Abstract
Mitoferrin-1 (Mfrn1; Slc25a37), a member of the solute carrier family localized in the mitochondrial inner membrane, functions as an essential iron importer for the synthesis of mitochondrial heme and iron–sulfur clusters in erythroblasts. The biochemistry of Mfrn1-mediated iron transport into the mitochondria, however, is poorly understood. Here, we used the strategy of in vivo epitope-tagging affinity purification and mass spectrometry to investigate Mfrn1-mediated mitochondrial iron homeostasis. Abcb10, a mitochondrial inner membrane ATP-binding cassette transporter highly induced during erythroid maturation in hematopoietic tissues, was found as one key protein that physically interacts with Mfrn1 during mouse erythroleukemia (MEL) cell differentiation. Mfrn1 was shown previously to have a longer protein half-life in differentiated MEL cells compared with undifferentiated cells. In this study, Abcb10 was found to enhance the stabilization of Mfrn1 protein in MEL cells and transfected heterologous COS7 cells. In undifferentiated MEL cells, cotransfected Abcb10 specifically interacts with Mfrn1 to enhance its protein stability and promote Mfrn1-dependent mitochondrial iron importation. The structural stabilization of the Mfrn1–Abcb10 complex demonstrates a previously uncharacterized function for Abcb10 in mitochondria. Furthermore, the binding domain of Mfrn1–Abcb10 interaction maps to the N terminus of Mfrn1. These results suggest the tight regulation of mitochondrial iron acquisition and heme synthesis in erythroblasts is mediated by both transcriptional and posttranslational mechanisms, whereby the high level of Mfrn1 is stabilized by oligomeric protein complexes.
Keywords: Abcb transporters, erythropoiesis, iron and heme metabolism, solute carriers, protein complexes
Iron is required for the function of many essential hemoproteins and iron–sulfur (Fe/S) proteins. Consequently, organisms ranging from prokaryotes to plants and mammals have developed elaborate, high-affinity uptake systems and corresponding cognate receptors to facilitate the acquisition of iron (1–6). In the circulating plasma of vertebrate organisms, Tf tightly binds extracellular ferric (Fe3+) iron until the Tf(Fe3+) complex binds its cognate receptor, transferrin receptor-1 (TfR1). The Tf–TfR1 complex internalizes into the cytoplasm through a receptor-mediated endocytosis in clathrin-coated pits, facilitated by Sec15l1 (7, 8). Trafficking of the Tf–TfR1 complex into acidified endosomes results in Fe3+ release from the complex and reduction by Steap3 (9). Reduced Fe2+ is transported out of the endosome by the divalent metal transporter-1 (10, 11) and then by unclear mechanisms is shuttled into mitochondria. Much less is known, however, about the iron trafficking required for mitochondrial biogenesis of heme and Fe/S clusters (12, 13).
In hemoglobin-synthesizing cells, the vast majority of the iron released from the endosome traverses both outer and inner mitochondrial membranes to reach ferrochelatase in the mitochondrial matrix (13). Pulse–chase experiments in mouse erythroleukemia (MEL) cells and primary reticulocytes have shown extremely efficient kinetics for the delivery of Tf-bound iron for heme synthesis (14, 15). Videomicrographs of confocal microscopy have shown the transient contact between the endosome and mitochondria, suggesting an efficient and direct delivery of iron into the mitochondria (16).
In yeast, mitochondrial iron acquisition is mediated by two transporters: MRS3/MRS4 (17–21). We previously described a zebrafish mutant, frascati (frs), which exhibits hypochromic anemia with erythroid maturation arrest. By using positional cloning, we identified mitoferrin1 (mfrn1, slc25a37), a member of the mitochondrial solute carrier family, as the gene disrupted in frs mutants (22). Mfrn1 is a mitochondrial inner membrane protein and functions as the principal iron importer in the mitochondria (22), which is orthologous to the role of yeast MRS3/MRS4. Furthermore, Mfrn1 is highly expressed in both fetal and adult hematopoietic tissues of zebrafish and mouse. Mfrn1 is essential for heme and Fe/S cluster synthesis in erythroblasts and is required for primitive and definitive erythropoiesis (22, 23).
Our current study focuses on how iron is transported into mitochondria through Mfrn1 from TfR1-containing endosomes for heme biosynthesis in erythroid cells. We showed previously that mouse Mfrn1, but not Mfrn2 (Slc25a28), complements the anemia of frs mutants, despite the 65% amino acid similarity between mouse Mfrn2 and Mfrn1 (22). Thus, Mfrn1 has a unique function in delivery of iron to rapidly dividing erythroid precursors. Recently, the prolongation of Mfrn1 protein half-life in differentiated MEL cells has been reported, which may explain its role in the efficient delivery of mitochondrial iron (24). To extend this observation, we postulated that specific Mfrn1-binding proteins contribute to these unique functions in the erythron. To test this, we used affinity purification and MS analysis to identify Mfrn1-binding proteins and thus further our understanding of Mfrn1-mediated mitochondrial iron metabolism. Here, we show that the mitochondrial inner membrane ATP-binding cassette transporter Abcb10, formerly known as ABC-me and whose functions is still unknown (25), physically interacts with Mfrn1 to enhance its protein stability and, as a consequence, increase iron import into the mitochondria.
Results
Recombinant BT-Mfrn1 Properly Targets to the Mitochondria and Has Biological Function.
