Abstract
The 22q11 deletion (or DiGeorge) syndrome (22q11DS), the result of a 1.5- to 3-megabase hemizygous deletion on human chromosome 22, results in dramatically increased susceptibility for “diseases of cortical connectivity” thought to arise during development, including schizophrenia and autism. We show that diminished dosage of the genes deleted in the 1.5-megabase 22q11 minimal critical deleted region in a mouse model of 22q11DS specifically compromises neurogenesis and subsequent differentiation in the cerebral cortex. Proliferation of basal, but not apical, progenitors is disrupted, and subsequently, the frequency of layer 2/3, but not layer 5/6, projection neurons is altered. This change is paralleled by aberrant distribution of parvalbumin-labeled interneurons in upper and lower cortical layers. Deletion of Tbx1 or Prodh (22q11 genes independently associated with 22q11DS phenotypes) does not similarly disrupt basal progenitors. However, expression analysis implicates additional 22q11 genes that are selectively expressed in cortical precursors. Thus, diminished 22q11 gene dosage disrupts cortical neurogenesis and interneuron migration. Such developmental disruption may alter cortical circuitry and establish vulnerability for developmental disorders, including schizophrenia and autism.
Keywords: psychiatric disease
The neurodevelopmental hypothesis for diseases of cortical connectivity, initially proposed for schizophrenia (1), and later extended to autism spectrum disorders (2), suggests that anomalous cortical development underlies behavioral pathology. Despite inferred relationships between suspect developmental mechanisms, neuroanatomical or functional changes in patients, and postmortem cortical pathology, to our knowledge, there are no known direct links between specific cortical developmental mechanisms and pathogenesis. The near impossibility of prospective analyses in at-risk human fetuses further complicates rigorous evaluation of the hypothesis. Thus, the hypothesis may be more effectively evaluated in animal models of genetic or environmental risk for relevant diseases. In humans, 22q11 deletion/DiGeorge syndrome (22q11DS) confers the highest known genetic risk for schizophrenia (≈30%) (3, 4), increased susceptibility for autism spectrum disorders (≈25%) (5), and vulnerability for additional behavioral and learning disabilities (>60%) (5). Brain imaging in 22q11DS patients shows consistent anatomical defects, including reduced cortical gray matter and polymicrogyria (6–8), and postmortem analysis indicates cellular pathology associated with developmental defects including periventricular heteropias (9). We found that diminished 22q11 gene dosage in a 22q11DS mouse model compromises specific cortical neural stem cells, basal progenitors, and alters frequency and distribution of cortical projection neurons and GABAergic interneurons. These phenotypes suggest a link between a genomic lesion, altered cortical development, and subsequent changes in cortical circuitry that likely intensify risk for behavioral disorders in 22q11DS patients.
Results
Diminished Dosage of 22q11 Genes Disrupts Basal Progenitor Proliferation.
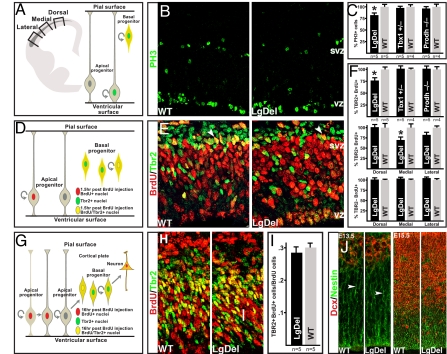
We first asked, using the LgDel mouse model of 22q11DS (10), whether 22q11DS cortical anomalies might reflect changes in cortical neurogenesis. We focused on two distinct classes of cortical progenitors: basal progenitors–transit amplifying progenitors in the cortical subventricular zone (SVZ)–and apical progenitors–self-renewing radial glial stem cells present in the cortical VZ. Each class can be recognized with combinations of proliferative and molecular markers (Fig. 1). We found reduced frequency of mitotic basal progenitors, identified by phosphohistone 3 labeling (PH3; a G2/M-phase cell-cycle marker) as well as SVZ location, throughout the embryonic day (E)13.5 LgDel cortex (79% of WT, P = 0.05, n = 5 per genotype) (Fig. 1 A–C; Fig. S1). Analysis of dual BrdU (90-min exposure; S-phase marker) and Tbr2 labeling (basal progenitors) confirms this deficit (76% of WT, t test, P = 0.049, n = 5) (Fig. 1 D–F). When systematically sampled at dorsal, medial, and lateral cortical locations, LgDel S-phase basal progenitor frequency was diminished by 32% in medial cortex (n = 5; t test, P = 0.045) (Fig. 1F Middle), 20% laterally (nonsignificant), and similar to normal dorsally.
Fig. 1.
