Abstract
Interkinetic nuclear migration (INM) is a hallmark of the polarized stem and progenitor cells in the ventricular zone (VZ) of the developing vertebrate CNS. INM is responsible for the pseudostratification of the VZ, a crucial aspect of brain evolution. The nuclear migration toward the apical centrosomes in G2 is thought to be a dynein-microtubule-based process. By contrast, the cytoskeletal machinery involved in the basally directed nuclear translocation away from the centrosome in G1 has been enigmatic. Studying the latter aspect of INM requires manipulation of the cytoskeleton without impairing mitosis and cytokinesis. To this end, we have established a culture system of mouse embryonic telencephalon that reproduces cortical development, and have applied it to explore a role of actomyosin in INM. Using the nonmuscle myosin II inhibitor blebbistatin at a low concentration at which neither cell cycle progression nor cytokinesis is impaired, we show that myosin II is required for the apical-to-basal (ap→bl), ab-centrosomal INM. Myosin II activity is also necessary for the nuclear translocation during delamination of subventricular zone (SVZ) cells, a second, telencephalon-specific type of neural progenitor. Moreover, the inhibition of ab-centrosomal INM changes the balance between VZ and SVZ progenitor cell fate. Our data suggest a unifying concept in which the actomyosin contraction underlying ab-centrosomal INM sets the stage for the evolutionary increase in VZ pseudostratification and for SVZ progenitor delamination, a key process in cortical expansion.
Keywords: cell cycle, cerebral cortex, cytoskeleton, neural stem cells, neurogenesis
Interkinetic nuclear migration (INM) is a hallmark of neuroepithelial and radial glial cells, the primary stem and progenitor cells of the vertebrate CNS (1–5). Here, the cell nuclei migrate in concert with the progression of the cell cycle, with the cells as a whole maintaining their overall position.
INM is linked to the apical–basal polarity of neuroepithelial and radial glial cells (3). These highly elongated cells extend across the entire wall of the neural tube, from the basal lamina to the luminal (apical) surface, and possess a single primary cilium at their apical plasma membrane (see ref. 6 and refs. therein), which can be regarded as a key determinant for INM for the following reasons. Given that one of the centrioles of the centrosome forms the basal body of the primary cilium, this localizes the centrosomes of these cells to their apical plasma membrane. As the primary cilium is disassembled only shortly before the onset of mitosis (7), the nucleus, to undergo mitosis (in which the centrosomes serve as mitotic spindle poles), needs to be in the vicinity of the centrosomes, that is, at the apical surface. Thus, mitosis of these stem and progenitor cells occurs apically, and they will collectively be referred to as apical progenitors (APs).
In INM, the nuclei of newborn APs move away from the apical surface toward the basal lamina during G1, undergo S phase at a basal location, and return to the apical centrosomes during G2 for the next mitosis (4, 5). Thus, INM is responsible for the pseudostratified appearance of the ventrical zone (VZ). Importantly, by moving the interphase nuclei of APs away from the apical surface, the apical-to-basal (ap→bl) leg of INM during G1 serves to reserve the apical space for mitosis, and thereby promotes the expansion of APs (see ref. 8 and refs. therein).
Hence, the molecular machinery mediating ap→bl INM is of prime importance. Remarkably, very little is known about this machinery, in contrast to that mediating the basal-to-apical (bl→ap) leg of INM, about which significant insight has been obtained recently. Thus, consistent with earlier studies implicating microtubules in INM (2), recent studies have established that the dynein-interacting proteins Lis1 and dynactin (9–11) and the centrosomal proteins Cep120 and TACC (12) are required for INM and specifically its bl→ap leg during G2, hence providing evidence for a role of microtubules and minus end-directed motor proteins in this process. Bl→ap INM of APs is thus highly related to nuclear positioning before mitosis, a ubiquitous microtubule-based process that evolved in single-cell eukaryotes (13).
In contrast to the bl→ap G2-phase INM, in which the nucleus moves toward the centrosomes, the nucleus moves away from the centrosome in the ap→bl G1-phase INM. One may envision that this ab-centrosomal INM is also a microtubule-based process, using for example plus end-directed kinesin-type motors (4). However, another possibility is that ap→bl INM is mediated by the actin–myosin system. The previously reported inhibition of INM by cytochalasin B (2, 14) does not constitute conclusive evidence for a role of actin microfilaments in INM because cytochalasin B not only interferes with actin microfilament function but also directly inhibits glucose uptake into cells (15).
In the present study, we have explored a possible role of the actin–myosin system in INM by interfering with nonmuscle myosin II function using the highly specific inhibitor blebbistatin (Bb) (16) in a newly characterized organ culture system of whole telencephalic hemispheres that reproduces cortical development. Importantly, this system has allowed us to apply a defined low concentration of Bb that does not block known actomyosin-dependent processes such as cytokinesis of APs and migration of neurons to the cortical layers but does suffice to inhibit INM.
Results
Rationale of Experimental Approach.