An in vivo epitope-tagging system was used for affinity purification of Mfrn1-associated protein complexes. The mouse Mfrn1 cDNA was cloned into the pEF1α-BT mammalian expression vector (26) to generate a FLAG-epitope fusion at the N terminus of Mfrn1. After generation of this Mfrn1-fusion construct (referred to as BT-Mfrn1; Fig. S1A), a series of control experiments were conducted to evaluate its mitochondrial targeting and functional activity.
BT-Mfrn1 was expressed in COS7 cells by transient transfection, and the mitochondrial and cytosolic fractions were prepared for Western blot analysis. Proper targeting of the BT-Mfrn1 protein to the mitochondrial compartment was assessed by both Western blot analysis and confocal fluorescence immunohistochemistry, which demonstrated localization of the BT-Mfrn1 protein within the mitochondria (Fig. S1 B and C). Furthermore, the engineered BT-Mfrn1 cRNA restored hemoglobinized cells in frs embryos with an efficiency similar to native mouse Mfrn1 cRNA [BT-Mfrn: 33% efficiency (three rescued/nine mutants); native Mfrn1: 58% efficiency (53 rescued/91 mutants); and uninjected control: 0% efficiency (0 rescued/47 mutants); Fig. S1D]. These data show BT-Mfrn1 functionally complements the anemia of frs embryos; therefore, the N-terminal epitope tag does not alter Mfrn1 protein localization and function. BT-Mfrn1 functions as well as native Mfrn1 to import mitochondrial iron; thus, we can safely assume it to interact properly with Mfrn1-associated proteins.
Stable MEL Clone Expressing BT-Mfrn1 Undergoes Normal Erythroid Differentiation.
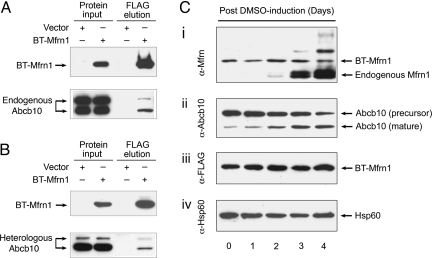
The BT-Mfrn1 cDNA construct was electroporated into an MEL DS19 clone, and subsequent stable clones were derived by antibiotic selection. One stable MEL clone expressing BT-Mfrn1 protein could undergo normal erythroid differentiation by chemical induction with DMSO (Fig. 1). The mitochondrial lysate from this differentiated stable MEL clone was analyzed for expression of the engineered BT-Mfrn1 and endogenous Mfrn1 proteins by Western blot analysis. Endogenous Mfrn1 is induced while BT-Mfrn1 is constitutively expressed during erythroid differentiation (Fig. 1). This analysis allowed us to assess the optimal time for harvesting the MEL cells, which is at 3 days after DMSO treatment, when putative interacting, erythroid-specific mitochondrial proteins were induced and the endogenous Mfrn1 protein remained relatively low so as not to compete with BT-Mfrn1 for binding of associating proteins.
Fig. 1.
Physical interaction between Mfrn1 and Abcb10 proteins is confirmed in MEL cells and transfected heterologous cells. (A) IP/Western blot analysis of interaction between Mfrn1 and endogenous Abcb10 in differentiated MEL cells stably expressing engineered BT-Mfrn1 or empty vector. (B) IP/Western blot analysis of Mfrn1–Abcb10 interaction from transient transfection of control vector or BT-Mfrn1 in heterologous COS7 cells. Protein input lysate is shown on the respective left columns. Only in the presence of Mfrn1 is Abcb10 selectively copurified from Mfrn1–protein complex. (C) Endogenous Mfrn1 and Abcb10 are induced in chemically differentiated stable MEL cells expressing BT-Mfrn1. Mitochondrial lysates collected from MEL cells exposed to 1.5% DMSO for varying numbers of days were serially probed with antisera against native mouse Mfrn1 (i), antisera against Abcb10 (ii), antisera against FLAG peptide (iii), and antisera against loading control Hsp60 (iv). BT-Mfrn1 protein is constitutively expressed from the EF1α promoter (i and iii), whereas endogenous Mfrn1 is induced during MEL erythroid maturation (i). Abcb10 is proteolytically processed to a mature isoform in MEL cells during erythroid differentiation (ii).
Mfrn1 Forms High-Order Protein Complexes.
A Blue Native gel was used to analyze the pattern of BT-Mfrn1 protein complexes from mitochondrial membranes. Mitochondria were fractionated from the MEL cells stably expressing BT-Mfrn1 at day 3 of DMSO-treated differentiation by using a differential hypotonic lysis method. Protein complexes were extracted from mitochondrial membrane with 1% nonionic detergent Nonidet P-40. Various nonionic detergents (Triton X-100, Tween-20, digitonin, and dodecyl maltoside) with different concentrations were empirically tested for their ability to maintain Mfrn1 protein complex integrity. Although other nonionic detergents can extract Mfrn1 higher-order protein complexes to some extent, Nonidet P-40 at 1% concentration was empirically determined to optimally solubilize mitochondrial proteins while maintaining their interaction by using a 2D gel system [first-dimension Blue Native gel (Fig. S2A), followed by a second-dimension Laemmli SDS/PAGE and Western blot analysis]. Although Western blot analysis after the first-dimension Blue Native gel showed four higher-order protein complexes containing BT-Mfrn1 protein, ranging in size from ≈250 to 800 kDa (Fig. S2B), additional protein complexes were more easily detected by adding a second-dimensional separation (Fig. S2C). The most abundant monomeric (≈37 kDa) and homodimeric (≈70 kDa) complexes of BT-Mfrn1 were detected as predicted for many SLC25 carrier proteins (27). The highly reproducible detection of higher-order Mfrn1 protein complexes (>70 kDa; Fig. S2C) is an additional demonstration that the SLC25 carrier proteins can form higher-order protein complexes within the mitochondria (28).
