenta) and DAPI (blue) images (Right Panels) are shown. Scale bars, 1 μm.
To test whether active translation is required for tmRNA-SmpB localization, translation was inhibited by addition of chloramphenicol before FISH. Under conditions in which translation was inhibited >90%, tmRNA was localized in the same pattern as in untreated wild-type cells (Fig. 5B). Taken together, these data suggest that tmRNA-SmpB is localized independently of translation and trans-translation activity.
RNase R Is Localized in a Structure Distinct from tmRNA.
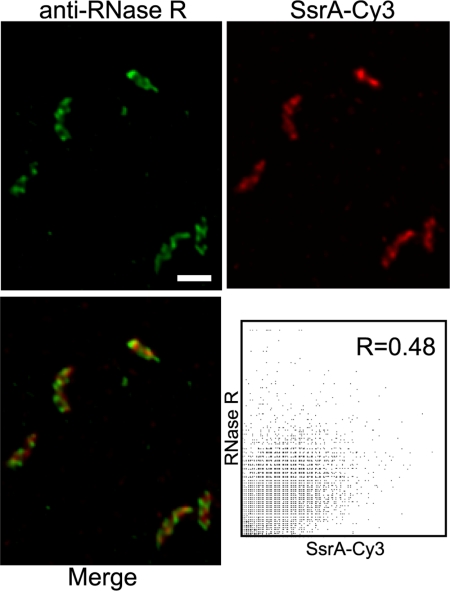
RNase R interacts with tmRNA in both C. crescentus and E. coli (10, 29). To determine whether RNase R is localized with the tmRNA-SmpB complex, IFM was performed using an antibody raised against RNase R. RNase R fluorescence signal was localized in a pattern similar to that of tmRNA and SmpB in 100% of cells observed (n = 125) (Fig. 6), and no signal was observed in cells deleted for the gene encoding RNase R. IFM experiments using synchronized C. crescentus cultures showed that the RNase R pattern did not change through the cell cycle (Fig. S3). RNase R localization was confirmed by IFM using cells expressing an RNase R variant with M2 epitope at the C terminus (RNase R-M2) and anti-M2 antibody conjugated to Cy3. The fluorescence pattern from RNase R-M2 in these cells was identical to that of wild-type RNase R (Fig. S4). To determine whether RNase R is colocalized with tmRNA-SmpB, fixed cells were probed with both SsrA-Cy3 and anti-RNase R antibody. Despite similar patterns of fluorescence from tmRNA and RNase R, a merged image showed red and green foci with little yellow, indicating that the signals were not coincident (Fig. 6). The overlap coefficient for these images was 0.48, confirming that there was little overlap between the tmRNA and RNase R patterns. A similar analysis on cells containing RNase R-M2 probed with anti-M2 antibody and anti-SmpB antibody showed that RNase R was not colocalized with SmpB (Fig. S4). RNase R was localized in ΔssrA and ΔsmpB cells, indicating that RNase R is localized independently of tmRNA-SmpB and trans-translation activity (Fig. S5). These results suggest that RNase R is not in a complex with tmRNA and SmpB. Instead, RNase R forms a helix-like pattern that is out of phase with the tmRNA-SmpB pattern.
Fig. 6.
tmRNA and RNase R are localized to distinct structures. Localization patterns of RNase R and tmRNA were detected in the same cells using IFM (green, Upper Left) and FISH (red, Upper Right). Merged image (Lower Left) shows regions of overlap (yellow). For each pixel, the intensity from the RNase R channel was plotted versus the intensity from the tmRNA channel (Lower Right), and the overlap coefficient (R) was calculated. Scale bar, 1 μm.
Discussion
Although localized RNAs have been identified in eukaryotes, tmRNA is the first example of an endogenous bacterial RNA that is localized. The localization of tmRNA and SmpB to a discrete structure within the cytoplasm demonstrates that bacteria have the capacity to localize RNA, and raises the possibility that other RNAs may be localized as well. How is tmRNA localized? Results showing that tmRNA localization depends on SmpB but SmpB is localized in the absence of tmRNA suggest that SmpB contains the localization information and that tmRNA is localized through binding to SmpB. Even though tmRNA-SmpB produces a pattern similar to proteins that polymerize to form helical filaments, such as MreB, it is unlikely that the tmRNA-SmpB complex polymerizes. trans-Translation requires that individual tmRNA-SmpB complexes pass through the ribosome, so tmRNA-SmpB would have to dissociate from the structure during trans-translation or be held with a long and flexible tether. The simplest model is that the tmRNA-SmpB complex is localized by binding of SmpB to another molecule that forms a helix in the cell. Several helical filaments have been observed in C. crescentus using electron cryotomography (30), but the only filament that has been examined in molecular detail is the actin-like protein MreB. The localization of tmRNA-SmpB was not disrupted by the addition of A22, an inhibitor that disrupts the MreB structure (Fig. S6), indicating that MreB is unlikely to anchor SmpB. Other potential interaction partners are under investigation.