Specific steps in INM, for example the bl→ap nuclear movement during G2, occur within a time frame of very few hours, in some cases even less than 1 h (see for example refs. 10, 12, 17). Moreover, both the microtubule and the actomyosin system mediate essential steps in mitosis and cytokinesis. These 2 considerations have critical implications as to the suitability of genetic approaches such as conditional gene ablation, RNA interference, and forced gene expression to dissect specific steps in INM. Specifically, the manipulation of the level or activity of a protein of the microtubule or the actomyosin system by genetic means is known to occur with a time course such that not only INM but also cell division will be affected (see for example ref. 10). Hence, these genetic approaches are suitable to investigate the steps in INM occurring before mitosis, i.e., the bl→ap nuclear migration in G2 (10–12), but not those initiated right after mitosis at the end of cytokinesis, i.e., the ap→bl nuclear translocation in G1.
We therefore decided to pursue an alternative approach to explore a role of actomyosin in INM, i.e., acute and dosed pharmacological interference such that progression through the cell cycle and, notably, cytokinesis are not affected. To this end, we established a culture system in which isolated mouse E14.5 hemispheres were maintained for up to 24 h in a defined medium, floating under rotation. This hemisphere rotation (HERO)-culture system allowed hemisphere development, notably neurogenesis, to proceed in a manner indistiguishable from hemispheres of embryos developed to E15.5 in utero (for details, see supporting information (SI) Fig. S1).
Weakening of the Actomyosin Cortex of Neural Progenitors in the VZ by the Nonmuscle Myosin II Inhibitor Blebbistatin.
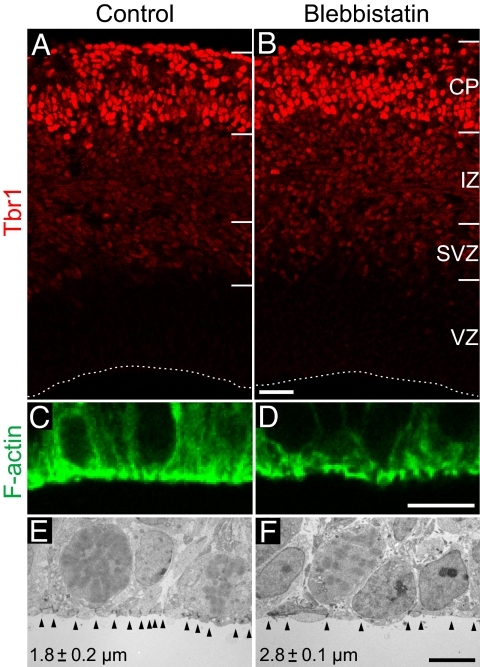
We first examined whether the potent and highly specific nonmuscle myosin II inhibitor Bb (16) can be applied to perturb the actomyosin cortex of APs without causing an overall impairment in cortical cytoarchitecture and cell dynamics such as apical–basal polarity and neuronal migration. When Bb was applied at concentrations ≥25 μM of the racemate, which were previously used to inhibit cell migration including that of neurons (18), we observed gross defects in cortical cytoarchitecture and morphology after 24 h of HERO culture (Fig. S2). By contrast, when the Bb concentration was lowered to 12.5 μM of the racemate [i.e., 6.25 μM of the effective (−)enantiomer], referred to from here onward as “low-Bb treatment,” no such defects were observed. Specifically, the cytoarchitecture of the cortical wall and, importantly, the radial morphology and apical–basal polarity of APs were unaffected by low-Bb treatment for 24 h (Fig. S3 and Fig. S4). Moreover, neuronal migration into the cortical plate was unperturbed by low-Bb treatment, as revealed by immunostaining for the neuron-specific transcription factor Tbr1 (19) (Fig. 1 A and B).
Fig. 1.
Low-blebbistatin treatment affects the actomyosin cortex of VZ progenitors, but does not block neuronal migration. Mouse E14.5 hemispheres subjected to HERO culture for 24 h in the absence (control; A, C, and E) or presence (B, D, and F) of 12.5 μM Bb. (A and B) Immunofluorescence for Tbr1 (3-μm optical sections). Dashed lines, ventricular surface; IZ, intermediate zone; CP, cortical plate. (C and D) Phalloidin fluorescence (1-μm optical sections). Ventricular surface is down. (E and F) Transmission EM. Arrowheads indicate junctional complexes; interjunctional distance is indicated (mean ± SEM of 3 hemispheres; P < 0.05; number of cells quantified is given in Table S2). [Scale bars, 50 μm (A and B), 10 μm (C and D), and 5 μm (E and F)].
Low-Bb treatment for 24 h did, however, alter the pattern of the apically enriched F-actin staining (Fig. 1 C and D). This effect was observed already at the earliest time point analyzed, i.e., after 1 h of HERO culture (Fig. S5). These observations suggested that low-Bb treatment impaired the apical actomyosin cell cortex. In further support of this, we observed a widening of the apical surface of 24-h Bb-exposed progenitors, as revealed by cadherin immunofluorescence (Fig. S3 E and F, arrows) and EM analysis (Fig. 1 E and F). We conclude that low-Bb treatment can be used to weaken the actomyosin cortex of neural progenitors in HERO culture, while maintaining the integrity and cytoarchitecture of the VZ as a pseudostratified epithelium and the overall structure of the cortical wall.