Diminished 22q11 gene dosage disrupts cortical basal progenitor proliferation. (A) Schematic representation of a coronal section through the mouse E13.5 forebrain with the cortical SVZ indicated in gray. Proliferating cells were counted at probe locations shown by boxes, or throughout the entire cortex (Left). Schematic representation showing that basal and apical progenitors, both labeled by PH3, are discerned by their positions in the SVZ versus VZ (Right). (B) PH3 immunolabeling in the E13.5 cortex of WT and Large Deletion mice (LgDel) (10). (C) PH3 labeled cell frequency in the SVZ throughout its entire lateral to dorsal extent is significantly reduced in the LgDel cortex (*, P ≤ 0.05). However, in Prodh−/− deficient or Tbx1+/− mutants, frequency is unchanged. (D) Schematic representation of short-pulse BrdU paradigm used to label the S-phase SVZ progenitors. Dual BrdU/Tbr2 immunolabeling allows assessment of S-phase basal (Tbr2+/BrdU+) as well as apical (Tbr2-/BrdU+) progenitor frequency. (E) BrdU (90-min exposure) and Tbr2 immunolabeling in the WT and LgDel E13.5 cortex (arrowheads: double-labeled cells). (F) Tbr2+/BrdU+ SVZ cells, throughout the entire cortex, are more frequent in WT versus LgDel (*, P ≤ 0.05). However, their frequency is not changed in Prodh−/− or Tbx1−/+ (Top). There is a significant decrease of Tbr2+/BrdU+ cells in the medial and a trend in the lateral E13.5 LgDel cortex (Middle). Tbr2-/BrdU+ cell frequency is unchanged at all WT and LgDel E13.5 cortical probe locations (Lower). (G) BrdU paradigm used to assess apical progenitor production of basal progenitors. The 16-h BrdU exposure provides enough time for a fraction of labeled apical progenitor progeny to migrate toward the SVZ, and express Tbr2. Concurrently, BrdU-labeled basal progenitors down-regulate Tbr2 and migrate toward the cortical plate. The fraction of BrdU+ cells that are also Tbr2+ reflect basal progenitor generation by apical progenitors (54). (H) Tbr2+/16 h BrdU+ cells (yellow) in WT and LgDel E13.5 cortex. (I) Apical progenitor genesis of basal progenitors (Tbr2+BrdU/BrdU cells) is not significantly different between E13.5 WT and LgDel cortex (medial probe). (J) Immunofluorescent labeling in E13.5 and E15.5 WT and LgDel cortex show no disruption of migrating neuroblasts (Dcx, doublecortin) or radial glial processes (nestin) (arrowheads).
Aberrant apical progenitor proliferation or radial migration is not the basis for the basal progenitor proliferation defect. There are comparable numbers of S-phase apical progenitors in LgDel and littermate controls at each cortical site (Fig. 1F Bottom). Also, apical progenitors generate basal progenitors at similar frequencies in each genotype (Fig. 1 G–I). Furthermore, apical progenitor radial processes, on which nascent basal progenitors as well as newborn neurons migrate, seem unperturbed in the mutant (Fig. 1J).
Of the 22 22q11 genes expressed in the developing cerebral cortex (11), two genes (Tbx1 and Prodh) have emerged as candidates for many 22q11DS phenotypes, including behavioral anomalies (12, 13). Nevertheless, we found no differences in basal progenitor frequency in either hemizygous Tbx1 or homozygous Prodh hypomorphic mice (Fig. 1 C and F) (embryonic gene expression; Fig. S2). Thus, diminished Tbx1 dosage or Prodh activity is unlikely to explain the proliferative phenotype associated with hemizygous 22q11 deletion.
A Subset of 22q11 Genes Is Associated with the Cortical Proliferative Zone.
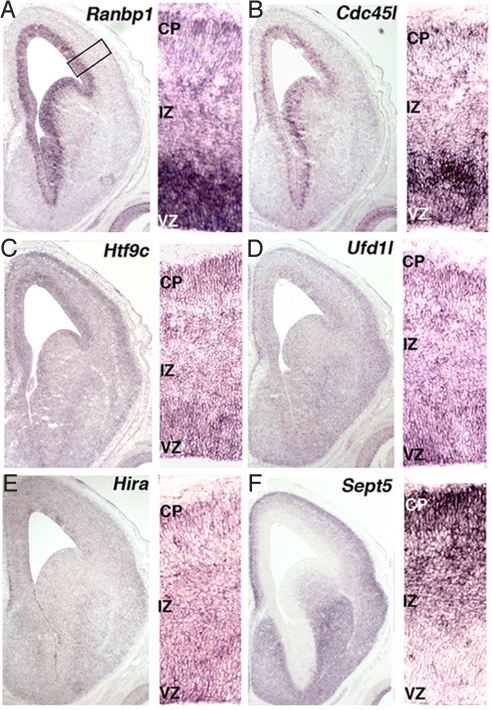
We asked whether other genes in the 22q11 region might contribute to apparent disruption of cortical basal progenitor proliferation in the LgDel mouse. We evaluated candidates by noting their putative cell-cycle function, selective expression in the cortical VZ/SVZ, and maximal expression during neurogenesis. Five 22q11 genes are thought to regulate the cell cycle: Ranbp1 (14), Cdc45l (15), Hira (16), Ufd1l (17), and Sept5 (18); a sixth gene with unknown function, Htf9c, shares a promoter with Ranbp1 (19), and thus, was also analyzed. Ranbp1 and Cdc45l are specifically and robustly expressed in the VZ/SVZ (Fig. 2 A and B). Htf9c and Ufd1l are enhanced in the VZ/SVZ and developing cortical plate (Fig. 2 C and D). Hira is weakly present throughout the developing cortex (Fig. 2E). Last, Sept5 is absent from the VZ/SVZ, instead localized to the developing cortical plate (Fig. 2F). Ranbp1, Cdc45l, and Htf9c are maximally expressed during cortical neurogenesis [E12-postnatal day (P)0] with a large decline thereafter (Fig. S3). In contrast, there is stability (Ufd1l), modest transient increase (Hira), or significant, sustained increase (Sept5) of the other genes (Fig. S3). Thus, expression localization, dynamics, and established cell-cycle function suggest candidate 22q11 genes for the basal progenitor phenotype.