What is the purpose for localization of tmRNA-SmpB? One possible explanation is provided by the localization of RNase R in a structure distinct from tmRNA-SmpB. tmRNA may be localized to sequester it from RNase R during portions of the cell cycle when trans-translation is required. Proteolysis of SmpB in early S phase would release tmRNA, allowing it to diffuse to localized RNase R and to be degraded. Consistent with this hypothesis, tmRNA was delocalized in early S phase cells in strains lacking RNase R (Fig. S2). Another possible rationale for localization of tmRNA-SmpB is to regulate trans-translation activity, either by ensuring access to substrate ribosomes or by limiting interaction with elongating translational complexes. Intriguingly, the enzymes that dispose of stalled translation complexes in eukaryotes are localized in structures called P-bodies (31). Stalled ribosomes move into P-bodies, where the ribosome is dissociated and the mRNA is degraded. trans-Translation serves a similar function in bacteria, so the tmRNA-SmpB helix may be analogous to P-bodies. These models are not mutually exclusive, and spatial regulation of tmRNA-SmpB might serve multiple functions in the cell.
Materials and Methods
Plasmids and Strains.
C. crescentus CB15N (32), and the mutant strains ΔssrA, ΔsmpB, and ΔvacB (33), were grown at 30 °C in PYE or M2G medium (34). Synchronized cultures were obtained by isolating swarmer cells from a Ludox density gradient (32, 35). To inhibit translation, chloramphenicol was added to cells for 10 min at a final concentration of 300 μg/ml. Translation was monitored by incorporation of [35S]methionine (15).
Plasmids producing SmpB-M2 and RNase R-M2 were constructed by cloning the relevant coding sequence between the xylose promoter sequence and the M2 sequence in plasmid pM2C (36). Plasmids were moved into C. crescentus cells by electroporation. Production of M2 fusion proteins was induced in C. crescentus cells containing pM2C-derived plasmids by addition of xylose to 0.3%. A plasmid producing tmRNANC was constructed by making a mutation in the ssrA sequence of pssrA (33) to change U217 of tmRNA to C, and by cloning the resulting gene into pMR20 (37).
Microscopy.
Images were obtained using an Eclipse 90i microscope (Nikon) with a 60X TIRF N. A. 1.4 objective, and a Nikon CoolSNAP HQ CCD camera controlled by Simple PCi (Compix, Inc.). Single microscopy images were deconvolved using the 2D Blind Deconvolution algorithm in AutoQuant software (Media Cybernetics).
The SsrA-Cy3 probe (5′-CGGCGAACTCTTCAGCGAAGTTATCGTTCGC-3′) was designed to anneal to the tmRNA tag reading frame. For FISH, cells were grown in M2G to OD660 = 0.3–0.4, fixed with 7.5% para-formaldehyde for 15 min at 30 °C, and incubated 30 min at 4 °C. Fixed cells were harvested by centrifugation, washed three times with phosphate-buffered saline (PBS) (140 mM NaCl, 3 mM KCl, 8 mM sodium phosphate, and 1.5 mM potassium phosphate [pH 7.5]), incubated with 10 μg/ml lysozyme for 4 min at 25 °C, and harvested by centrifugation. Cells were resuspended in GTE (50 mM glucose, 20 mM Tris-HCl, and 10 mM EDTA), and deposited on polylysine coated coverslips. After 5 min, excess liquid was removed and coverslips were allowed to dry. Dried coverslips were washed once with 0.1% Triton X-100 for 5 min, washed twice with 2 X SSCT (300 mM NaCl, 30 mM sodium citrate [pH 7.0], and 0.1% Tween-20) for 5 min in a humid chamber at 25 °C, and washed once in 2 X SSCT plus 50% formamide for 30 min at 37 °C in a humid chamber. Liquid was removed, coverslips were incubated 4 min at 95 °C in hybridization solution (450 mM NaCl, 45 mM sodium citrate [pH 7.0], 55% formamide, 11% dextran sulfate, and 20 ng/μl SsrA-Cy3), and incubated for 8–12 h at 42 °C in a humid chamber. Coverslips were washed with 2 X SSCT plus 25% formamide for 30 min at 37 °C in a humid chamber, stained with 2 X SSCT plus 150 μg/ml DAPI for 5 min at 25 °C, washed twice with 2 X SSCT for 5 min at 25 °C, washed with PBS, and mounted on slides with Cytoseal60 (Richard-Allan Scientific).
For IFM, cells were grown at 30 °C in PYE or M2G to OD660 = 0.3–0.4, and prepared as previously described (38). Cells were stained with 150 μg/ml of DAPI and probed with rabbit anti-SmpB antibody (10) or rabbit anti-RNase R antibody (10), followed by goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Invitrogen). Alternatively, for M2-tagged proteins, cells were probed with anti-FLAG M2-Cy3 antibody (Sigma).