Myosin II Inhibition by Low-Blebbistatin Treatment Selectively Impairs Apical-to-Basal Nuclear Migration of Neural Progenitors.
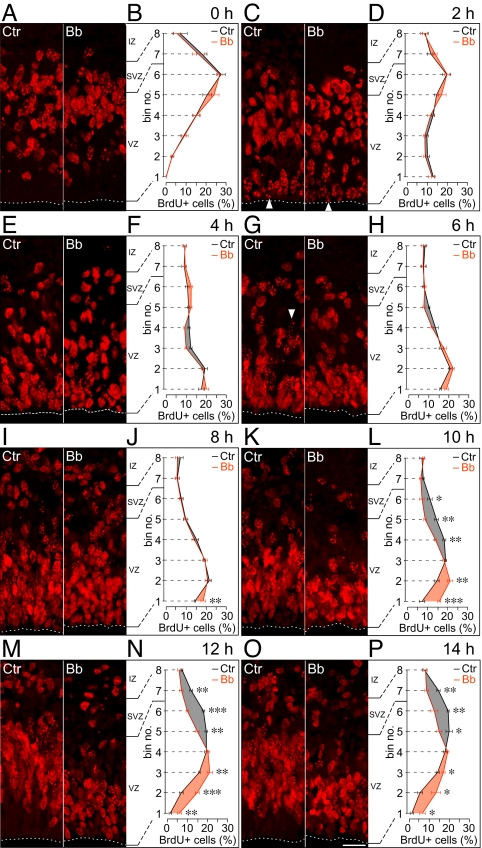
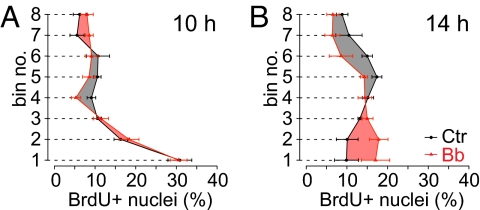
To investigate the possible role of myosin II in INM, we analyzed neural progenitors in HERO culture with and without low-Bb treatment by pulse labeling with BrdU, a thymidine analog incorporated during S phase, followed by chase for up to 14 h (Fig. 2). In the control (Fig. 2, Left immunofluorescence panel of each pair and black curves), consistent with the known location of neural progenitor nuclei in S phase (20), nuclei labeled by a 15-min BrdU pulse at the beginning of HERO culture (0 h of chase) were found mostly in the basal half of the VZ (presumably nuclei of APs) and in the SVZ (presumably nuclei of basal progenitors, see below) (Fig. 2 A and B). During chase in the absence of Bb, BrdU-labeled VZ nuclei first migrated in the apical direction toward the ventricular surface, as would be expected for INM of APs in G2. This bl→ap INM was detectable already after 1 and 2 h of chase and continued until about 6 h of chase (Fig. 2 C–H), consistent with a G2 phase of <2 h and an S phase of ≈4 h (20, 21). Interestingly, the first nuclei to reach the ventricular surface showed a sparse, spotty BrdU-labeling pattern (Fig. 2C, arrowheads), as would be expected for nuclei that had been leaving S phase and entering G2 during the 15-min BrdU pulse (the “forefront” nuclei). Importantly, although low-Bb treatment had affected cortical actomyosin already by 1 h of chase (Fig. S5), it did not significantly affect the bl→ap, G2-phase INM of BrdU-labeled VZ nuclei (Fig. 2 C–H).
Fig. 2.
Low-blebbistatin treatment selectively impairs apical-to-basal nuclear migration of neural progenitors. E14.5 hemispheres in HERO culture were subjected to 15-min BrdU pulse labeling followed by chase for up to 14 h as indicated, in the absence (control, Ctr) or presence (Bb) of 12.5 μM Bb. (A, C, E, G, I, K, M, and O) BrdU immunofluorescence (3-μm optical sections). Arrowheads indicate nuclei with sparse, spotty BrdU labeling; dashed lines, ventricular surface; IZ, intermediate zone. (Scale bar, 30 μm.) (B, D, F, H, J, L, N, and P) Distribution of BrdU-labeled nuclei across the VZ plus SVZ, divided into 8 bins as indicated, with bin no. 1 being at the ventricle and bin no. 8 at the SVZ–IZ boundary. Nuclei in a given bin are expressed as percentage of the total number of BrdU-labeled nuclei in all 8 bins. Black curves, control (Ctr); red curves, Bb; differences between control and Bb values are highlighted by black (control > Bb) and red (Bb > control) shading. Data are the mean of 4–6 hemispheres; bars indicate SEM; (L, N, and P) *, P < 0.05; **, P < 0.01; ***, P < 0.001; number of BrdU-labeled nuclei counted are given in Table S2.