Affinity Purification and MS Identification of Mfrn1-Interacting Complexes.
By using an affinity purification scheme with anti-FLAG M2 monoclonal beads, we were able to efficiently bind BT-Mfrn1 proteins, which were eluted with a competing FLAG peptide. Eluted sample was submitted for MS analysis in triplicate runs and compared to control FLAG-eluted lysate from MEL cells expressing empty vector. Fragmentation peptides found to be unique in samples enriched by the BT-Mfrn1 bait protein were analyzed (Table S1). The prominent proteins in Table S1 specifically appear only in purified extracts from MEL cells that stably express the “bait protein” BT-Mfrn1. In addition to Mfrn1 itself, Abcb10 was consistently identified by MS analysis of triplicate, independent pulldowns (Table S1). Other members of the SLC25 carrier family, such as Slc25A12 (Aralar1) and Slc25A11 (OGCP), were also identified, but these may reflect the nonphysiological heterodimerization between these proteins and Mfrn1.
Mfrn1 Physically Interacts with Abcb10.
The physical interaction between Mfrn1 and Abcb10 was rigorously confirmed by serial immunoprecipitation (IP) and Western blot analysis using endogenous proteins in differentiated MEL cells and in transiently cotransfected COS7 cells (Fig. 1). MEL clones either stably expressing BT-Mfrn1 or empty vector were induced to undergo erythroid differentiation by the addition of DMSO. Protein complexes interacting with Mfrn1 were affinity-purified with anti-FLAG beads and analyzed by Western blot analysis with anti-Abcb10 antisera. The presence of the input bait BT-Mfrn1 was detected by reprobing with anti-FLAG antisera. The endogenous Abcb10 was enriched as an oligomeric complex by BT-Mfrn1 in differentiated MEL cells (Fig. 1A). Interestingly, the proteolytically processed mature isoform of Abcb10 was also induced in MEL cells with erythroid differentiation, similar to Mfrn1 (Fig. 1Cii). Furthermore, the physical interaction between Mfrn1 and Abcb10 was validated in transiently cotransfected COS7 cells (Fig. 1B). Myc-tagged Abcb10 (Abcb10-Myc) and FLAG-tagged Mfrn1 (BT-Mfrn1) or control vector were transiently cotransfected into COS7 cells. The results in heterologous COS7 cells confirm the interaction of endogenous Abcb10 with Mfrn1 observed in MEL cells (Fig. 1).
Pulse–Chase Assay Shows Abcb10 Stabilizes Mfrn1.
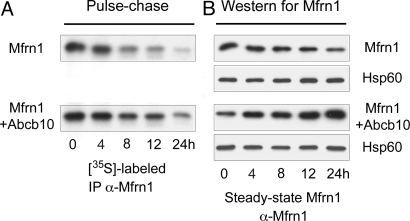
Mfrn1 was recently shown to have a longer protein half-life in differentiated MEL cells compared with undifferentiated cells (24). To test the hypothesis that the Mfrn1–Abcb10 interaction enhances Mfrn1 protein stability, a pulse–chase assay was performed to measure the half-life of expressed Mfrn1 alone or cotransfected with Abcb10 in COS7 cells.
COS7 cells transfected with FLAG-tagged Mfrn1 alone or cotransfected with an Myc-tagged Abcb10 were starved of amino acids for 1 h at 24 h after transfection. After metabolic labeling with [35S]methionine (pulse), cells were harvested at selected times from 0 to 24 h after labeling (chase), and the mitochondrial lysate was immunoprecipitated with anti-FLAG antisera. The pulse–chase results show that cotransfection with Abcb10 greatly enhanced the half-life of Mfrn1 (Fig. 2A and Fig. S3A). Similar results were obtained when steady-state Mfrn1 protein was evaluated by Western blot analysis to compare lysates from Mfrn1 and Abcb10 cotransfected cells and Mfrn1 alone transfected cells during the time course of the chase (Fig. 2B and Fig. S3B). Mitochondrial hsp60 was used as protein input control with anti-hsp60 by Western blot analysis (Fig. 2). These results demonstrate the specificity of the Abcb10–Mfrn1 interaction and protein stability.
Fig. 2.
Pulse–chase assay and steady-state Western blot analysis of Mfrn1 show Abcb10 stabilizes Mfrn1 protein half-life. (A) COS7 cells were metabolically pulse-labeled with [35S]methionine and chased for various amounts of time after cotransfection with Mfrn1-FLAG and Abcb10 or Mfrn1-FLAG and empty vector. At 24 h after chase, there is more labeled Mfrn1 IP Mfrn1 and Abcb10 cotransfected samples compared with Mfrn1 alone. (B) Steady-state Mfrn1 proteins were detected by Western blot analysis. More Mfrn1 proteins accumulated in the presence of Abcb10 at 24 h after chase compared with Mfrn1 alone. Equal levels of Hsp60 were shown as loading protein input control.
