Locating the 104 damaged nucleotides in a cell's DNA poses a significant challenge to DNA repair enzymes on a daily basis (1). Base excision repair (BER) glycosylases are responsible for finding subtle atomic-level changes in a base amidst a sea of undamaged bases, and they must do so before replication leads to improper nucleotide insertion causing a mutation (2). Many years ago it was recognized that random on-off binding of proteins to duplex DNA was an inefficient way to locate the sites of damage, leading to proposals of protein sliding on the duplex in addition to short hopping or longer-range jumping to different sections of the genome (3). In a recent issue of PNAS, new data from Boal et al. (4) provided evidence of an alternative mechanism in which the bacterial BER enzymes EndoIII and MutY communicate over a long segment of base-paired DNA by tapping into the ability of a perfectly complementary duplex to transport charge along the helix. Such a mechanism would greatly exceed the rate of 1D scanning by procession of a macromolecule along the DNA duplex.
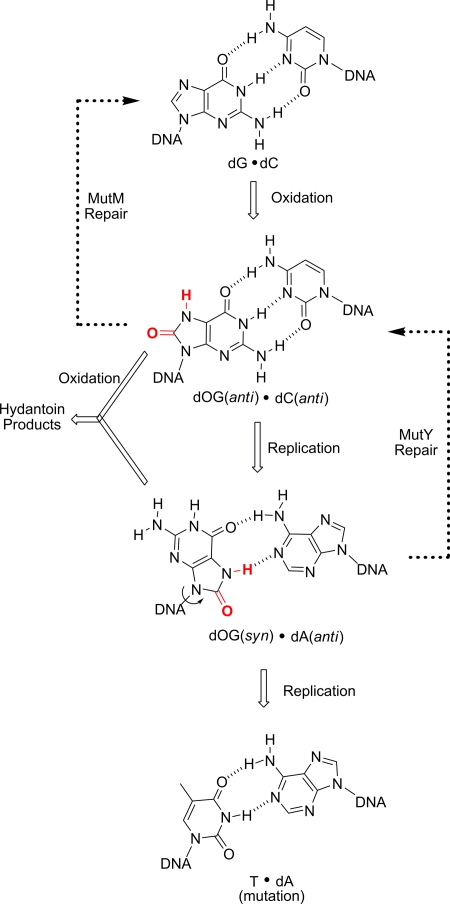
The case under study concerns oxidative DNA damage, for which there exists an elaborate system of repair (5). Oxidation of guanine to 8-oxoguanine (OG) must be located and OG must be excised, by MutM in bacteria or Ogg1 in eukaryotes, before replication because DNA polymerases often misinsert A opposite OG (Fig. 1). As a backup for the initial chance at repair, the BER enzyme MutY removes the misinserted A opposite OG after first locating the OG·A base pair. All of this must happen before further oxidative damage converts OG to more highly mutagenic hydantoin lesions (6, 7). The debate about the mechanism by which DNA damage sites are recognized is fueled by the fact that BER glycosylases, particularly MutY, are present in very low copy number in the cell.
Fig. 1.
Oxidation of G to OG leads to a cascade of errors requiring the BER enzymes MutM and MutY.
Adding to the curiosity of how damage is recognized is the fact that the changes in the DNA base can be extremely subtle; only two atoms are changed in the case of G oxidation to OG (Fig. 1), and the lack of a lone CH3 group is the basis for distinguishing U vs. T by uracil DNA glycosylase (UDG). For the latter problem, recent data point to duplex dynamics as the starting point of damage recognition in which thermal opening of base pairs initiates the recognition process (8). For human Ogg1 (hOgg1) recognition of OG, fast sliding has been observed by single-molecule imaging techniques for the protein in contact with duplex DNA (9), and it is proposed that hOgg1 then probes the base pair dynamics as it slides, extruding OG from the duplex at a higher rate than undamaged bases. Theoretical analysis of the process of target location suggests that a combination of sliding over short distances and jumping (via dissociation/recombination) over large distances provides the most kinetically efficient pathway (10).