Analysis of later chase time points (from 6 h onward) in the absence of Bb revealed that, following their arrival at the ventricular surface and mitosis (see Fig. 3), BrdU-labeled VZ nuclei migrated in the basal (pial) direction (Fig. 2 G–P). This ap→bl, G1-phase nuclear migration was detectable at 8 h of chase (Fig. 2, compare I and J with G) and even more so thereafter (Fig. 2 K–P), resulting, by 14 h, in a basally enriched distribution of BrdU-labeled nuclei across the VZ, similar to that observed at the beginning of chase (Fig. 2, compare B with P, black curves). In fact, ap→bl migration of BrdU-labeled nuclei could be inferred to have occurred already at 6 h from the appearance of nuclei with sparse, spotty BrdU staining (presumably the forefront nuclei) in the basal portion of the VZ (Fig. 2G, Left, arrowhead). The coexistence of the subpopulation of BrdU-labeled nuclei showing bl→ap migration and that showing ap→bl migration at ≈6 h of chase is expected, given that the 15-min BrdU pulse labeled nuclei randomly at any time point during the ≈4-h-long S phase (20, 21).
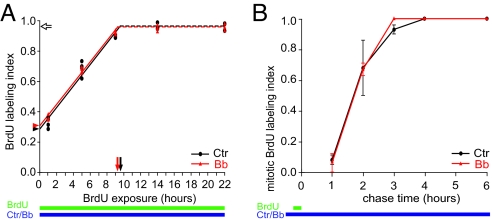
Fig. 3.
Low-blebbistatin treatment does not alter cell cycle parameters of neural progenitors in the VZ. (A) Cumulative BrdU labeling (green bar) of VZ progenitors in E14.5 hemispheres subjected to HERO culture for up to 22 h (blue bar) in the absence (control, Ctr, black circles) or presence (Bb, red triangles) of 12.5 μM Bb. The ratio of BrdU-labeled nuclei over total, DAPI-stained nuclei in the VZ is depicted; the data from 4 (22 h, 3) hemispheres each are shown. Dashed lines and open arrow, growth fraction; arrowheads, proportion of progenitors in S phase; arrows, time interval between 2 consecutive S phases (TC–TS). Number of DAPI-stained nuclei counted are given in Table S2. (B) BrdU pulse labeling (15 min, green bar) followed by chase into mitotic neural progenitors in E14.5 hemispheres subjected to HERO culture for up to 6 h (blue bar) in the absence (control, Ctr, black circles) or presence (Bb, red triangles) of 12.5 μM Bb. Data show the ratio of BrdU-labeled, phosphohistone 3-positive mitoses over total mitoses in the ventricularmost bins no. 1 and 2 and are the mean of 2 hemispheres; bars indicate deviation of the individual values from the mean. Number of mitoses counted are given in Table S2.
In contrast to the lack of effect of low-Bb treatment on the bl→ap, G2-phase INM (Fig. 2 C–H), myosin II inhibition markedly impaired the ap→bl, G1-phase nuclear migration, as was obvious from the apical shift in the distribution of BrdU-labeled nuclei across the VZ at 10 h of chase (Fig. 2 K and L) and thereafter (Fig. 2 M–P) when compared to control. In fact, impairment of ap→bl nuclear migration by low-Bb treatment was detectable already after 6 h and 8 h of chase, as was evident from the relative accumulation of BrdU-labeled nuclei in the ventricularmost bin (Fig. 2 H and J, bin no. 1) when compared to control.
Given that myosin II inhibition by low-Bb treatment impaired the ap→bl, G1-phase nuclear migration of neural progenitors, one would expect their S phase to occur in a location less basal than normal. Analysis of neural progenitors in HERO culture subjected to low-Bb treatment for 24 h and a 15-min BrdU pulse at the end of the culture period showed that this was indeed the case (Fig. S6).
Myosin II Inhibition by Low-Blebbistatin Treatment Does Not Alter Cell Cycle Parameters of Apical Progenitors.
To investigate whether the present condition of myosin II inhibition affected progression of neural progenitors through the cell cycle including M phase, we performed cumulative BrdU labeling (20) and determined the labeling index of the cells in the VZ. This yielded essentially identical data for control and low-Bb-treated progenitors in the VZ (Fig. 3A). Importantly, the present myosin II inhibition did not reduce the proportion of progenitors incorporating BrdU, which was nearly 100% (Fig. 3A, open arrow; Table S1, growth fraction). Given that alterations in the F-actin staining at the apical cell cortex upon low-Bb treatment were observed at the earliest time point studied (1 h) (Fig. S5), the unaltered BrdU incorporation (Fig. 3A) implies that the low-Bb treatment did not interfere with actomyosin-dependent processes in M phase, notably cytokinesis.
We also analyzed the time interval between S phase and M phase by determining the appearance of BrdU label in mitotic (phosphohistone 3-positive) APs following a 15-min BrdU pulse. Again, essentially identical data for control and Bb-treated progenitors were obtained (Fig. 3B). The first BrdU-labeled mitotic cells were observed at 1 h of chase, and their proportion increased to nearly 100% by 3 h of chase, with 50% of mitotic cells showing BrdU label at 1.7 h. These data indicate that for both control and Bb-treated APs, G2 ranged from 1 h to 3 h and on average lasted 1.7 h.