Fig. 2.
Expression localization of 22q11 cell-cycle genes during cortical neurogenesis. In all images, the entire cortical hemisphere from an E14.5 WT embryo is shown (Left), whereas higher magnification of VZ, intermediate zone (IZ), and cortical plate (CP) is shown (Right). ISH shows that 22q11 cell-cycle genes are enhanced in the VZ [Ranbp1 (A); Cdc45l (B)] enhanced in both the VZ and CP [Htf9c (C); Ufd1l (D)] lightly, but broadly expressed [Hira (E)] or enhanced in the CP [Sept5 (F)].
Altered Expression of Cell-Cycle Genes in the LgDel Cortex.
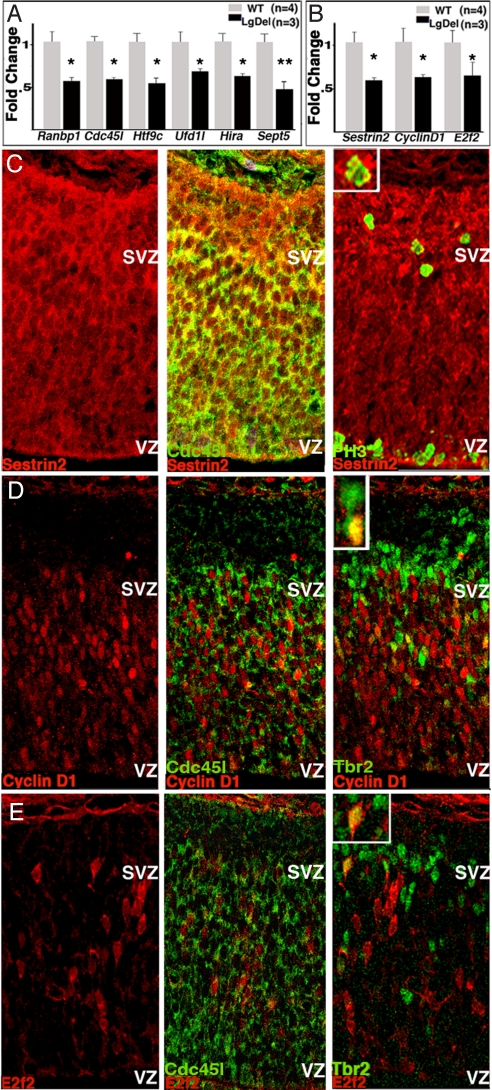
We next asked whether expression of these six 22q11 genes or other genes beyond the 22q11 interval is diminished in the embryonic LgDel mouse cortex. Expression of all six 22q11 genes is diminished by ≈50% in E13.5 LgDel cortex (Fig. 3A). Diminished cortical expression of these 22q11 genes, as well as altered basal progenitor proliferation, suggests there may be further changes in cell-cycle regulatory genes beyond the 22q11 region. We probed a cell-cycle specific array (SABiosciences) with cDNA from E13.5 LgDel and WT cortex. We found and confirmed reduced expression in LgDel cortex of 3/84 array genes (Fig. 3B): Cyclin D1, a key component of the G1/S cell-cycle transition (20, 21); E2f2, involved in S-phase progression (20); and Sestrin2, thought to regulate proliferation in response to cellular stress (22). Local expression of each protein is enhanced in the SVZ, and each is variably seen in basal progenitors, sometimes in combination with the 22q11 protein, Cdc45l (Fig. 3 C–E). Thus, expression of cell-cycle regulatory genes beyond the 22q11 region is disrupted in the cortical VZ/SVZ in concert with aberrant basal progenitor proliferation.
Fig. 3.
