Abstract
Rapid genome-wide identification of genes required for infection would expedite studies of bacterial pathogens. We developed genome-scale “negative selection” technology that combines high-density transposon mutagenesis and massively parallel sequencing of transposon/chromosome junctions in a mutant library to identify mutants lost from the library after exposure to a selective condition of interest. This approach was applied to comprehensively identify Haemophilus influenzae genes required to delay bacterial clearance in a murine pulmonary model. Mutations in 136 genes resulted in defects in vivo, and quantitative estimates of fitness generated by this technique were in agreement with independent validation experiments using individual mutant strains. Genes required in the lung included those with characterized functions in other models of H. influenzae pathogenesis and genes not previously implicated in infection. Genes implicated in vivo have reported or potential roles in survival during nutrient limitation, oxidative stress, and exposure to antimicrobial membrane perturbations, suggesting that these conditions are encountered by H. influenzae during pulmonary infection. The results demonstrate an efficient means to identify genes required for bacterial survival in experimental models of pathogenesis, and this approach should function similarly well in selections conducted in vitro and in vivo with any organism amenable to insertional mutagenesis.
Keywords: Illumina, mariner, mutagenesis, pathogenesis, transposon
Whole-genome analytic techniques have been developed to identify bacterial genes essential for growth or survival in vitro or during infection of model hosts. The most direct of these approaches can be classified as “negative selection” strategies, in which large pools of diverse mutants are analyzed to identify mutations that reduce fitness under a particular condition. “Signature-tagged mutagenesis” utilizes DNA arrays representing unique hybridization tags that are introduced into each mutant within a library of strains to be evaluated (1). The “transposon-site hybridization” and “microarray tracking of transposon mutants” methods use microarrays displaying each gene of the target organism to monitor the relative abundance of transposon insertions in these genes under varied selection conditions (2–4). Each of these methods has been effectively used to identify virulence genes in diverse bacteria. For many pathogens, however, generation of large banks of uniquely tagged mutants is impractical and whole-genome microarrays may be unavailable, particularly for newly recognized organisms or genetically diverse species. In both microarray-based methods, hybridization is used to detect the abundance of a given mutation within the library of mutants. Therefore, quantification is limited by background hybridization levels and the dynamic range of signal detection. A method that generates an output that allows precise noise filtering and a broad dynamic range would represent a significant advancement of the negative selection strategy.
In this study we report a technique termed “high-throughput insertion tracking by deep sequencing” (HITS) that uses a whole-genome transposon mutant bank in combination with massively parallel sequencing to efficiently analyze bacterial genes involved in pathogenesis. HITS allowed analysis of genes required by Haemophilus influenzae to resist clearance from the lung, a site colonized during pneumonia and chronic obstructive pulmonary disease (5, 6). Because deep sequencing is used for detection, background signal is easily identified and removed during data analysis, and the dynamic range of detection is limited only by the number of sequencing reads, which can be readily increased. The results highlight the utility of HITS in systematic discovery and analysis of virulence genes required in environments encountered by bacteria during pathogenesis.
Results
Overview of the HITS Technique.
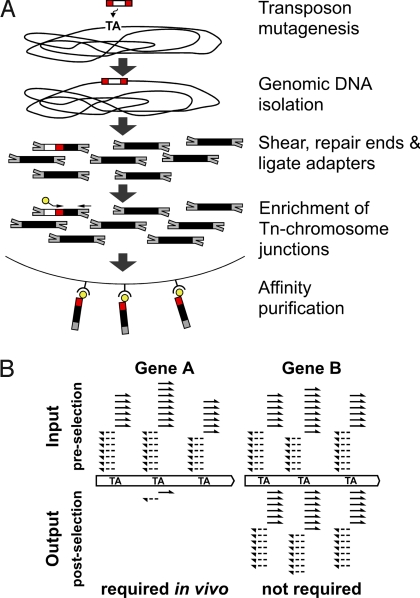
HITS is outlined schematically as two steps in Fig. 1 A and B. The first step involves fragmentation and ligation of adapters to sheared genomic DNA prepared from a high-density mutant bank carrying random transposon insertion mutations. In this study, mutagenesis was performed with a minitransposon derived from the Himar1-mariner transposon, which inserts efficiently in the genomes of H. influenzae and other bacteria, with only the dinucleotide TA as the apparent insertion site specificity (7–9). Selective amplification of transposon/chromosome junction regions is performed by PCR, and the resulting amplicons are purified by affinity capture. Sequencing is performed en masse on the Illumina next-generation sequencing platform. The second step identifies the genomic location of each transposon insertion site within the bank by mapping chromosomal sequences adjacent to inverted terminal repeats of the transposon to the reference genome. The fitness of insertion mutants containing disruptions in a given gene is reflected in both the relative number of insertion sites detected within the gene and the number of times each site is detected by the sequence analysis.