The cumulative BrdU labeling (Fig. 3A) and BrdU pulse-chase (Fig. 3B) data were used to determine the various cell cycle parameters of the progenitors in the VZ (Table S1). No significant differences between control and Bb-treated progenitors were found, corroborating that the present myosin II inhibition by low-Bb treatment did not affect their cell cycle progression. Our data confirm and extend the notion (2, 4, 14) that INM is not required for cell cycle progression.
Myosin II Is Required for the Apical-to-Basal Nuclear Translocation of Basal Progenitors.
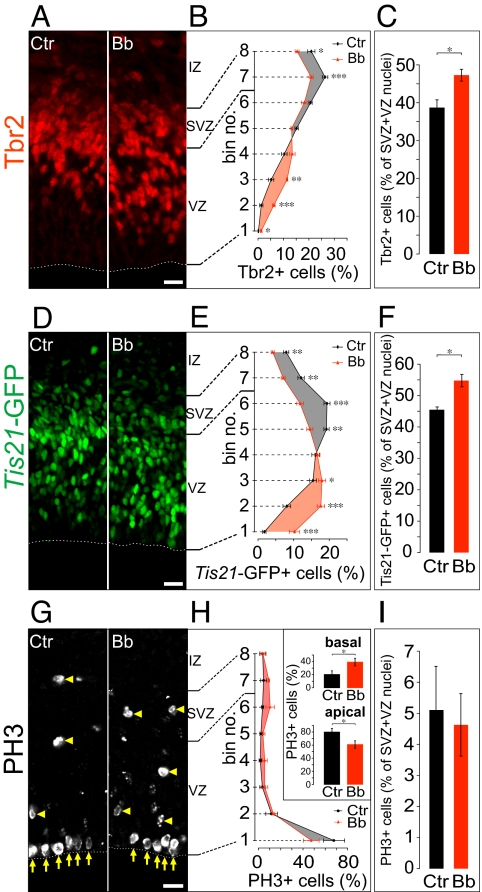
In the mammalian cerebral cortex, APs generate a second type of progenitor that shows ap→bl nuclear translocation in conjunction with delamination from the VZ to the SVZ [called basal (BP), intermediate, nonsurface, or SVZ progenitor] (17, 22, 23). It was therefore important to investigate whether the selective impairment of ap→bl nuclear migration upon low-Bb treatment reflected an effect of the myosin II inhibitor on APs, BPs, or both. To identify BP nuclei, we immunostained for the transcription factor Tbr2, a marker of these cells (19). After 24 h of HERO culture under control conditions, Tbr2-immunoreactive nuclei were found mostly in the basal VZ and in the SVZ (Fig. 4A, Left), consistent with previous observations (19). Low-Bb treatment for 24 h resulted in a shift in the distribution of Tbr2-immunoreactive nuclei to a less basal localization, with a relative decrease in the SVZ and an increase in the VZ (Fig. 4 A, Right, and B). We conclude that myosin II is required for the ap→bl nuclear translocation of BPs.
Fig. 4.
Effects of low-blebbistatin treatment on the distribution and proportion of neurogenic progenitor nuclei in the VZ and SVZ. E14.5 hemispheres from heterozygous Tis21-GFP mice (A–F) or C57BL/6 mice (G–I) were subjected to HERO culture for 24 h in the absence (control, Ctr) or presence (Bb) of 12.5 μM Bb, followed by immunofluorescence for Tbr2 (A), analysis of intrinsic Tis21-GFP fluorescence (D) and phosphohistone 3 (PH3) immunofluorescence (G). Dashed lines in A, D, and G, ventricular surface; yellow arrows and arrowheads in G, ventricular (apical) and abventricular (basal) mitoses, respectively; IZ, intermediate zone. (Scale bars, 20 μm.) (B, E, and H) Distribution of Tbr2-positive (B) and Tis21-GFP-positive (E) nuclei and phosphohistone 3-positive mitotic figures (H) across the VZ plus SVZ (bin analysis as in Fig. 2). Tbr2-positive and Tis21-GFP-positive nuclei and mitotic figures in a given bin are expressed as percentage of the total number of labeled nuclei/mitotic figures in all 8 bins. Black curve and circles, control (Ctr); red curve and triangles, Bb (shading as in Fig. 2). Inset in H: apical, sum of the percentage values of bins no. 1 plus 2; basal, sum of the percentage values of bin nos. 3 to 8. Number of nuclei and mitoses counted are given in Table S2. (C, F, and I) Quantification of Tbr2-positive (C) and Tis21-GFP-positive (F) nuclei and phosphohistone 3-positive mitotic figures (I), expressed as percentage of total nuclei in the VZ plus SVZ as revealed by DAPI staining. Number of DAPI-stained nuclei counted are given in Table S2. (B, C, E, F, H, and I) Data are the mean of 4–7 hemispheres; bars indicate SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Myosin II Is Required for the Apical-to-Basal Nuclear Migration of Neurogenic Progenitors.