Changes in cell-cycle gene expression in embryonic LgDel cortex. (A) Six 22q11 putative cell-cycle genes show diminished expression by ≈50% (*, P ≤ 0.05; **, P ≤ 0.001) relative to WT E13.5 cortex. (B) Quantitative PCR verifies that three cell-cycle gene transcripts are diminished by ≈50% (*, P ≤ 0.05) in E13.5 LgDel cortex relative to WT as suggested by cell-cycle array. (C–E) Protein products of these three genes are detected in cells that also express Cdc45l, PH3, or Tbr2. (C) Sestrin2 (Left), colabeled with Cd45l (Center), or PH3 (Right). (D) CyclinD1 in E13.5 cortex (Left), colabeled with Cdc45l (Center), or Tbr2 (Right). (E) E2f2 in E13.5 cortex (Left), colabeled with Cdc45l (Center), or Tbr2 (Right).
Altered Layer 2–4 Neurons in the Maturing LgDel Cortex.
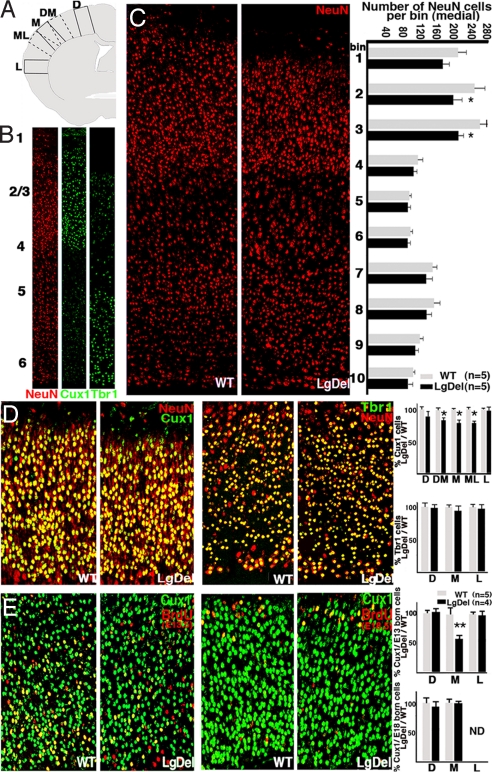
Basal progenitors give rise primarily to layer 2–4 projection neurons. Therefore, diminished proliferation of these cells in the embryonic LgDel cortex might be accompanied by changes in neuronal composition of the supragranular cortical layers at later ages. Thus, we determined the frequency of supra- and infragranular cortical neurons in P5 LgDel cortex. Using a pan-neuronal marker, NeuN, we found no significant change in the total frequency of neurons located in radial probes systematically located at lateral, medial, or dorsal cortical sites (Fig. 4 A and B). However, there was a trend toward diminished NeuN cell frequency in the medial probe location. Therefore, we determined NeuN cell frequency at this location in 10 equivalent bins between the pial surface and white matter. We found reduced NeuN cell frequencies in the medial supragranular LgDel cortex (Fig. 4C). We further evaluated the laminar specificity of this change using markers for molecularly distinct cortical neurons. There was a 20% decrease in the frequency of neurons labeled for Cux1, a layer 2–4 selective marker (23), in the medial LgDel cortex (n = 5 per genotype; t test, P = 0.02) (Fig. 4D). In contrast, the frequency of neurons labeled for Tbr1, a layer 5/6 selective marker (24), is unchanged (Fig. 4D). Also, Cux1 neuron frequency is unchanged at all cortical sites in Tbx1+/− mice and Prodh−/− deficient mice (data not shown).
Fig. 4.
Altered frequency of supragranular projection neurons accompanies disrupted basal progenitor proliferation. (A) Schematic representation of probe locations for quantifying frequency of (B) pan-neuronal (NeuN), layer 2–4 (Cux1), and layer 5/6 (Tbr1) cortical neurons in the WT and LgDel cortex at P5. (C) NeuN labeled cells in medial WT and LgDel probes (Left). The frequency of NeuN labeled cells across 10 equal bins from pia to white matter at the medial probe (two-way ANOVA, P ≤ 0.01, n = 5) (Right). Posthoc LSD test indicates significant reductions at LgDel bins 2 and 3 (*, P ≤ 0.05), which include supragranular cortical layers (layer 2–4). (D) Layer 2–4 neurons double-labeled for NeuN and Cux1 in P5 WT and LgDel cortex (Left). Layer 5/6 neurons double-labeled for NeuN and Tbr1 in P5 WT and LgDel cortex (Right). There is a significant decrease in Cux1 labeled neurons at dorsomedial, medial, and mediolateral cortical locations in the LgDel mutant (Upper; *, P ≤ 0.05) Tbr1 cells are not significantly altered at any cortical location (Lower). (E) Layer 2–4 neurons in P5 mouse cortex BrdU birth-dated at E13.5 (Left) and E18.5 (Right) and double-labeled for Cux1. There is a significant decrease in frequency of E13.5 generated Cux1 neurons only at the medial location in the LgDel cortex (Upper; **, P ≤ 0.01). There is no significant difference in E18.5 generated Cux1 cells (Lower; ND, not detected). D, dorsal; DM, dorsomedial; M, medial; ML, mediolateral; L, lateral.