Fig. 1.
HITS and comparison of selected libraries. (A) HITS sample preparation and enrichment of transposon/chromosome junctions. After transposon mutagenesis, chromosomal DNA is purified from H. influenzae mutant library. Red, ITRs of himar1 transposon; white, contents of transposon, including the kanamycin resistance gene. Illumina oligonucleotide adapters (gray) are ligated to sheared genomic DNA. Fragments of the transposon/chromosome junctions are enriched via PCR using transposon- and adapter-specific primers. The biotinylated transposon-specific primer (yellow) anneals to the ITRs of the transposon and includes the Illumina sequencing primer site. The adapter-specific primer anneals to only 1 oligonucleotide of the partially complementary adapter. Enriched fragments are collected using streptavidin-coated paramagnetic beads. After washing, single-stranded DNAs are eluted from the beads and used for cluster formation on Illumina flow cells. (B) Comparison of lung-selected output library to input library. After sequencing, reads are mapped to the reference genome (solid arrows, plus strand; dashed arrows, minus strand) to identify the transposon insertion sites. The number of insertion sites detected per gene and the number of sequencing reads per site are used to determine the relative abundance of the mutant within the library before and after selection. The examples depict insertion patterns at TA sites in hypothetical gene A, in which insertion mutations confer attenuated growth or survival during infection, and gene B that is not required for growth in vitro or in vivo. Insertions in genes that are essential for growth on rich culture media are absent in the input library and are not detected by HITS.
Generation of a Mutant Bank Selected for Growth or Survival in Vivo in the Murine Lung Model.
The mechanisms that allow H. influenzae to persist in the lung are not well understood. Mouse pulmonary infection provides a well-established model for investigation of mechanisms used by H. influenzae and other bacteria to persist and resist host defenses during lung pathogenesis (10–12), yet there have been no comprehensive studies to identify H. influenzae genes needed at this site. To evaluate the utility of HITS for virulence gene identification using the mouse lung model of infection, we inoculated 5 mice with 107 cfu of a ≈75,000 member insertion mutant library of H. influenzae generated with a Himar1 mariner–derived minitransposon. At 24 h after inoculation an average of 9.2 × 105 cfu were recovered from the lungs of each mouse. Chromosomal DNA was isolated for analysis from both the inoculum and from the ex vivo bacterial populations. The numbers of cfu in the inoculum and recovered from mice suggested that the mutant library was likely to be sufficiently represented in both populations and that mutants had been subjected to in vivo selection.
Analysis of Genomic Mutant Banks by HITS and Application to Genome-Wide Identification of H. influenzae Genes Required in the Lung.
We conducted the HITS procedure (Fig. 1) on the input library and mapped the insertions to their chromosomal positions (Fig. S1). Insertions were evenly distributed around the chromosome, and ≈44% of the 131,960 total possible chromosomal TA target sites for mariner were found to have sustained insertions in this library. Before passage in vivo, a total of 534,567 sequencing reads mapped to nonrepetitive chromosomal regions immediately flanking 55,935 unique sites, with 44,270 in predicted protein coding genes and 11,665 in intergenic regions or structural RNAs. Of 1,657 annotated genes, no insertions were detected within 268 genes and 90 sustained insertions in <5% of their possible TA insertion sites, implicating at least 358 genes as essential for growth or viability on laboratory medium in vitro. Twenty-five genes with <8 possible TA insertion sites were excluded from analysis on the basis of an estimated probability of 0.05 that they could fail to sustain insertions owing to chance at this level of transposon insertion density in the library. Thirty-five genes could not be analyzed because they either contained extensive repetitive sequences or were duplicated in the genome. After subtraction of essential genes, genes containing repetitive sequence, or genes with very few possible insertion sites, there were 1,239 genes that could be analyzed.
For fitness analysis of mutants after in vivo selection, we considered the number of transposon insertions detected in the first 5–80% of each gene, the region in which insertions are expected to abrogate gene function. To exclude genes in which insertion mutations led to potential in vitro growth defects, we set a threshold requiring that candidate virulence genes sustain insertions in at least 40% of their possible mariner transposon target sites before in vivo selection, and 201 genes sustained densities of insertions below this threshold (Fig. 2 and Table S1). In the 1,038 annotated protein-coding genes that were dispensable in vitro, an average of 287 sequencing reads detecting insertions in the 5′ 5–80% region of each gene was observed (Fig. S2). HITS analysis of these genes was quite reproducible. When two preparations of genomic DNA from the transposon mutant bank were independently analyzed, the number of insertions detected in each gene was similar, with the majority (82%) of genes having <20% variation in insertion density between samples (Fig. S2). Therefore, both the complexity of the transposon bank and the detection of mutations by sequencing seemed to be sufficiently saturating for reproducible analysis of the relative abundance of mutants in the library.