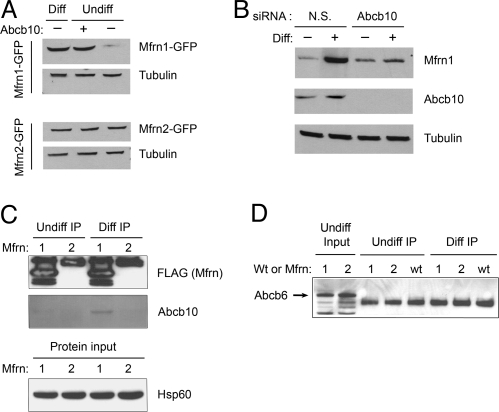
The affect of Abcb10 on Mfrn1 stability was also evaluated in MEL cells. Abcb10 was coexpressed in undifferentiated MEL cells along with either Mfrn1-GFP or Mfrn2-GFP, and the cell lysates were probed with anti-GFP. The results showed that coexpression of Abcb10 stabilized Mfrn1-GFP, but not Mfrn2-GFP, in undifferentiated MEL cells (Fig. 3A). In differentiated MEL cells, the normal induction of endogenously expressed Abcb10 contributed to the stability of Mfrn1-GFP. These results in MEL cells are similar to those obtained in COS7 (Fig. 2). As further confirmation, the stability of Mfrn1-GFP was evaluated in MEL cells with Abcb10 expression reduced by siRNA knockdown. When Abcb10 was silenced by using siRNA in differentiated MEL cells coexpressing Mfrn1-GFP, the Mfrn1-GFP protein was not stabilized compared with cells coexpressing Abcb10 but transfected with nonspecific siRNA control (Fig. 3B). This series of experiments conclusively demonstrates that Abcb10 protein is required for Mfrn1 protein stability.
Fig. 3.
The enhanced stability of Mfrn1 half-life by Mfrn1–Abcb10 interaction is specific. (A) Even though Mfrn1 and Mfrn2 have structural similarity and analogous function in transporting mitochondrial iron, Mfrn1 protein is uniquely stabilized by the coexpression of Abcb10 in MEL cells. In undifferentiated MEL cells, cotransfected Abcb10 does not confer protein stability to Mfrn2. Enhanced Mfrn1 stability in differentiated MEL cells is shown as a positive control. Tubulin levels are also shown as a loading control. (B) Abcb10 knockdown with siRNA destabilizes Mfrn1 protein in differentiated MEL cells. Mfrn1 protein accumulates in differentiated MEL cells treated with nonspecific (N.S.) siRNA control. Loss of Abcb10 by siRNA results in lower steady-state Mfrn1 accumulation, validating the demand for Abcb10 to stabilize Mfrn1 half-life. The absence of Abcb10 protein in cells treated with Abcb10 siRNA confirms effective knockdown of the target protein. (C) Abcb10 interacts only with Mfrn1 in differentiated MEL cells. MEL cells stably expressing Mfrn1-FLAG or Mfrn2-FLAG were anti-FLAG IP. Abcb10 was copurified by Mfrn1 in differentiated MEL cells. No Abcb10 was copurified from undifferentiated MEL cells or with coexpression of Mfrn2. (Lower) Equal hsp60 levels as protein input control. (D) Abcb6 does not physically interact with Mfrn1 or Mfrn2 in MEL cells. Endogenous Abcb6 proteins (arrow) are present in undifferentiated MEL cells as input control (first two lanes). Abcb6 is not copurified as an associated protein in MEL cells coexpressing either Mfrn transporters (lanes 1 and 2) or empty vector (wt), regardless of erythroid maturation stage.
Mfrn1, Not Mfrn2, Selectively Interacts with Abcb10 in Differentiated MEL Cells.
Mouse Mfrn1 mRNA is highly expressed in hematopoietic organs, fetal liver, adult bone marrow, and spleen, whereas Mfrn2 mRNA is ubiquitously expressed at low levels (22). To test the specificity and selectivity of the Mfrn1–Abcb10 interaction in MEL cells, stable MEL clones expressing either Mfrn1-FLAG or Mfrn2-FLAG were cultured and harvested under undifferentiated and differentiated conditions for anti-FLAG IP/Western blot analysis. Anti-Abcb10 antibody was used to detect the endogenous Abcb10 protein in the immunoprecipitated samples. Western blotting results show that only Mfrn1 interacted with endogenous mature Abcb10 in differentiated MEL cells (Fig. 3C).
An analogous test was performed for unrelated mitochondrial, ATP-binding cassette protein Abcb6, previously shown to be the coproporphyrin importer (29). By using antisera against the endogenous Abcb6 transporter in these samples, we demonstrated that neither Mfrn1 nor Mfrn2 interacts with Abcb6 (Fig. 3D), which further confirms the specificity of the Mfrn1–Abcb10 interaction in erythroid cells.
Abcb10 Enhances the Importation of Mitochondrial Iron by Mfrn1.
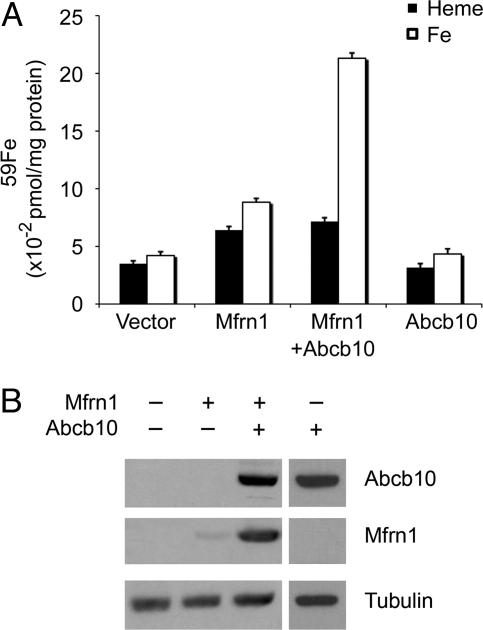
To determine whether the stabilization of Mfrn1 by Abcb10 has functional consequences, empty vector, Mfrn1-GFP alone, and Mfrn1-GFP plus Abcb10 were transfected into MEL cells. The quantity of total mitochondrial 59Fe and 59Fe-heme incorporation from 59Fe-transferrin was assayed in MEL cells transfected with these various constructs. In undifferentiated MEL cells, expression of Mfrn1 alone increased both mitochondrial total 59Fe and 59Fe-heme above vector control background (Fig. 4). This is not surprising, given our knowledge that Mfrn1 is the mitochondrial iron importer (22). Coexpression of Abcb10 with Mfrn1 greatly increased the accumulation of mitochondrial 59Fe but not the incorporation of 59Fe into heme (Fig. 4A). To exclude the contribution of Abcb10 per se to the influx of mitochondrial 59Fe or the synthesis of 59Fe-heme, Abcb10 alone was overexpressed in undifferentiated MEL cells to show that no measurable differences were observed in comparison with vector control cells. Western blot analyses of the corresponding lysates confirmed the efficiency of transfection and again showed that Abcb10 greatly stabilized Mfrn1 protein (Fig. 4B). Therefore, Abcb10 enhances the function of Mfrn1 to import more 59Fe into the mitochondria as a functional consequence of Mfrn1 stabilization.