To determine whether this specific change in layer 2–4 neuron frequencies reflects altered basal progenitor proliferation at E13.5 (Fig. 1), we analyzed the frequency of E13.5 birth-dated Cux1 or Tbr1 neurons in the P5 cortex. The frequency of E13.5 BrdU birth-dated/Cux1 double-labeled neurons was diminished by 50% only at the medial location (n = 4 LgDel, n = 5 WT; P = 0.01) (Fig. 4E). However, there was no significant difference in E13.5 BrdU injected/Tbr1 labeled layer 5/6 cells (Fig. S4). There was no evidence of compensation by accelerated neurogenesis later in development. We found no significant difference in E18.5 BrdU/Cux1 cell frequency between genotypes (Fig. 4E). Last, we did not see prolonged neurogenesis. E19.5 birth-dated neurons were not present in LgDel or WT P5 cortex (data not shown). Apparently, proliferative changes in E13.5 basal progenitors prefigure a corresponding change in the frequency of supragranular projection neurons in the medial cortex of LgDel mice.
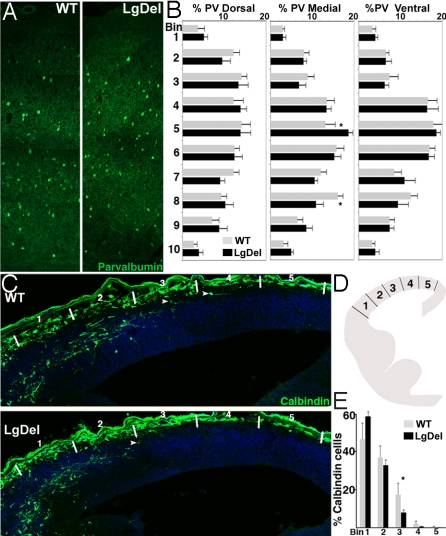
Altered Distribution of Interneurons in the LgDel Cortex.
Changes in interneurons, as well as projection neurons, have been associated with schizophrenia and other disorders of cortical connectivity (25, 26). Thus, we analyzed the distribution of parvalbumin-labeled interneurons, a proposed target for pathologic change (25, 26), in the LgDel postnatal cortex. There was no significant difference in parvalbumin cell frequency at P21. Nevertheless, their laminar distribution changes in the medial, but not lateral or dorsal cortex (two-way ANOVA, P = 0.03, n = 5 per genotype) (Fig. 5 A and B). We asked whether this change reflects altered migration of embryonic GABAergic neuroblasts. Parvalbumin is not expressed in mouse cortex until the second postnatal week (27). Therefore, we labeled the embryonic GABAergic cohort with calbindin (Fig. 5C), which is expressed in migrating interneurons (28). The frequency of calbindin-labeled cells did not differ in the two genotypes at E13.5. However, their distribution from the corticostriatal boundary to the cortical hem was altered in LgDel embryos (two-way ANOVA, P = 0.05, n = 5 per genotype) (Fig. 5E). The proportion of calbindin labeled cells in progressively medial locations diminishes more rapidly (Fig. 5 D and E), suggesting disrupted or delayed interneuron migration in the embryonic LgDel cortex.
Fig. 5.
Interneuron distribution is disrupted in the LgDel cortex. (A) Immunolabeling for parvalbumin in P21 WT and LgDel cortex (medial probe) shows altered location of cells in the LgDel cortex. (B) Parvalbumin cell distribution across 10 equivalent bins from pia to white matter in WT and LgDel P21 cortex. Cells were counted in dorsal, medial, and lateral probes. A significant interaction was observed between genotype and bin location medially (two-way ANOVA, P = 0.03, n = 5 per genotype). LSD test records a significant difference in frequency between genotypes at bins 5 and 8 (*, P ≤ 0.05). (C) Calbindin-labeled migrating interneurons in E13.5 WT and LgDel cortex. White lines mark bin locations for counting. (D) The E13.5 cortex was divided into five equidistant bins from the corticostriatal boundary to cortical hem and calbindin cell numbers in each bin determined. (E) The distribution of calbindin cells across five bins in LgDel and WT E13.5 cortex. Genotype has a significant effect on calbindin cell distribution (two-way ANOVA, *, P ≤ 0.05, n = 5 per genotype). A significant difference in calbindin frequency between genotypes at bin 3 was seen (LSD; *, P ≤ 0.05).