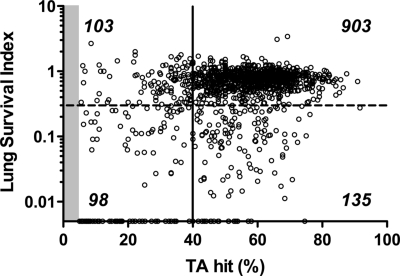
Fig. 2.
Comparison of transposon insertions in the mutant library before and after selection in the lung model. Insertion sites in the 5′ 5–80% protein coding sequence of the gene and reads associated with these sites were considered for fitness analysis. The saturation of transposon insertions within 1,239 genes in the input library is shown on the x axis. Saturation was calculated as the percentage of sites within a gene sustaining transposon insertions to the total number of possible of insertion sites (TA dinucleotides). The lung survival index (s.i.) is represented on the y axis as the number of reads mapped to a gene in the output library divided by the reads identified in the input library (points on the x axis represent s.i. values of zero). Essential genes, those sustaining insertions in <5% of possible sites, are not shown (shaded); the majority of these sustained no insertions, and the remaining 25% averaged 1 insertion per gene. The threshold for an inferred in vitro growth defect (solid line) was set at a saturation of 40% of the possible TA insertion sites within a gene. The threshold for in vivo attenuation (dashed line) was set at a lung s.i. of <0.30. Numbers of genes falling within each quadrant are indicated.
To identify genes required during infection, we analyzed the relative number of insertions in each gene in the output library obtained after lung infection vs. insertions in the library before in vivo selection. The results are shown graphically in Fig. 2, and the complete data are listed in Table S1. A total of 903 genes had similar numbers of insertions before and after selection in the lung, indicating that they were not required in this model. This large number of genes with insertion patterns in the output library that were similar to those in the input library indicated that significant stochastic loss of mutations had not occurred in the infection model. The 135 genes that sustained insertions in >40% of their possible TA insertion sites and in which the number of insertions decreased by at least 3.3-fold after selection in the lung were considered candidate virulence genes (Table S2). Representative insertion patterns for genes detected as being required during infection (galU and orfH) vs. those that are not required in vivo (xylA) are shown in Fig. S3. In summary, HITS implicated 8.1% of the 1,657 annotated genes in the genome in survival or growth of H. influenzae in the lung.
Genetic footprinting provides a means for analyzing insertions in discrete genes to verify results obtained with HITS. Genetic footprinting uses PCR with a specific chromosomal primer paired with a transposon primer for physical mapping of insertions to the chromosome in a bank of mutants (13). For a given gene, PCR results in a set of products varying in size that correspond to the distance between the chromosome-specific primer and each transposon mutation within that gene. Specificity is further assured by conducting the procedure with a primer 5′ of the gene and independently with a primer 3′ of the gene. For this validation we chose genes of LPS biosynthesis (opsX, rfaF, orfH, and galU) in which mutations resulted in pronounced attenuation relative to wild-type according to HITS data (Table S2). H. influenzae produces a short chain carbohydrate on its LPS (also called lipooligosaccharide, LOS) and lacks the repeating O-antigen carbohydrate typical of some bacterial LPS. The LOS of H. influenzae consists of a conserved “inner core” usually composed of 3 heptose residues and an “outer core” composed of variable-length oligosaccharide extensions from the heptose residues. The opsX, rfaF, and orfH genes encode heptosyltransferases I, II, and III, respectively, and generate the chain of 3 heptose residues initiating at a single 3-deoxy-D-manno-octulosonic acid, which is attached to lipid A (14–16). The galU gene encodes a UDP-glucose pyrophosphorylase that catalyzes the UTP-dependent conversion of D-glucose-1-phosphate into UDP-glucose, the activated form of the sugar required for biosynthesis of various carbohydrates, and galU is required for addition of glucose and galactose residues to the LPS of diverse pathogenic bacteria (17–19).
Representative genetic footprinting results are shown for galU and orfH and compared with insertion patterns detected by HITS in Fig. 3 A and B. The decrease in insertion mutations detected in these genes after in vivo passage of the bank provided verification of selection against mutants with disruptions in these genes, and band intensities on genetic footprinting gels were in good agreement with the abundance of insertions at each site as detected by HITS. Genetic footprinting also detected in vivo attenuation of mutants with insertions in opsX, rfaF, and galE, and similar results were obtained in reactions with primers positioned either 5′ or 3′ of each gene (Fig. S4). In contrast, xylA, a gene of D-xylose metabolism that is not required for bacteremia (20), exhibited similar mutational profiles in both the input and output banks (Fig. 3C), indicating that it is dispensable in the lung model. Therefore, these results provided a verification of HITS results by an independent method, identifying virulence factors previously implicated in bacteremia.