Fig. 4.
The physical interaction of Mfrn1–Abcb10 enhances the function of Mfrn1 to import mitochondrial iron. (A) Mitochondrial total 59Fe (white bars) and 59Fe-heme (black bars) were measured in undifferentiated MEL cells transfected with various constructs. Mfrn1 alone minimally increases both total mitochondrial 59Fe and 59Fe-heme (column 2). The coexpression of Abcb10 with Mfrn1 greatly enhances the accumulation of mitochondrial 59Fe but not heme (column 3). (B) Western blot analyses of different transfections show that Mfrn1 is stable in the presence of Abcb10 in undifferentiated MEL cells. Control tubulin levels are also shown.
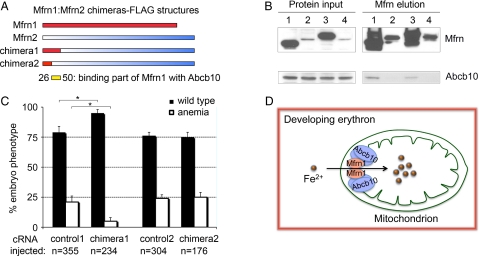
Amino Acid Residues 26–50 at the N Terminus of Mfrn1 Bind to Abcb10.
To map the domain required for Mfrn1–Abcb10 interaction, Mfrn1–Mfrn2 chimeric constructs (chimeras 1–2) fused in frame with the FLAG epitope at the C terminus were generated (Fig. 5A). MEL stable cell lines expressing these proteins were induced to differentiate with DMSO. The binding of these Mfrn chimeras, Mfrn1, and Mfrn2, to Abcb10 was directly tested by IP/Western blot analysis. Mfrn1 and chimera1 bound to Abcb10, but Mfrn2 and chimera2 proteins were not able to interact with Abcb10 (Fig. 5B). These studies conclusively show that the Mfrn1–Abcb10 interaction domain in Mfrn1 is localized to the N terminus between the first 26 to 50 amino acids, as indicated by the yellow bar in Fig. 5A. Mfrn1 and chimera1 proteins were stabilized, whereas Mfrn2 and chimera2 showed low steady-state stable protein expression as a consequence of their instability in differentiated MEL cells (Fig. 5B).
Fig. 5.
The interaction domain of Mfrn1–Abcb10 is localized to N-terminal amino acids 26–50 of Mfrn1. (A) Different Mfrn1:Mfrn2 chimeras were generated with a FLAG epitope at the respective C termini to map the Mfrn1–Abcb10 interacting domain. (B) IP/Western blot analysis of the interaction between Mfrn chimeras and endogenous Abcb10 in differentiated MEL cells stably expressing Mfrn chimeras, Mfrn1, or Mfrn2. Mfrn1 and chimera1 bind to Abcb10, but Mfrn2 and chimera2 failed to bind Abcb10. Lanes 1–4 are full-length Mfrn1, Mfrn2, chimera1, and chimera2, respectively. (C) Enumeration of wild-type and anemic embryos in the clutches of frs embryos uninjected (control) or injected with chimeras 1 and 2 cRNA. Complementation was scored by the restoration of hemoglobinized cells stained with o-dianisidine. *, Significance with P < 0.001 by one-way ANOVA analysis using compiled data from three independent experiments. The biochemical and genetic complementations map the Mfrn1–Abcb10 interaction domain to the N terminus of Mfrn1 between amino acid residues 26 and 50, as indicated by yellow bar in A. (D) A model for Mfrn1 and Abcb10 forming a protein complex, which facilitates the importation of mitochondrial iron in the developing red blood cells. The presence of Abcb10 stabilizes the protein half-life of Mfrn1, thereby facilitating more iron to be shuttled into the mitochondria for heme and Fe/S cluster synthesis during erythroid maturation.
We functionally tested the ability of chimeras 1 and 2 to complement the anemia in frs embryos by cRNA injection. Previously, only Mfrn1, not Mfrn2, was functionally able to complement the anemia (22). Consistent with our biochemical data, only chimera1, containing the Abcb10-interacting domain, complemented the anemia of frs embryos (Fig. 5C). A significant measurable difference was noted in the number of anemic frs embryos with restored hemoglobinized cells when injected with chimera1 cRNA. Genotyping with allele-specific probes rigorously verified that frstq223 mutant embryos were robustly complemented by chimera1 cRNA microinjection (81% efficiency = 50 rescued/62 mutants). Therefore, there is full concordance among chimeric protein stability, protein interaction with Abcb10, and functional in vivo complementation of anemia.