To further distinguish between the various subpopulations of neural progenitors with regard to the effects of myosin II inhibition on nuclear migration, we extended our analyses to Tis21, which is specifically expressed in the neurogenic subpopulations of neural progenitors. These subpopulations comprise (i) neuron-generating BPs, (ii) neuron-generating APs, and (iii) APs that give rise to neuron-generating BPs (17, 24).
We used the Tis21-GFP knock-in mouse line, in which nuclear GFP is expressed under the control of the Tis21 promoter (17), to identify Tis21-expressing progenitors. Compared to Tbr2-positive nuclei (Fig. 4A, Left), Tis21-GFP-positive nuclei in the control condition showed a broader distribution, with relatively more nuclei found in the VZ (Fig. 4D, Left), as observed previously (17). Low-Bb treatment during 24 h of HERO culture resulted in a marked shift in the distribution of Tis21-GFP-positive nuclei to a less basal localization (Fig. 4 D, Right, and E). As this shift was more pronounced than that of the Tbr2-positive nuclei (Fig. 4, compare B and E), we conclude that myosin II is required for the ap→bl nuclear translocation/migration not only of neurogenic BPs, but also of neurogenic APs. This conclusion was corroborated by the observation that upon BrdU pulse labeling and chase for 12 h in HERO culture, i.e., when ap→bl INM of neurogenic APs and delamination of BPs is ongoing, analysis of the distribution of the Tis21-GFP-positive nuclei revealed that low-Bb treatment significantly impaired this ap→bl nuclear migration/translocation (Fig. S7).
Myosin II Is Required for the Apical-to-Basal INM of Proliferative Apical Progenitors.
In light of these observations, we also examined APs that were not committed to the neuronal lineage for a requirement of myosin II in ap→bl INM, using the lack of Tis21-GFP expression to identify these progenitors and BrdU pulse-chase labeling as described in Fig. 2 to study their nuclear migration. Upon BrdU pulse labeling and 10 h chase in HERO culture, the distribution of the BrdU-labeled nuclei of Tis21-GFP-negative APs was very similar for the control and low-Bb treatment (Fig. 5A), consistent with the notion that the present myosin II inhibition did not impair the bl→ap, G2-phase INM of proliferative apical progenitors. In either condition, the distribution of the labeled nuclei showed a peak in the ventricularmost bin (Fig. 5A, bin no. 1), reflecting the residence of the BrdU label-inheriting nuclei of newborn Tis21-GFP-negative APs in G1 at the ventricular surface after 10 h of chase.
Fig. 5.
Low-blebbistatin treatment impairs apical-to-basal INM of proliferative neural progenitors. E14.5 hemispheres from heterozygous Tis21-GFP mice in HERO culture were subjected to 15-min BrdU pulse labeling followed by chase for 10 h (A) and 14 h (B), in the absence (control, Ctr; black curves and circles) or presence (Bb; red curves and triangles) of 12.5 μM Bb. Data show the distribution of BrdU-labeled Tis21-GFP-negative nuclei across the VZ plus SVZ (bin analysis and shading as in Fig. 2) and are the mean of 3–4 hemispheres; bars indicate SEM. Ctr and Bb curves in B are significantly different with a KS P value of 0.001, whereas this is not the case for the curves in A (KS P value of 0.94). Number of BrdU-labeled Tis21-GFP-negative nuclei counted are given in Table S2.
In contrast, after another 4 h of chase in HERO culture, the distribution of the BrdU-labeled nuclei of Tis21-GFP-negative APs was significantly different for the control and low-Bb treatment (Fig. 5B). Whereas in the control the labeled nuclei had migrated away from the ventricular surface, peaking in the basal region of the VZ (Fig. 5B, black curve), this nuclear migration was markedly impaired upon low-Bb treatment, with the peak of labeled nuclei still being observed in the apical region of the VZ (Fig. 5B, red curve). We conclude that myosin II is required for the ap→bl, G1-phase INM of proliferative APs.
Myosin II Inhibition Leads to an Increase in Neurogenic Basal Progenitors.
It has been proposed that the residence time of neural progenitor nuclei in the apical versus basal region of the VZ influences their cell fate, i.e., their proliferation versus differentiation (4, 11, 14). We therefore investigated whether concomitant with the impaired ap→bl nuclear translocation/migration of neural progenitors upon low-Bb treatment, there was a shift in the proliferative versus neurogenic progenitor subpopulations. Indeed, low-Bb treatment in HERO culture for 24 h resulted in an increase in BPs, as revealed by Tbr2 immunostaining (Fig. 4C), in an increased proportion of neurogenic progenitors in general, as revealed by analysis of Tis21-GFP (Fig. 4F), and in an increase in basal mitoses at the expense of apical mitoses, as revealed by phosphohistone 3 immunostaining (Fig. 4 G and H). Despite the increase in the proportion of Tbr2- and Tis21-GFP-positive nuclei in the VZ (Fig. 4 A and D), all VZ nuclei remained positive for the transcription factor Pax6, a marker of cortical APs (19) (Fig. S8). Consistent with the notion that the present myosin II inhibition did not block cytokinesis, the total number of mitotic figures as identified by phosphohistone 3 immunostaining was not increased by the low-Bb treatment (Fig. 4I). Taken together, our data indicate that inhibition of myosin II by low-Bb treatment changes neural progenitor fate toward a more differentiated state.