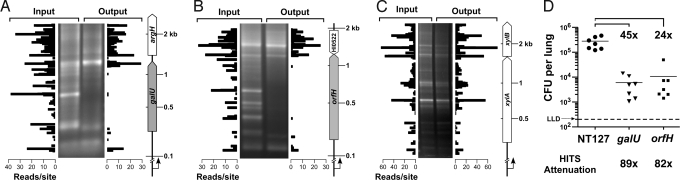
Fig. 3.
Comparison of HITS analysis, genetic footprinting, and single-strain infections. Genetic footprinting of input and output libraries for (A) galU, encoding UDP-glucose pyrophosphorylase, (B) orfH, encoding heptosyltransferase III, and (C) xylA, encoding xylose isomerase, are shown in the gel images. PCR analysis was conducted using the transposon-specific primer marout and chromosomal primers galU_F, orfH_F, or xylA_F that anneal 278 bp, 202 bp, and 279 bp upstream of the respective genes. In the plots, HITS data correspond to regions analyzed by footprinting. (D) H. influenzae NTHi wild-type (NT127) and deletion mutants of galU and orfH recovered from lungs of C57BL/6 mice (7 mice per strain) 24 h after intranasal inoculation with each strain. Bars represent the mean cfu per lung. Comparisons between wild-type and mutants were statistically significant via one-way ANOVA with Tukey's multiple comparison test (P < 0.001). LLD, lower limit of detection. Fold differences in mean cfu recovered for NT127 wild-type strain vs. the galU or orfH mutants in individual mutant infections (brackets) are compared with HITS results (below the chart). HITS survival indices were 0.011 for galU and 0.012 for orfH, corresponding to in vivo attenuations (calculated as the reciprocal of the s.i.) of 89-fold and 82-fold, respectively. In A–C, genome coordinates of transposon insertion sites detected via HITS analysis were reoriented with respect to the chromosomal primer positions used in footprinting. The y axis was modeled to the migration of the molecular weight (MW) standard of footprinting gels using nonlinear regression, and the x axis represents the number of sequencing reads mapped to insertion sites. The scale of the MW standards on the right of each panel applies to both the genetic footprints and the HITS analysis plots. White, nonessential genes; gray, genes required for growth or survival in the lung.
To assess whether genes identified by HITS as being required in vivo are also required in single-strain infections, we generated nonpolar mutations removing the complete coding sequences of galU or orfH, genes implicated as being required in the lung model by HITS. To address mutant phenotypes with a recent clinical isolate, mutations were constructed in the nontypeable H. influenzae strain, NT127 (21). In agreement with HITS and genetic footprinting results, both mutants were attenuated for survival in the lung. Moreover, the degree of attenuation calculated by HITS falls within the variation in fold difference observed between single-strain infections of individual mice. (Fig. 3D). The galU and orfH genes were previously shown to be essential for survival of H. influenzae in bloodstream models of infection (22, 23). A requirement for these genes in the lung supports the view that H. influenzae utilizes structures of the LPS inner and outer core in virulence strategies to combat clearance mechanisms of the host found in both of these environments.
Discussion
The genes implicated in bacterial growth or survival in the lung were functionally diverse, although several general categories were notable (Table S3). On the basis of Clusters of Orthologous Groups (COG) classifications, categories that were overrepresented in the attenuated gene set relative to their representation in the genome overall were “cell wall/membrane/envelope biosynthesis,” “amino acid transport and metabolism,” and “nucleotide transport and metabolism” (Tables S2 and S3). The genes identified provided insight into the selection conditions encountered by H. influenzae in the lung model.
Components of the bacterial cell surface are frequently the most direct participants in host–pathogen interactions. A major class of genes related to the cell envelope that was identified as markedly attenuated in vivo consisted of genes of LPS synthesis. LPS is essential in models of H. influenzae pathogenesis in the middle ear and blood and contributes to numerous aspects of NTHi infection, including evasion of complement and antimicrobial peptides (24–26). Genes needed for extension of the LPS inner-core structures (opsX, rfaF, and orfH) were required in vivo in the lung [survival index (s.i.) ≤0.012] (Table S2), in agreement with the requirement for these genes for bacteremia (22, 23). Genes required for precursor production for LPS carbohydrate outer-core hexose extensions (galU and galE) were also required (s.i. ≤ 0.025), suggesting that unmodified inner-core LPS results in enhanced clearance of H. influenzae from the lung. Genes required for hexose extensions from the first heptose, lgtF (s.i. = 0.152), or the terminal heptose of the inner core, lpsA (s.i. = 0.258), were partially required (16, 27), and a trend of moderate attenuation (≈1.5-fold) was also observed in single-strain infections with an lpsA mutant (Fig. S5). Distal modifications of the LPS outer-core structure mediated by genes such as lic3A, which adds sialic acid or the lic1 locus responsible for addition of phosphorylcholine seemed to be nonessential in vivo in these experiments. The lic1D gene was previously implicated in the lung model at a late time during infection, but not at 24 h (28), and it is possible that other distal modifications also are more important at later times.