Discussion
Iron is a required element but can be toxic in high concentrations. The level of iron within cells is tightly regulated by either iron transport or storage. The regulation of iron transport to organelles might also be expected to be subject to regulation. In the mitochondria, iron is required for the synthesis of heme and Fe/S clusters. Mfrn1 mRNAs are induced highly in erythroid tissue (22, 30). Developing erythrocytes show an enhanced need for iron that is met by Mfrn1. The levels of Mfrn1 mRNA are transiently increased during erythroid differentiation, which constitutes one level of regulation of iron transport (22). As shown here, once synthesized, Mfrn1 levels are regulated at the level of protein stability by the expression of Abcb10, which by binding to Mfrn1 increases its half-life, and thus its concentration.
Abcb10 was identified as an interacting protein of Mfrn1 during MEL differentiation. Induced by the transcription factor GATA-1, Abcb10 protein is a mitochondrial, inner membrane, ATP-binding cassette transporter that is highly expressed in hematopoietic tissues (25). The expression pattern of Mfrn1 is similar to Abcb10 in terms of cell types. Besides Abcb10, Mfrn1 is also a mitochondrial inner membrane protein and is highly expressed in fetal and adult hematopoietic tissues. Forced expression of Abcb10 in differentiating MEL cells enhanced hemoglobinization, suggesting its participation in heme synthesis, although its precise biochemical role is not known (25). The predicted yeast ortholog for vertebrate Abcb10 is MDL1, which is involved in the export of peptides derived from the mitochondria (31); however, the physiological substrate for the vertebrate Abcb10 still remains unclear (25).
Abcb10 possesses an unusually long 105-amino acid, mitochondrial-targeting presequence. Like other ABC transporters, Abcb10 is extensively processed by posttranslational proteolysis during its maturation and targeting to the inner mitochondrial membrane (32). Our data show that during erythroid differentiation of MEL cells, Abcb10 is proteolytically processed to a mature isoform (Fig. 1Cii) in parallel with the induction of Mfrn1 protein. The coinduction of Mfrn1 with erythroid maturation to permit more mitochondrial iron assimilation explains why forced expression of Abcb10 in differentiated MEL cells, but not undifferentiated MEL cells, induced more hemoglobinization (25).
Induction kinetics of mature Abcb10 is similar to Mfrn1 at the protein level during DMSO-treated MEL cell differentiation. Furthermore, Abcb10 mRNA was induced within 12 h after DMSO treatment to induce MEL cell maturation (25). Mfrn1 mRNA showed similar induction kinetics in MEL cells induced with DMSO (22). Consistent with their parallel expression pattern during erythroid maturation at the RNA and protein levels, the physical interaction between Mfrn1 and Abcb10 occurs only in differentiated MEL cells. Another mitochondrial ABC transporter family member, Abcb6, does not interact with Mfrn1. Furthermore, Abcb10 does not interact with the paralog Mfrn2 in MEL cells, regardless of erythroid maturation stage. All of these data demonstrate the specificity of Mfrn1–Abcb10 interaction during erythroid cell differentiation.
The physiological consequence of the Mfrn1–Abcb10 protein complex during differentiation is the stabilization of Mfrn1 protein and, as a result, increased mitochondrial iron importation, as summarized in our proposed model (Fig. 5D). The half-transporter Abcb10 functioning as a homodimer (32) is required for Mfrn1 protein stability and mitochondrial iron import in erythroid cells. In differentiated erythrocytes, heme synthesis depends on the availability of protoporphyrin IX (PPIX) besides imported iron, which is catalytically incorporated into PPIX by ferrochelatase to produce heme. In our undifferentiated MEL cell system, components of PPIX synthesis pathway are poorly induced. Thus, coexpression of Abcb10 with Mfrn1 in undifferentiated MEL cells greatly increased the accumulation of mitochondrial iron, but not the amount of heme, because of the rate-limiting synthesis of protoporphyrins.
The biochemical and in vivo functional binding domain of Mfrn1–Abcb10 was mapped to between amino acids 26 and 50 at the N terminus of Mfrn1, which is N terminal to the first predicted transmembrane-spanning segment. Amino acid analysis comparing Mfrn1 and Mfrn2 with SIM Alignment Tool (http://us.expasy.org/tools/sim-prot.html) revealed no similarity within this 25-aa stretch, in contrast to the overall ≈65% similarity in general between these two transporters, lending additional support to the unique interaction between Mfrn1 and Abcb10. Further refinements in defining the critical amino acids within this 25-aa stretch by site-directed mutagenesis would shed light on the biochemistry of the Mfrn1–Abcb10 interaction.
The loss-of-function phenotype for Mfrn1 in zebrafish (22) and mouse (23) is early embryonic lethality due to profound anemia. Given the role of Abcb10 in stabilizing Mfrn1 and enhancing its ability to import mitochondrial iron, we predict that the deficiency of Abcb10 in mouse would result in a similar hypochromic anemia phenotype. Targeted disruption of murine Abcb10 has recently been reported showing anemia from a failure of embryonic erythropoiesis (33). This posttranslational regulation of Mfrn1 function through interaction with Abcb10 provides a previously undescribed mechanism for regulating mitochondrial iron homeostasis and heme biosynthesis in erythroid development.