Discussion
We show that a genomic lesion associated with significant risk for schizophrenia, autism, and other behavioral disorders disrupts neurogenesis as well as frequency and distribution of projection neurons and migration of interneurons in a restricted region of the cerebral cortex. Our results suggest local changes in gray matter and neuropil, deduced from imaging and pathological assessment in 22q11DS patients (6–9), may reflect altered identity, abundance, and connections of projection neuron classes due to disrupted basal progenitor proliferation, and parallel changes in interneuron migration and placement during development. These phenotypes provide support for the neurodevelopmental hypotheses of disorders of cortical connectivity associated with 22q11DS.
The basal progenitor deficit in the LgDel model of 22q11DS apparently leads to altered frequency of supragranular neurons later in life. Basal progenitors may generate projection neurons for all cortical layers (29). Nevertheless, several observations suggest that they give rise primarily to supragranular pyramidal cells (30). Such projection neurons, present in altered numbers in LgDel cortex, may fail to elaborate appropriate local and long distance projections. The resulting disruptions in circuitry may lead to pathological changes, including altered interneuron distribution and frequency, as well as altered projection neuron connection in diseases of cortical connectivity that accompany 22q11DS (31–33). In contrast, apical progenitors are spared. Their ability to generate basal progenitors is unimpaired; their capacity to support radial migration is not noticeably altered, and their primary neuronal progeny (infragranular projection neurons) do not seem affected. Thus, our results identify a specific cortical progenitor class as a target for pathologic change, and specific cortical layers as subsequent loci for potentially altered circuitry in 22q11DS.
These alterations in the 22q11DS mouse model are restricted to medial cortical regions. Regionally selective basal progenitor and supragranular neurons deficits may reflect divergent activity of local signals and transcription factors (34). 22q11 genes may normally modulate the mode and tempo of basal progenitor division in the context of regional signals, resulting in appropriate local differences in cortical thickness, surface area, and gyral patterns (35). At present, one postmortem study of three 22q11DS schizophrenic brains (9) and a limited number of global MRI studies in 22q11DS patients (6–8) have been completed. The periventricular heterotopias and subtle differences in cortical thickness and gyral complexity in frontal and temporal cortical regions are generally consistent with region-selective developmental changes. Anterior frontal motor/association areas (36) appear most compromised in LgDel mice; nevertheless, it is difficult to predict cortical regions that might be similarly altered in 22q11DS patients.
Secondary effects due to cardiovascular phenotypes associated with Tbx1 (10, 37) or metabolic deficiency due to mitochondrial Prodh function (38, 39) do not appear to compromise basal progenitor proliferation and cortical development independent of other 22q11 genes. A recent study indicates that congenital heart defects may contribute to cortical anomalies in 22q11DS (40). However, the largest defects in cortical gyral patterns are independent of cardiovascular malformations. Thus, our results are consistent with current clinical evidence that suggests some dissociation between brain and cardiovascular phenotypes in 22q11DS. Nevertheless, additional 22q11 genes that influence either cardiovascular development or metabolic function (41, 42) may secondarily contribute to cortical phenotypes in 22q11DS.
Recent evidence suggests that at least two 22q11 genes, Zddhc8 (43) and Dgcr8 (44), can compromise forebrain circuitry, particularly in the hippocampus. Our data suggests additional candidates (particularly Ranbp1 and Cdc45l) as potential contributors to altered cortical development in 22q11DS. RANBP1 polymorphisms have been associated with schizophrenia vulnerability in non-22q11 deleted individuals (45) and restricted CDC45L, as well as UFD1L, deletion has been associated with 22q11DS clinical features including heart, craniofacial, and thymic anomalies (42). The cell-cycle functions of both Ranbp1 and Cdc45l, their diminished expression in LgDel VZ/SVZ, and parallel diminished expression of CyclinD1 and E2f2, major regulators of the G1/S check point, indicate that local activity of 22q11 genes may influence cortical cell-cycle dynamics. This influence may extend beyond cortical development. These genes continue to be expressed in adult forebrain neurons and precursors (11), and may be part of larger cell-cycle regulatory networks that are differentially disrupted in schizophrenic and bipolar patients (46). Last, altered cell-cycle control in cortical precursors due to diminished 22q11 gene dosage may also contribute to increased tumorigenesis in 22q11 patients (47).
There are several potential explanations for altered placement, but not frequency, of parvalbumin interneurons, the most numerous cortical interneuron subtype (48), most often associated with pathology in disorders of cortical connectivity (25, 26). Diminished 22q11 gene dosage may disrupt interneuron specification in the medial ganglionic eminence, where most of these cells are generated, or their migration to the cortex. Such changes may compromise the ability of subsets of interneurons to recognize appropriate regional or laminar positions. Alternately, altered migratory interneurons might perturb basal progenitor integrity (49) due to the close proximity of these cell types; thus, disrupting subsequent cortical proliferation as well as projection neuron and interneuron distribution. Last, changes in interneuron migration or distribution may reflect disrupted basal progenitor proliferation. Basal progenitors secrete chemoattractants for migrating interneurons (50), and diminished numbers in medial cortical regions may make them less effective migratory targets.