Numerous genes involved in transport of proteins or other substrates were implicated in the lung model, including the complete twin-arginine translocation system (tatA, tatB, and tatC), which translocates folded proteins that lack Sec-dependent signal sequences across the plasma membrane and contributes to virulence in multiple pathogens (29). An intriguing set of genes with recently predicted functions in maintenance of outer-membrane lipid asymmetry (30) was implicated in pathogenesis in the lung. These genes included vacJ and a set of 5 genes annotated as “hypothetical genes” that are putative orthologs of an ABC transport system of Escherichia coli encoded by the mlaA and mlaBCDEF loci (for clarity, E. coli names of these genes are noted in Table S2). Orthologs of these genes were implicated in virulence of enteroinvasive E. coli, Shigella flexneri, and Burkholderia pseudomallei (31–33). The mla gene orthologs were required late during the intracellular life cycle for escape from the phagocytic vacuole (31). A role for the mla genes in both intracellular pathogens and H. influenzae suggests that H. influenzae may encounter membrane-damaging host defenses, such as cationic peptides or stress conditions in the lung, that are similar to those found in the phagocytic vacuole.
Additional overrepresented COG groups included genes involved in nutrient acquisition and interrelated adaptations to physiologic stress. Pathways of amino acid metabolism were required in the lung and included enzymes for synthesis or interconversion of methionine, asparagine, aspartate, serine, tryptophan, and branched-chain amino acids. Consistent with amino acid limitation, genes predicted to encode regulators involved in the stringent response were implicated in vivo, including RelA, which synthesizes (p)ppGpp in response to amino acid starvation (34), DksA, which modulates rRNA expression in response to (p)ppGpp (35), and Lon protease, which is activated by polyphosphate generated from (p)ppGpp (36) and has been implicated in proteolytic control of virulence factors (reviewed in ref. 37). Genes of nucleotide uptake and metabolism included those required for synthesis of purines and pyrimidines, in addition to genes involved in NAD uptake, nadN and hel. These genes mediate sequential conversion of NAD to NMN and nicotinamide riboside for uptake of this nucleotide that H. influenzae is unable to synthesize de novo (38). The complete set of genes for phosphate uptake (pstS, pstB, pstA, and pstC) was implicated in pathogenesis, as was the gene predicted to encode PhoB, a conserved response regulator protein that becomes active under low-phosphate conditions and controls diverse virulence functions in bacterial pathogens (reviewed in ref. 39). PhoR, a sensor kinase that activates PhoB, was not implicated in the lung. In other species, PhoB can be activated by “cross-talk” with other signaling systems independently of PhoR (40), and therefore H. influenzae PhoB may be required for responses to alternative signals. Resistance to oxidative stress is important for many pathogens. Genes involved in adaptations to oxidative stress conditions were identified, including pgdX, encoding a glutathione-dependent peroxidase (41), oxyR, which regulates genes critical for oxidative stress resistance, including pgdX (42), and genes of recombination pathways (ruvA, ruvB, ruvB, recR, recC, xerC, and xerD) required to repair DNA damaged by oxidative stress (43). Several genes implicated in the lung model (nadN, hel, and pdgX) are dispensable for bloodstream colonization by H. influenzae type b (41, 44). It is possible that H. influenzae strains differ in their requirements for these genes in vivo, or that these genes are specifically needed in the lung, where nucleotide sources and levels of oxidative stress may differ from those in the blood.
Conclusion
HITS provides a massively parallel system to simultaneously monitor the relative fitness of thousands of individual mutants undergoing a selection condition of interest. In this report, a large library of ≈75,000 H. influenzae mutants was subjected to selection in a murine pulmonary model of pathogenesis to identify genes required for prolonging survival of H. influenzae in the lung. Analysis of the mutant library by HITS was easily performed, highly reproducible, and remarkably comprehensive. Sequencing of transposon/chromosome junctions revealed independent insertions in nearly 56,000 genomic sites. More than 96% of H. influenzae protein coding genes were analyzed using a conservative cutoff that excluded 35 genes that were duplicated or contained repetitive sequences and 25 genes that had <8 TA dinucleotides available for mariner insertion. It is anticipated that with improvements in high-throughput sequencing technology, the depth of sequencing coverage will substantially increase to allow an even greater level of resolution and dynamic range. The results provide a genome-wide assessment of the genetic requirements of this bacterium for growth or survival in the lung, and also represent the most comprehensive fitness analysis that has been applied to H. influenzae mutants in any animal model. The profile of genes required in this environment provides a view of the host–pathogen interactions occurring during pulmonary pathogenesis and will provide insight into potential strategies for the design of vaccines or therapeutics to specifically target H. influenzae in this site of disease.