By using a 2D gel system, we detected not only monomeric (≈37 kDa) and dimeric (≈70 kDa) complexes of Mfrn1, but also higher-order protein complexes that include other mitochondrial proteins. Mfrn1–Abcb10 interaction is the previously uncharacterized demonstration of functional complex involving proteins of the SLC25 mitochondrial solute carrier family. Because higher-order multimeric protein complexes were identified from our 2D gel system, we predict additional interacting proteins besides Abcb10 might be identified by using milder nonionic detergents or solubilization conditions able to better preserve the integrity of oligomeric protein complexes. The identification of additional proteins residing in the outer mitochondrial membrane or late endosome that physically interact with Mfrn1, which is located in the inner mitochondrial membrane, would give us further insight into the mechanism of iron delivery to the mitochondria. Likewise, the identification of other inner membrane and mitochondrial matrix proteins would provide significant information on the trafficking of imported iron throughout the mitochondria for Fe/S and heme biogenesis and mitochondrial storage in the form of mitochondrial ferritin (34).
Materials and Methods
Tissue Culture.
COS7 cells were maintained in DMEM (Invitrogen) containing heat-inactivated 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Stable MEL cells expressing BT-Mfrn1 were maintained in DMEM with 10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, and selection puromycin (5 mg/mL). For erythroid differentiation, MEL cells were induced through incubation in culture medium with 1.5% DMSO for 3 days.
Transfection.
COS7 cells were transfected with pEF1α-BT-Mfrn1 constructs by using Lipofectamine 2000 (Invitrogen) or FuGENE6 Transfection Reagent (Roche) as recommended by the manufacturer. GenBank accession numbers for clones used in this paper are NM_026331.3 for Mfrn1, NM_145156.1 for Mfrn2, and NM_019552.2 for Abcb10.
Confocal Microscopy, Construction of Mfrn Chimeras, cRNA Microinjections, and Genotyping.
These assays were performed as described in SI Materials and Methods.
Western Blot Analysis.
Cellular proteins were extracted with 1% Nonidet P-40 lysis buffer and denatured with 1 volume of 2× Laemmli SDS sample buffer (Bio-Rad) at 95 °C for 5 min. Protein samples were separated on 10% acrylamide SDS/PAGE gel and transferred on Immun-Blot PVDF membrane (Bio-Rad). The membranes were probed by using anti-Mfrn1, anti-FLAG, anti-Myc, and anti-Abcb10 antibodies with peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG as the secondary antibody. As a loading control, membranes were reprobed by using anti-hsp60 antibody with donkey anti-goat IgG as the secondary antibody. Chemiluminescent method was used for detection (SuperSignal West Pico Substrate; Pierce). The hsp60 antibody, goat anti-rabbit IgG-HRP, and donkey anti-goat IgG-HRP were from Santa Cruz Biotechnology. Anti-Abcb6 antiserum was a gift from John Schuetz (St. Jude Research Children's Hospital, Memphis, TN).
Mitochondrial Extraction.
Mitochondrial and cytosolic fractions from COS7 cells were performed by using the Mitochondria Isolation Kit for Cultured Cells (Pierce). MEL cells were lysed in a hypotonic buffer (10 mM NaCl, 1.5 mM MgCl2, and 10 mM Tris·HCl, pH 7.5), and mitochondria were extracted in a Dounce homogenizer in mitochondrial buffer (1 mM EDTA, 210 mM mannitol, 70 mM sucrose, and 5 mM Tris·HCl, pH 7.5), followed by centrifugation at 1,300 × g for 10 min at 4 °C. The supernatant was further centrifuged at 17,000 × g for 15 min at 4 °C to pellet the mitochondria. The crude mitochondrial fraction was resuspended for washing and centrifuged at 17,000 × g for 15 min at 4 °C. The pellets were collected as the mitochondrial fraction.
Affinity Purification and MS Identification.
MEL cells stably expressing BT-Mfrn1 or BT vector alone were differentiated with 1.5% DMSO. Cells were harvested at day 3 after DMSO treatment for mitochondrial preparation. The mitochondrial proteome was extracted with 1% Nonidet P-40 lysis buffer (1 mM EDTA, 150 mM NaCl, 1% Nonidet P-40, and 50 mM Tris·HCl, pH 7.5, plus protease inhibitor mixture tablet from Roche). Mfrn1 protein complexes were affinity-purified with ANTI-FLAG M2 Affinity Gel (Sigma) and eluted with FLAG peptide (Sigma). Affinity-purified protein samples were reduced with DTT and cysteine-alkylated with iodoacetamide before running on SDS/PAGE gel. Proteins on gel were stained with Colloidal Coomassie G250 dye (Invitrogen), and the whole gel lane was submitted for MS analysis to Taplin Biological Mass Spectrometry Facility at Harvard Medical School (Boston) for MS analysis.
Metabolic 35S Labeling and Pulse–Chase Assay.
COS7 cells were transfected with pCMV-Tag5-Abcb10 and pEF1α-Mfrn1-FLAG or pEF1α-Mfrn1-FLAG alone. Metabolic [35S]methionine labeling and pulse–chase assay were performed as described previously (24). l-[35S]methionine (>1,000 Ci/mmol specific activity) was from Perkin–Elmer.
59Fe Flux Assay.
Transfected MEL cells were labeled with 59Fe-saturated transferrin and assayed for total mitochondrial 59Fe and 59Fe-incorporated heme as described previously (24). 59FeCl3 (54.56 mCi/mg specific activity) was purchased from Perkin–Elmer.
Supplementary Material
Acknowledgments.