Altered placement of interneurons, as well as frequency of layer 2–4 projection neurons due to diminished 22q11 gene dosage, may establish vulnerability for any of several behavioral disorders associated with 22q11DS (3–5). Although our observations are unlikely to provide a singular explanation for the range of psychiatric illness found in 22q11DS, disrupted corticogenesis due to basal progenitor defects establish a likely contributor to circuit vulnerability (and behavioral disturbances) in the LgDel mouse model of 22q11DS (51), and provide some guidance for subsequent analyses of brain abnormalities in 22q11DS.
Materials and Methods
Experimental Animals.
Mice carrying a hemizygous deletion from Idd to Hira were maintained on a C57BL/6J background (LgDel mice) (10). The Pro/ReJ line (C57BL/6J) contains a base substitution that reduces, but not abolishes Prodh function (13). The Tbx1 line (129S) replaces Tbx1 with a neomycin cassette deleting all but the first 22 aa (52). Animal procedures met the University of North Carolina, Chapel Hill Institutional Animal Care and Use Committee requirements.
RNA Isolation, cDNA Synthesis, and qPCR.
Tissues were dissected and homogenized in TRIzol (Invitrogen). RNA was isolated, and cDNA synthesized as described previously (53). Primers for qPCR are listed in Maynard et al. (11) or Table S1. Sybr green reagent (Applied Biosystems), with 200 μM forward and reverse primer, was used to amplify cDNA using the ABI 7500 system.
Tissue Preparation and Immunofluorescence.
Embryonic brains were dissected in PBS and immersion-fixed overnight in 4% paraformaldehyde (PFA). Postnatal animals were perfused through the heart with 4% PFA after anesthetization (urethane 2 mg/kg). Fixed brains were sectioned and stained as described previously (11). Primary antibodies are listed in Table S2. For fluorescent detection, species appropriate Alexafluor 488 and 546 nm secondary antibodies were used. To generate a novel avian anti-Cdc45l antibody, chickens were immunized with the peptide CZERNRKRRGRREWEARRKD and antisera were affinity purified and tested by Western blotting (Aves Labs).
BrdU Labeling.
Timed pregnant mice (E12.5 and 13.5 for proliferation, and E13.5, 18.5, and 19.5 for birth-dating) were injected i.p. with BrdU (50 mg/kg body weight). Standard BrdU immunolabeling techniques were used after sodium citrate steam treatment (antigen retrieval), and only heavily labeled cells were counted.
In Situ Hybridization (ISH).
The 22q11 genes were cloned into PBluescript for DNA templates. Digoxigenin-labeled riboprobes were made and used for ISH as described previously (53). A Nikon 80i microscope and Retiga CCD camera was used for brightfield imaging.
Cell Counts.
The 1-μm optical sections were collected on a Zeiss LSM-510 laser scanning confocal microscope coded, and counted blind. Values reflect averages across at least two adjacent sections. In embryonic brains, cells were counted at the level of mid-ganglionic eminences across the whole cortex from the cortical/striatal boundary to the cortical hem. Also, counting boxes (200 μm wide) were placed at dorsal, medial, and lateral sites. For migration analysis, five equidistant bins were defined to assess distribution from the corticostriatal boundary to the cortical hem. In postnatal cortex, cells were counted in coronal sections at the level of the anterior commissure. Counting boxes (400 μm wide) were placed at dorsal, medial, and lateral sites.
Statistics.
We used two-tailed Student t tests to evaluate significance of mean cell number differences between LgDel and WT samples, and two-way ANOVA to compare cell distributions. When differences were observed using ANOVA, posthoc least significant difference (LSD) tests were used to determine significant difference between specific bins.
Supplementary Material
Acknowledgments.
A.-S.L. was supported by National Institute of Child Health and Human Development Grants HD042182 and HD029178, and the Silvio M. Conte Grant MH64065. Confocal microscopy and ISH analysis used University of North Carolina Neurosciences Center core facilities (NS031768).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905696106/DCSupplemental.