The HITS procedure was demonstrated using a mariner transposon bank in H. influenzae; however, none of the procedures are organism specific, and the approach should be applicable to any organism amenable to mutational analysis with transposons. A major advantage of the approach we present in this report is that it can be applied to existing mutant libraries and does not require use of a specifically engineered transposon. In fact, the procedure should be readily adaptable to libraries generated with any insertion mutation capable of providing a primer-binding site. Although a complete genome sequence is useful for HITS, mapping insertions to annotated contigs of draft genome sequences should yield much of the same information. Although HITS was used in this report to obtain a genome-wide assessment of the requirements for lung pathogenesis, the procedure should be equally effective for analysis of requirements for growth or survival under any selective condition that can be applied to large populations of mutants en masse. Because of the speed and resolution of HITS, it will be possible to efficiently conduct fitness analyses in diverse contexts of host–microbe interactions. Application of this approach is expected to generate multifaceted views of the genetic requirements of pathogens in the environments they encounter in diverse stages of pathogenesis.
Materials and Methods
High-Density Mutagenesis of H. influenzae by in Vitro Transposition.
H. influenzae Rd strain BA042 and clinical isolate nontypeable strain NT127 (21) were grown in brain heart infusion broth (BHI) supplemented with 10 μg/mL hemin and 10 μg/mL NAD (sBHI) or on sBHI agar plates at 35 °C. Media contained kanamycin sulfate at 20 μg/mL (sBHI-Kan) where indicated. The mini-mariner transposon mmTrcK (carried on plasmid pENTtrcK) was derived from magellan1 (8) by replacement of the endogenous promoter for the kanamycin resistance gene, aphI, with the trc promoter. Transposition reactions were performed in vitro as described in ref. 45. Transposition products were transformed into H. influenzae as described previously (8, 46). After selection on sBHI-Kan plates, the insertion library (≈75,000 colonies) was harvested in BHI with 20% glycerol and stored at −80 °C.
Selection of Transposon Insertion Mutant Library in the Lung Model.
The H. influenzae insertion library (3.1 × 1010 cfu) was inoculated in 50 mL sBHI and grown with shaking at 225 rpm to a final OD600 of 0.45. For representation of the input library, cells from 35 mL of culture were collected by centrifugation and stored at −80 °C. Inoculum for murine lung infection was prepared by pelleting 5 mL of the culture, washing in 1× Hank's buffered salt solution, and dilution to concentration of 2.5 × 108 cfu/mL. Forty microliters (107 cfu) was inoculated into the nares of 5 female C57BL/6 mice (7 to 8 weeks old) anesthetized with ketamine (50 mg/kg) and xylazine (5 mg/kg) by i.p. injection. At 24 h of infection, lungs were harvested and homogenized using a Fisher Tissuemiser. Dilutions of homogenates were plated on sBHI to enumerate total cfu per lung. To recover the output library, homogenates from each mouse lung were plated on 12 sBHI agar plates, and resulting colonies were collected for chromosomal DNA isolation via phenol chloroform extraction (8). All experiments with mice were conducted with prior approval of the University of Massachusetts Institutional Animal Use and Care Committee (IACUC).
Genetic Footprinting.
Genetic footprinting was conducted on H. influenzae genomic DNA from input and output libraries as described elsewhere (45) with transposon-specific primer, marout, and gene-specific primers that bind 5′ or 3′ of each gene. Primer design, PCR conditions, and image analysis are described in SI Methods, and footprinting primers are listed in Table S4.
Illumina Sequencing of Transposon–Chromosome Junctions from Mutant Libraries.
Genomic DNA from mutant libraries prepared before and after in vivo selection was sheared using a Covaris S2 device. Paired-end Illumina libraries were created by ligation of adaptors to sheared DNA as described by Bentley et al. (47) and size selected between 200 and 400 bp. Enrichment of transposon/chromosomal junction regions was performed by PCR amplification with a 5′ biotinylated transposon enrichment primer, PE1MAR, and adapter-specific PCR PE2.0 enrichment primer (Table S4). Thermocycler settings were as follows: 30 s, 98 °C; 18 cycles of 10 s, 98 °C, 30 s, 65 °C, 30 s, 72 °C; 5 min, 72 °C. Fragments between 250 and 300 bp were gel purified and added to Dynal MyOne C1 beads (Invitrogen) to capture biotinylated templates containing transposon insertions. The beads were washed according to the manufacturer's instructions, and the nonbiotinylated strand was eluted with 125 mM NaOH. Supernatants were recovered from beads, neutralized, and templates purified with MinElute PCR purification columns (Qiagen). The resulting transposon libraries were quantified on an Agilent Bioanalyzer 2100 RNA Pico6000 chip (Agilent Technologies). Single-stranded templates were cluster amplified and sequenced on an Illumina GAII, as described in ref. 47.