FISH/IFM colocalization assays were performed by fixing and probing cells using the FISH procedure described above, except that DAPI staining was omitted. Before mounting, coverslips were washed with 2% bovine serum albumin (BSA) in PBS for 30 min at 25 °C in a humid chamber, and incubated with 2% BSA in PBS with anti-SmpB antibody or anti-RNase R antibody for 8–12 h at 25 °C in a humid chamber. Coverslips were then washed 10 times with PBS and incubated with 2% BSA in PBS with goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Invitrogen) plus 150 μg/ml DAPI for 1 h at 25 °C in a humid chamber, washed 10 times with PBS, and mounted on slides with Cytoseal60 (Richard-Allan Scientific). Colocalization analysis for tmRNA, SmpB, and RNase R was performed using Image Pro software (Media Cybernetics).
tmRNA Charging Assay.
Cells expressing tmRNANC and tmRNA were grown in PYE at 30 °C and harvested at an OD660 = 0.3–0.4. Total RNA was harvested and prepared for base-labile mobility shift on Northern blots as previously described (39). Northern blots were probed with the genes encoding tmRNA and tRNAAla to determine charging.
Supplementary Material
Acknowledgments.
The authors thank Elaine Kunze and Nicole Zembower, of the Huck Institutes of the Life Sciences Cytometry Facility at University Park, for assistance with imaging software. This work was supported by National Institutes of Health grant GM68720.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904904106/DCSupplemental.
References
- 1.Shapiro L, Losick R. Dynamic spatial regulation in the bacterial cell. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 2.Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 3.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 4.Paquin N, et al. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Felden B, Gesteland RF, Atkins JF. Eubacterial tmRNAs: Everywhere except the alpha-proteobacteria? Biochim Biophys Acta. 1999;1446:145–148. doi: 10.1016/s0167-4781(99)00085-8. [DOI] [PubMed] [Google Scholar]
- 6.Gueneau de Novoa P, Williams KP. The tmRNA website: Reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob Y, Seif E, Paquet PO, Lang BF. Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA. 2004;10:605–614. doi: 10.1261/rna.5227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keiler KC, Shapiro L, Williams KP. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proc Natl Acad Sci USA. 2000;97:7778–7783. doi: 10.1073/pnas.97.14.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- 10.Hong SJ, Tran QA, Keiler KC. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okan NA, Bliska JB, Karzai AW. A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2006;2:e6. doi: 10.1371/journal.ppat.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells. 2002;7:629–638. doi: 10.1046/j.1365-2443.2002.00549.x. [DOI] [PubMed] [Google Scholar]
- 14.Ebeling S, Kundig C, Hennecke H. Discovery of a rhizobial RNA that is essential for symbiotic root nodule development. J Bacteriol. 1991;173:6373–6382. doi: 10.1128/jb.173.20.6373-6382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keiler KC, Shapiro L. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J Bacteriol. 2003;185:1825–1830. doi: 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen RB, Wang SC, Shapiro L. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat Rev Mol Cell Biol. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- 18.Gitai Z, Shapiro L. Bacterial cell division spirals into control. Proc Natl Acad Sci USA. 2003;100:7423–7424. doi: 10.1073/pnas.1332806100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 21.Carballido-Lopez R, Errington J. The bacterial cytoskeleton: In vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- 22.Mignot T, Merlie JP, Jr, Zusman DR. Two localization motifs mediate polar residence of FrzS during cell movement and reversals of Myxococcus xanthus. Mol Microbiol. 2007;65:363–372. doi: 10.1111/j.1365-2958.2007.05789.x. [DOI] [PubMed] [Google Scholar]
- 23.Taghbalout A, Rothfield L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci USA. 2007;104:1667–1672. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manders EMM, Verbeek FJ, Aten JA. Measurement of colocalization of objects in dualcolor confocal images. J Microsc. 1993:169375–169382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 25.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation, and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 27.McClain WH, Gabriel K, Bhattacharya S, Jou YY, Schneider J. Functional compensation by particular nucleotide substitutions of a critical G*U wobble base-pair during aminoacylation of transfer RNA. J Mol Biol. 1999;286:1025–1032. doi: 10.1006/jmbi.1999.2542. [DOI] [PubMed] [Google Scholar]
- 28.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 29.Karzai AW, Sauer RT. Protein factors associated with the SsrA. SmpB tagging and ribosome rescue complex. Proc Natl Acad Sci USA. 2001;98:3040–3044. doi: 10.1073/pnas.051628298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briegel A, et al. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- 31.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keiler KC, Shapiro L. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol. 2003;185:573–580. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 35.Evinger M, Agabian N. Caulobacter crescentus nucleoid: Analysis of sedimentation behavior and protein composition during the cell cycle. Proc Natl Acad Sci USA. 1979;76:175–178. doi: 10.1073/pnas.76.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell JH, Keiler KC. Screen for localized proteins in Caulobacter crescentus. PLoS ONE. 2008;3:e1756. doi: 10.1371/journal.pone.0001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts RC, et al. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 39.McClain WH, Jou YY, Bhattacharya S, Gabriel K, Schneider J. The reliability of in vivo structure-function analysis of tRNA aminoacylation. J Mol Biol. 1999;290:391–409. doi: 10.1006/jmbi.1999.2884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.