Discussion
Myosin II Is Required for Apical-to-Basal INM of Apical Progenitors.
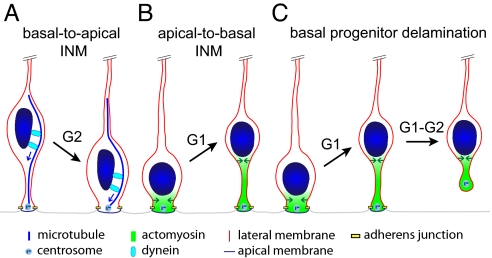
Our study shows that the ap→bl, ab-centrosomal INM of neuroepithelial and radial glial cells in G1, which is the primary cause for the pseudostratification of the VZ, requires myosin II. It should be emphasized that our data do not exclude a role of actomyosin in bl→ap INM during G2, as it is conceivable that this process may be impaired upon myosin II inhibition using higher concentrations of Bb. In this context, it is worth noting that the small GTPase Rac, a well-known regulator of the actin cytoskeleton, is required for bl→ap nuclear translocation, although this is thought to reflect the regulation of centrosomes and microtubules (25). Nonetheless, given the requirement for Lis1 (10), dynactin (10, 11), and the centrosomal proteins Cep120 and TACC (12) in bl→ap INM, the picture emerges that this ad-centrosomal leg of INM in G2 is a microtubule-based process using minus end-directed dynein motors (Fig. 6A), whereas the ap→bl, ab-centrosomal leg of INM in G1 is an actin microfilament-based process using myosin II motors (present study) (Fig. 6B). The observations that the speed of bl→ap INM is substantially greater than that of ap→bl nuclear translocation (10, 12, 17, 23–25) are consistent with this notion.
Fig. 6.
Model depicting the distinct roles of the microtubule-based dynein system and actomyosin in apical progenitor INM and basal progenitor delamination. (A) Dynein-mediated ad-centrosomal transport of the nucleus along microtubules (blue arrows) results in bl→ap INM in G2. (B) Myosin II-mediated contraction of the actin cortex, starting apically and progressing in the basal direction (green arrows), leads to constriction of the apical process, which in turn results in ap→bl INM in G1. (C) Similar ap→bl nuclear translocation as in B occurs in newborn BPs and continues in the course of their delamination.
Myosin II Is Required for Apical-to-Basal Nuclear Translocation in Delaminating Basal Progenitors.
We found that not only the ap→bl INM of APs, but also the nuclear translocation of delaminating BPs, requires myosin II. This similarity of machineries is highly intriguing, given that BPs originate from APs. Specifically, the use of the actomyosin system for both, the ap→bl INM of APs (Fig. 6B) and the ap→bl nuclear translocation in the course of BP delamination (Fig. 6C), is likely to facilitate the transition from the former to the latter type of progenitor. In this context, it is interesting to note that newborn BPs may retain their apical process during ap→bl nuclear translocation and retract it only thereafter (22, 23) (Fig. 6C).
Dosed Inhibition of Actomyosin-Dependent Processes in Cortical Progenitors Cycling in Brain Organ Culture.
We perturbed nonmuscle myosin II function by using a highly specific inhibitor, Bb, rather than by genetic manipulations such as RNA interference. The crucial advantage of our approach lies in the fact that by using a low concentration of Bb, we could inhibit myosin II function such that only INM, but not cell division, of APs was impaired. Given that myosin II is known to function in cytokinesis (16), this was an essential precondition to explore the possible function of myosin II in ap→bl INM. To be able to apply a defined, low-Bb concentration, we established a hemisphere culture system, which reproduced in utero cortical development and in which the kinetics of INM were consistent with previous observations (10, 12, 17, 22–25).
Basal-to-Apical Nuclear Migration Versus Apical-to-Basal Nuclear Translocation.
What is the mechanism by which myosin II causes ap→bl nuclear movement? Extrapolating from the role of myosin II with regard to nuclear movement in the context of neuronal migration (18) and other systems (26), we find it most likely that the ap→bl nuclear translocation in INM of APs and delamination of BPs results from the directional ap→bl myosin II-mediated constriction of the apical process, which pushes the nucleus in the basal direction (Fig. 6 B and C). The increase in the average interjunctional distance at the apical surface upon myosin II inhibition likely reflects impairment of this constriction, consistent with our model. In mechanistic terms, ap→bl nuclear translocation is therefore principally different from the bl→ap nuclear migration in which the nucleus is presumably actively transported as cargo along microtubules by dynein (4, 10, 12) (Fig. 6A).
Myosin II, Nuclear Residence, and Neural Progenitor Fate.
Besides inhibiting ap→bl nuclear translocation, myosin II inhibition by low-Bb treatment also resulted in a cell fate change of neural progenitors, yielding more Tis21-GFP- and Tbr2-expressing BPs at the expense of Tis21-GFP-negative APs, and more basal mitoses at the expense of apical mitoses. It is conceivable that this cell fate change may be the consequence of the altered residence of AP nuclei in the apical versus basal region of the VZ, in line with previous proposals (4, 11, 14).