Analysis and Mapping of Illumina Sequencing Data.
The Illumina sequencing reads that contained the Himar1 inverted terminal repeat (ITR) sequence and the adjacent TA insertion site were identified in the raw fasta files and trimmed of the ITR sequence. The processed sequencing reads are provided as multifasta files for Input Library Sample1 (Dataset S1), Input Library Sample2 (Dataset S2), and Lung Output Library (Dataset S3). Processed reads, typically 53 bp in length, were aligned to the H. influenzae Rd KW20 genome sequence (48) (GenBank accession no. L42023) using SOAPv1.11 alignment software using default settings (2 mismatches allowed per read) (49). A custom PERL script was used to parse insertion site coordinates from the SOAP output file to report the number of reads mapped per site and strand orientations of aligned reads (SI Computer Script). The data were imported into Microsoft Excel, and insertion site coordinates were mapped to positions within protein coding genes annotated in protein table RefSeq file NC_000907.ptt (from the National Center for Biotechnology Information: ftp://ftp.ncbi.nih.gov/). For each gene, the number of insertion sites identified and the total number of sequencing reads in the internal 5–80% of the gene were determined using Excel functions. Additional details are provided in SI Methods. A draft version of H. influenzae strain RdAW (also referred to as BA042) genome sequence was generated and is at least 99.98% identical to strain Rd KW20 (48).
Single-Strain Infections in the Pulmonary Clearance Model.
Nonpolar mutations deleting the galU and orfH genes were introduced into nontypeable H. influenzae strain, NT127 (SI Methods). Each H. influenzae strain was used to inoculate C57BL/6 mice by the intranasal route as described above. At 24 h of infection, mice were killed and bacterial cfu in the lungs were enumerated as described above. The number of cfu recovered from the lungs of each mouse was compared by one-way ANOVA with Tukey's multiple comparison test. Blood samples obtained immediately before killing revealed no detectable cfu. Procedures were approved by the University of Massachusetts IACUC.
Supplementary Material
Acknowledgments.
We thank John Leong for his helpful comments; and David Lapointe (UMass Medical School) and David Borenstein, Philip Montgomery, and Carsten Russ (Broad Institute) for analytic support and technical input that contributed to the development of HITS methodology. This project has been funded in part by the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN266200400001C (Broad Institute), and by National Institutes of Health Grant 1RO1-AI49437 (to B.J.A.).
Note Added in Proof.
During review of this manuscript we learned of an independent report submitted to Nature Methods by Tim van Opijnen, Kip L. Bodi, and Andrew Camilli, in which transposon junction sequencing was successfully applied to study genetic networks (personal communication).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Draft genome sequences for H. influenzae strains have been submitted to the National Center for Biotechnology Information: Rd BA042 (RdAW) (accession no. ACSN01000000) and NT127 (accession no. ACSL01000000).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906627106/DCSupplemental.
References
- 1.Hensel M, et al. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 2.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badarinarayana V, et al. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat Biotechnol. 2001;19:1060–1065. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- 5.Vila-Corcoles A, et al. Epidemiology of community-acquired pneumonia in older adults: A population-based study. Respir Med. 2009;103:309–316. doi: 10.1016/j.rmed.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Sethi S, Murphy TF. Bacterial infection in chronic obstructive pulmonary disease in 2000: A state-of-the-art review. Clin Microbiol Rev. 2001;14:336–363. doi: 10.1128/CMR.14.2.336-363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 8.Akerley BJ, et al. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin EJ, et al. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wieland CW, et al. The MyD88-dependent, but not the MyD88-independent, pathway of TLR4 signaling is important in clearing nontypeable Haemophilus influenzae from the mouse lung. J Immunol. 2005;175:6042–6049. doi: 10.4049/jimmunol.175.9.6042. [DOI] [PubMed] [Google Scholar]
- 11.Toews GB, Hart DA, Hansen EJ. Effect of systemic immunization on pulmonary clearance of Haemophilus influenzae type b. Infect Immun. 1985;48:343–349. doi: 10.1128/iai.48.2.343-349.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toews GB, Viroslav S, Hart DA, Hansen EJ. Pulmonary clearance of encapsulated and unencapsulated Haemophilus influenzae strains. Infect Immun. 1984;45:437–442. doi: 10.1128/iai.45.2.437-442.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh IR, Crowley RA, Brown PO. High-resolution functional mapping of a cloned gene by genetic footprinting. Proc Natl Acad Sci USA. 1997;94:1304–1309. doi: 10.1073/pnas.94.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gronow S, Brabetz W, Lindner B, Brade H. OpsX from Haemophilus influenzae represents a novel type of heptosyltransferase I in lipopolysaccharide biosynthesis. J Bacteriol. 2005;187:6242–6247. doi: 10.1128/JB.187.17.6242-6247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichols WA, et al. Identification of the ADP-L-glycero-D-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect Immun. 1997;65:1377–1386. doi: 10.1128/iai.65.4.1377-1386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hood DW, et al. Genetic basis for expression of the major globotetraose-containing lipopolysaccharide from H. influenzae strain Rd (RM118) Glycobiology. 2001;11:957–967. doi: 10.1093/glycob/11.11.957. [DOI] [PubMed] [Google Scholar]