We thank Dr. Alan Cantor (Children's Hospital, Boston) for the pEF1α-BT expression vector, Dr. Ross Tomaino for the MS analysis, Dr. John Schuetz for the anti-Abcb6 antibody, Dr. Nancy Kedersha and Tyler Hickman for helping with the immunofluorescence assay, and Christian Lawrence and Jason Best for the zebrafish animal husbandry. We thank Drs. David Traver, Nikolaus Trede, Seth Alper, Hartmut Wohlrab, Prem Ponka, and members of our labs for reviewing this manuscript. This work was supported by grants from the March of Dimes Foundation (to B.H.P.) and National Institutes of Health Grants R01 DK052380 (to J.K.) and R01 DK070838 and P01 HL032262 (to B.H.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 16012.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904519106/DCSupplemental.
References
- 1.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 2.Andrews NC. Iron metabolism: Iron deficiency and iron overload. Annu Rev Genomics Hum Genet. 2000;1:75–98. doi: 10.1146/annurev.genom.1.1.75. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan J. Mechanism of cellular iron acquisition: Another iron in the fire. Cell. 2002;111:603–606. doi: 10.1016/s0092-8674(02)01164-9. [DOI] [PubMed] [Google Scholar]
- 4.Roy CN, Andrews NC. Recent advances in disorders of iron metabolism: Mutations, mechanisms and modifiers. Hum Mol Genet. 2001;10:2181–2186. doi: 10.1093/hmg/10.20.2181. [DOI] [PubMed] [Google Scholar]
- 5.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 6.Andrews NC. Forging a field: The golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim JE, et al. A mutation in Sec15l1 causes anemia in hemoglobin deficit (hbd) mice. Nat Genet. 2005;37:1270–1273. doi: 10.1038/ng1659. [DOI] [PubMed] [Google Scholar]
- 8.White RA, et al. Iron metabolism mutant hbd mice have a deletion in Sec15l1, which has homology to a yeast gene for vesicle docking. Genomics. 2005;86:668–673. doi: 10.1016/j.ygeno.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Ohgami RS, et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming MD, et al. Microcytic anemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 11.Gushin H, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 12.Ponka P. Tissue-specific regulation of iron metabolism and heme synthesis: Distinct control mechanisms in erythroid cells. Blood. 1997;89:1–25. [PubMed] [Google Scholar]
- 13.Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondria: Novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 14.Zhang AS, Sheftel AD, Ponka P. Intracellular kinetics of iron in reticulocytes: Evidence for endosome involvement in iron targeting to mitochondria. Blood. 2005;105:368–375. doi: 10.1182/blood-2004-06-2226. [DOI] [PubMed] [Google Scholar]
- 15.Richardson DR, Ponka P, Vyoral D. Distribution of iron in reticulocytes after inhibition of heme synthesis with succinylacetone: Examination of the intermediates involved in iron metabolism. Blood. 1996;87:3477–3488. [PubMed] [Google Scholar]
- 16.Sheftel AD, Zhang AS, Brown C, Shirihai OS, Ponka P. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood. 2007;110:125–132. doi: 10.1182/blood-2007-01-068148. [DOI] [PubMed] [Google Scholar]
- 17.Foury F, Roganti T. Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J Biol Chem. 2002;277:24475–24483. doi: 10.1074/jbc.M111789200. [DOI] [PubMed] [Google Scholar]
- 18.Mühlenhoff U, et al. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem. 2003;278:40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Kaplan J. A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J Biol Chem. 2004;279:33653–33661. doi: 10.1074/jbc.M403146200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Lyver ER, Knight SA, Lesuisse E, Dancis A. Frataxin and mitochondrial carrier proteins, Mrs3p and Mrs4p, cooperate in providing iron for heme synthesis. J Biol Chem. 2005;280:19794–19807. doi: 10.1074/jbc.M500397200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Mrs3p, Mrs4p, and frataxin provide iron for Fe-S cluster synthesis in mitochondria. J Biol Chem. 2006;281:22493–22502. doi: 10.1074/jbc.M604246200. [DOI] [PubMed] [Google Scholar]
- 22.Shaw GC, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 23.Paw BH, et al. Targeted disruption of the mouse mitoferrin (Slc25A37) mitochondrial solute carrier results in defective primitive and definitive erythropoiesis. Blood. 2006;108:82A–83A. [Google Scholar]
- 24.Paradkar PN, et al. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol Cell Biol. 2009;29:1007–1016. doi: 10.1128/MCB.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirihai OS, Gregory T, Yu C, Orkin SH, Weiss MJ. ABC-me: A novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 2000;19:2492–2502. doi: 10.1093/emboj/19.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo AJ, et al. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol Cell Biol. 2008;28:2675–2689. doi: 10.1128/MCB.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmieri F. The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflugers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, et al. Mitochondrial ATP synthasome: Three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP. J Biol Chem. 2004;279:31761–33178. doi: 10.1074/jbc.M401353200. [DOI] [PubMed] [Google Scholar]
- 29.Krishnamurthy PC, et al. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443:586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- 30.Amigo J, et al. Mitoferrin1 transgenic zebrafish line serves as a model to study erythroid cell fate during hematopoiesis. Blood. 2008;112(11):1223. (abstract 3576) [Google Scholar]
- 31.Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- 32.Graf SA, Haigh SE, Corson ED, Shirihai OS. Targeting, import, and dimerization of a mammalian mitochondrial ATP binding cassette (ABC) transporter, ABCB10 (ABC-me) J Biol Chem. 2004;279:42954–42963. doi: 10.1074/jbc.M405040200. [DOI] [PubMed] [Google Scholar]
- 33.Hyde BB, et al. ABC-Me (ABCB10) is required for erythroid development in the mouse embryo and is protective against mitochondrial oxidative stress. Blood. 2008;112(11):199. [Google Scholar]
- 34.Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.