References
- 1.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 2.Geschwind DH, Levitt P. Autism spectrum disorders: Developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Bassett AS, et al. Clinical features of 78 adults with 22q11 deletion syndrome. Am J Med Genet A. 2005;138:307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 5.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Bearden CE, et al. Mapping cortical thickness in children with 22q11.2 deletions. Cereb Cortex. 2007;17:1889–1898. doi: 10.1093/cercor/bhl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaer M, et al. Abnormal patterns of cortical gyrification in velo-cardio-facial syndrome (deletion 22q11.2): An MRI study. Psychiatry Res. 2006;146:1–11. doi: 10.1016/j.pscychresns.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 8.van Amelsvoort T, et al. Brain anatomy in adults with velocardiofacial syndrome with and without schizophrenia: Preliminary results of a structural magnetic resonance imaging study. Arch Gen Psychiatry. 2004;61:1085–1096. doi: 10.1001/archpsyc.61.11.1085. [DOI] [PubMed] [Google Scholar]
- 9.Kiehl TR, Chow EW, Mikulis DJ, George SR, Bassett AS. Neuropathologic features in adults with 22q11.2 deletion syndrome. Cereb Cortex. 2009;19:153–164. doi: 10.1093/cercor/bhn066. [DOI] [PubMed] [Google Scholar]
- 10.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 11.Maynard TM, et al. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc Natl Acad Sci USA. 2003;100:14433–14438. doi: 10.1073/pnas.2235651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paylor R, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: Implications for 22q11 deletion syndrome. Proc Natl Acad Sci USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gogos JA, et al. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat Genet. 1999;21:434–439. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- 14.Tedeschi A, et al. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J Cell Sci. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- 15.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall C, et al. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol Cell Biol. 2001;21:1854–1865. doi: 10.1128/MCB.21.5.1854-1865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 18.Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 19.Guarguaglini G, et al. Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem J. 1997;325:277–286. doi: 10.1042/bj3250277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukas J, Petersen BO, Holm K, Bartek J, Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 22.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieto M, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 24.Bulfone A, et al. T-brain-1: A homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron. 1995;15:63–78. doi: 10.1016/0896-6273(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 25.Benes FM. Emerging principles of altered neural circuitry in schizophrenia. Brain Res Brain Res Rev. 2000;31:251–269. doi: 10.1016/s0165-0173(99)00041-7. [DOI] [PubMed] [Google Scholar]
- 26.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 27.del Rio JA, de Lecea L, Ferrer I, Soriano E. The development of parvalbumin-immunoreactivity in the neocortex of the mouse. Brain Res Dev Brain Res. 1994;81:247–259. doi: 10.1016/0165-3806(94)90311-5. [DOI] [PubMed] [Google Scholar]
- 28.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 29.Kowalczyk T, et al. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009 doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold SJ, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benes FM. Neural circuitry models of schizophrenia: Is it dopamine, GABA, glutamate, or something else? Biol Psychiatry. 2009;65:1003–1005. doi: 10.1016/j.biopsych.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Benes FM, Vincent SL, Todtenkopf M. The density of pyramidal and nonpyramidal neurons in anterior cingulate cortex of schizophrenic and bipolar subjects. Biol Psychiatry. 2001;50:395–406. doi: 10.1016/s0006-3223(01)01084-8. [DOI] [PubMed] [Google Scholar]
- 33.Lewis DA, Glantz LA, Pierri JN, Sweet RA. Altered cortical glutamate neurotransmission in schizophrenia: Evidence from morphological studies of pyramidal neurons. Ann N Y Acad Sci. 2003;1003:102–112. doi: 10.1196/annals.1300.007. [DOI] [PubMed] [Google Scholar]
- 34.O'Leary DD, Nakagawa Y. Patterning centers, regulatory genes and extrinsic mechanisms controlling arealization of the neocortex. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]
- 35.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 36.Caviness VS., Jr Architectonic map of neocortex of the normal mouse. J Comp Neurol. 1975;164:247–263. doi: 10.1002/cne.901640207. [DOI] [PubMed] [Google Scholar]
- 37.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 38.Maynard TM, et al. Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes. Mol Cell Neurosci. 2008;39:439–451. doi: 10.1016/j.mcn.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raux G, et al. Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum Mol Genet. 2007;16:83–91. doi: 10.1093/hmg/ddl443. [DOI] [PubMed] [Google Scholar]
- 40.Schaer M, et al. Congenital heart disease affects local gyrification in 22q11.2 deletion syndrome. Dev Med Child Neurol. doi: 10.1111/j.1469-8749.2009.03281. [DOI] [PubMed] [Google Scholar]
- 41.Gogos JA, et al. Catechol-O-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc Natl Acad Sci USA. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- 43.Mukai J, et al. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, et al. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:16859–16864. doi: 10.1073/pnas.232186099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benes FM, Lim B, Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc Natl Acad Sci USA. 2009;106:11731–11736. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald-McGinn DM, et al. Malignancy in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Am J Med Genet A. 2006;140:906–909. doi: 10.1002/ajmg.a.31199. [DOI] [PubMed] [Google Scholar]
- 48.Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex. 1997;7:347–358. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- 49.LoTurco JJ, Owens DF, Heath MJ, Davis MB, Kriegstein AR. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 50.Tiveron MC, et al. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long JM, et al. Behavior of mice with mutations in the conserved region deleted in velocardiofacial/DiGeorge syndrome. Neurogenetics. 2006;7:247–257. doi: 10.1007/s10048-006-0054-0. [DOI] [PubMed] [Google Scholar]
- 52.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 53.Meechan DW, et al. Gene dosage in the developing and adult brain in a mouse model of 22q11 deletion syndrome. Mol Cell Neurosci. 2006;33:412–428. doi: 10.1016/j.mcn.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: Effect of increased p21 expression. Cereb Cortex. 2008;18:1865–1875. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.