- 17.Nesper J, et al. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun. 2001;69:435–445. doi: 10.1128/IAI.69.1.435-445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weissborn AC, Liu Q, Rumley MK, Kennedy EP. UTP: Alpha-D-glucose-1-phosphate uridylyltransferase of Escherichia coli: Isolation and DNA sequence of the galU gene and purification of the enzyme. J Bacteriol. 1994;176:2611–2618. doi: 10.1128/jb.176.9.2611-2618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhury B, Carlson RW, Goldberg JB. The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr Res. 2005;340:2761–2772. doi: 10.1016/j.carres.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Rosadini CV, Wong SM, Akerley BJ. The periplasmic disulfide oxidoreductase DsbA contributes to Haemophilus influenzae pathogenesis. Infect Immun. 2008;76:1498–1508. doi: 10.1128/IAI.01378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington JC, et al. Resistance of Haemophilus influenzae to reactive nitrogen donors and gamma interferon-stimulated macrophages requires the formate-dependent nitrite reductase regulator-activated ytfE gene. Infect Immun. 2009;77:1945–1958. doi: 10.1128/IAI.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood DW, et al. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]
- 23.Wong SM, Alugupalli KR, Ram S, Akerley BJ. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol. 2007;64:1375–1390. doi: 10.1111/j.1365-2958.2007.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueira MA, et al. Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect Immun. 2007;75:325–333. doi: 10.1128/IAI.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho DK, et al. lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. J Immunol. 2007;178:1002–1012. doi: 10.4049/jimmunol.178.2.1002. [DOI] [PubMed] [Google Scholar]
- 26.Lysenko ES, et al. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect Immun. 2000;68:1664–1671. doi: 10.1128/iai.68.3.1664-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hood DW, et al. Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology. 2004;150:2089–2097. doi: 10.1099/mic.0.26912-0. [DOI] [PubMed] [Google Scholar]
- 28.Pang B, et al. Lipooligosaccharides containing phosphorylcholine delay pulmonary clearance of nontypeable Haemophilus influenzae. Infect Immun. 2008;76:2037–2043. doi: 10.1128/IAI.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Buck E, Lammertyn E, Anne J. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 2008;16:442–453. doi: 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, et al. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol. 1994;11:31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 32.Hong M, Gleason Y, Wyckoff EE, Payne SM. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect Immun. 1998;66:4700–4710. doi: 10.1128/iai.66.10.4700-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuccui J, et al. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun. 2007;75:1186–1195. doi: 10.1128/IAI.01240-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul BJ, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Kuroda A, et al. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 37.Butler SM, Festa RA, Pearce MJ, Darwin KH. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- 38.Kemmer G, et al. NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J Bacteriol. 2001;183:3974–3981. doi: 10.1128/JB.183.13.3974-3981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamarche MG, Wanner BL, Crepin S, Harel J. The phosphate regulon and bacterial virulence: A regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev. 2008;32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 40.Wanner BL. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- 41.Vergauwen B, Herbert M, Van Beeumen JJ. Hydrogen peroxide scavenging is not a virulence determinant in the pathogenesis of Haemophilus influenzae type b strain Eagan. BMC Microbiol. 2006;6:3. doi: 10.1186/1471-2180-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison A, et al. The OxyR regulon in nontypeable Haemophilus influenzae. J Bacteriol. 2006;189:1004–1012. doi: 10.1128/JB.01040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stohl EA, Seifert HS. Neisseria gonorrhoeae DNA recombination and repair enzymes protect against oxidative damage caused by hydrogen peroxide. J Bacteriol. 2006;188:7645–7651. doi: 10.1128/JB.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt-Brauns J, et al. Is a NAD pyrophosphatase activity necessary for Haemophilus influenzae type b multiplication in the blood stream? Int J Med Microbiol. 2001;291:219–225. doi: 10.1078/1438-4221-00122. [DOI] [PubMed] [Google Scholar]
- 45.Akerley BJ, Lampe DJ. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 2002;358:100–108. doi: 10.1016/s0076-6879(02)58082-4. [DOI] [PubMed] [Google Scholar]
- 46.Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 47.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 49.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.