Developmental and Evolutionary Implications.
A key step in the development of the CNS is the actomyosin-mediated apical constriction by which the neural plate invaginates to form the neural tube. Our data imply that the same principal machinery (actomyosin) and cell biological process (constriction of the entire apical process) are responsible for 2 hallmarks of CNS development, the pseudostratification of the VZ, which increases in vertebrate evolution (5, 27), and the generation of a second neurogenic progenitor layer basal to the VZ, the SVZ, which is characteristic of the mammalian telencephalon (28). Interestingly, an increase in the SVZ is thought to underly cortical expansion (8, 27–29), and the major type of primate SVZ progenitor has been proposed to retain certain features of APs (8, 27). Given these considerations, and our findings showing that the ap→bl nuclear translocation in the course of SVZ formation is highly related to the ap→bl leg of INM of VZ progenitors, the actomyosin machinery determining the extent of ap→bl nuclear translocation is likely to be a key target of evolutionary change.
Materials and Methods
HERO Culture, Blebbistatin Treatment, BrdU Labeling, Immunofluorescence, and Transmission EM.
C57BL/6 mouse embryos and, when indicated, heterozygous Tis21-GFP knock-in mouse embryos (17) (C57BL/6 background) were used. Details of the HERO culture system, of Bb treatment, of BrdU labeling (50 μM), and of sample processing are described in SI Text. Immunofluorescence on 10-μm coronal cryosections and transmission EM of 70-nm plastic sections were performed according to standard procedures (see SI Text).
Quantitations.
Analysis was restricted to the dorsal cortex (800–900 μm caudally from the olfactory bulb). For each hemisphere, 2 fields (each from a different cryosection) taken with a 40× objective were analyzed for the markers indicated, and the mean per hemisphere was calculated. For quantification of the distribution of a given nuclear marker across the progenitor layers, the VZ plus SVZ were divided into 8 equidistant bins. Nuclei were identified by DAPI staining, and the nuclei in each bin expressing the marker under study were counted. Cell cycle parameters were determined as described (20). For any given dataset, each of the hemispheres used was from a different litter, and typically 2 or more independent experiments were pooled. Statistical significance was determined using Student's t test or according to Kolmogorov-Smirnov (KS).
Supplementary Material
Acknowledgments.
We thank the animal facility and the light microscopy facility of the Max Planck Institute of Molecular Cell Biology and Genetics for excellent support, Jula Peters and Christiane Haffner for superb technical assistance and advice, Dr. Yannis Kalaidzidis for generously helping with statistical analysis, and Drs. Lilla Farkas, Joe Howard, Ewa Paluch, Elena Taverna, and Iva Tolic-Norrelykke for helpful comments on the manuscript. J.S. was a member of the International Max Planck Research School for Molecular Cell Biology and Bioengineering. W.B.H. was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) (SFB 655, A2), by the DFG-funded Center for Regenerative Therapies Dresden, and by the Fonds der Chemischen Industrie.
Footnotes
Conflict of interest statement: S.P. works together with the W.B.H. group on other projects that have to do with brain development.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908928106/DCSupplemental.
References
- 1.Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- 2.Messier PE. Microtubules, interkinetic nuclear migration and neurulation. Experientia. 1978;34:289–296. doi: 10.1007/BF01922992. [DOI] [PubMed] [Google Scholar]
- 3.Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 4.Baye LM, Link BA. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyata T. Development of three-dimensional architecture of the neuroepithelium: Role of pseudostratification and cellular ‘community’. Dev Growth Differ. 2008;50(Suppl 1):S105–112. doi: 10.1111/j.1440-169X.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- 6.Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos N, Reiter JF. Building it up and taking it down: The regulation of vertebrate ciliogenesis. Dev Dyn. 2008;237:1972–1981. doi: 10.1002/dvdy.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fish JL, Kennedy H, Dehay C, Huttner WB. Making bigger brains: The evolution of neural-progenitor-cell division. J Cell Sci. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- 9.Gambello MJ, et al. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Z, et al. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinsch S, Gönczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111(Pt 16):2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 14.Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- 15.Klip A, Paquet MR. Glucose transport and glucose transporters in muscle and their metabolic regulation. Diabetes Care. 1990;13:228–243. doi: 10.2337/diacare.13.3.228. [DOI] [PubMed] [Google Scholar]
- 16.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 17.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19(7):347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi T, Nowakowski RS, Caviness VS., Jr Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 23.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 24.Attardo A, Calegari F, Haubensak W, Wilsch-Bräuninger M, Huttner WB. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS ONE. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minobe S, et al. Rac is involved in the interkinetic nuclear migration of cortical progenitor cells. Neurosci Res. 2009;63:294–301. doi: 10.1016/j.neures.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Charras G, Paluch E. Blebs lead the way: How to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 27.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Mannan O, Cheung AF, Molnar Z. Evolution of cortical neurogenesis. Brain Res Bull. 2008;75:398–404. doi: 10.1016/j.brainresbull.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 29.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.