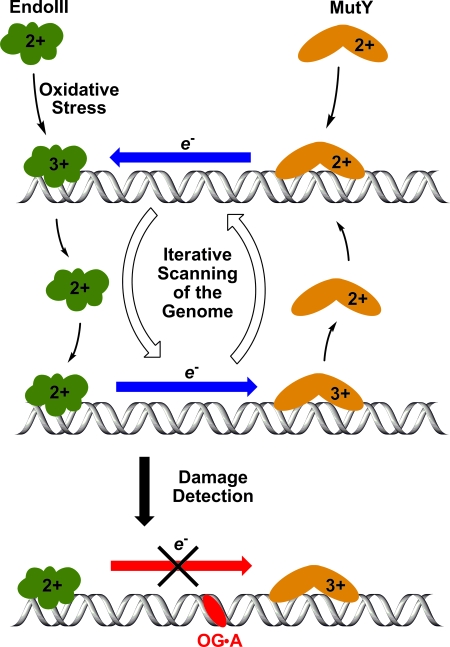
The charge transport mechanism proposed by the Barton laboratory (4) adds an alternative search process to the pathways described above (11). In this mechanism, two BER enzymes containing [4Fe-4S]3+/2+ clusters collaborate by probing the integrity of a long stretch of the DNA duplex (Fig. 2). Transport of an electron (or conversely, an electron hole) is fast and efficient through undamaged duplex DNA allowing one enzyme to remotely reduce another. Previous studies have shown that when the [4Fe-4S] cluster of EndoIII or MutY is in the +3 state, the enzyme is more tightly bound; in the +2 state, the enzyme can more readily dissociate from DNA (12). Thus, efficient charge transport through an undamaged duplex results in MutY being reduced, and therefore released from that segment of DNA. Such a mechanism allows the repair protein to detect the absence of DNA damage in a large region, potentially a few hundred base pairs long. Successful electron transfer then leads to release of the bound repair protein from a section of DNA where it is not needed, so that it is ready to diffuse to a new site and begin the search process again. This process can continue cycling until a segment of DNA is found that does not support charge transport; at this point (Fig. 2 Bottom), MutY remains in the +3 state, tightly bound to DNA, and can engage in 1D scanning to find the exact site of the lesion (13).
Fig. 2.
The [4Fe-4S] clusters of EndoIII and MutY can shuttle electrons through the DNA π stack, leading to dissociation of the protein in the reduced +2 state. When DNA damage interrupts the charge transport, a protein in the +3 state remains tightly bound to home in on the damage site.
New in the current paper (4) is evidence from the Barton laboratory that the [4Fe-4S]-containing proteins preferentially relocate to strands containing a base mismatch that interrupts charge transport in duplex DNA. The study was conducted by using long mismatched vs. short matched duplexes, and the bound proteins were detected by using atomic force microscopy. The observation of a higher density of EndoIII bound to the long mismatched strands is consistent with the blockage of DNA charge transport to reduce and release the protein.
In a second part of the study, the researchers examined cooperativity between EndoIII and MutY inside Escherichia coli. Using a MutY reporter strain for the readout, they inactivated the gene encoding EndoIII, which led to a decrease in the in vivo activity of MutY, although the decrease was modest. A further study used the Y82A mutant of EndoIII, a protein that was shown to contain the [4Fe-4S] cluster and maintain glycosylase activity toward a natural substrate, 5-hydroxyuracil; however, electrochemical studies suggest that it has attenuated charge transfer properties when bound to DNA. In accord with these properties, Y82A was unable to participate in the in vivo helper function to assist MutY.
Taken in sum, the experiments lend support to the collaboration between the two [4Fe-4S] proteins EndoIII and MutY in which EndoIII can assist the low-copy-number partner MutY to find lesions that disrupt the charge transport properties of the duplex. Iron–sulfur clusters are also found in the human homologs of these proteins hNTH1 and MUTYH, with deficiencies in the latter protein being implicated in certain types of inherited colorectal cancers (14). Thus, the possibility exists that the preservation of the [4Fe-4S] cluster in many BER enzymes throughout evolution has a functional, in addition to a structural, basis.
Questions about the charge transport mechanism remain, however. Why would a repair system for oxidative damage use a redox process? This is particularly curious in light of the fact that OG is extremely sensitive to further oxidation [E1/2 ≈ 700 mV vs. the normal hydrogen electrode (NHE)], and oxidants as mild as ferricyanide (E1/2 ≈ 400 mV vs. NHE) are able to convert OG to highly mutagenic hydantoin lesions (15). The answer must lie with the significantly lower redox potential of MutY [≈100 mV when bound to DNA (12)]; indeed the [4Fe-4S]2+ state of MutY should be a potent reductant of the OG+• radical cation (16). The lifetime of OG+• in duplex DNA can be on the order of seconds (17), raising the possibility that another role of the iron–sulfur cluster could be to reduce this highly mutagenic species back to an OG·A base pair in the vicinity of a now-oxidized [4Fe-4S]3+ form of MutY that remains tightly bound and ready to begin the excision process.
Unfortunately, mechanisms can never be proven, only disproven, and so we are limited by our imaginations as to what alternative search algorithms proteins might select to locate their target sites on DNA. There may be additional features of lesion dynamics or electronics that attract a repair enzyme to the damage site. For example, the presence of OG in a strand increases the energy of the highest occupied molecular orbital (HOMO) of the DNA and increases the density of states, which may result in higher efficiency of electron trapping by oligomers containing OG (18).
Sliding, hopping, jumping, wandering…or zapping? The data of Boal et al. (4) support the imaginative proposal of DNA damage location by redox-active glycosylases that tap the charge transport properties of the duplex to remotely detect, and then home in on, the culprit base.
Acknowledgments.
Work on DNA damage in our laboratory is supported by research grants from the National Institutes of Health (R01 CA090689) and the National Science Foundation (0809483).
Footnotes
The authors declare no conflict of interest.
See companion article on page 15237 in issue 36 of volume 106.
References
- 1.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 2.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 4.Boal AK, et al. Redox signaling between DNA repair proteins for efficient lesion detection. Proc Natl Acad Sci USA. 2009;106:15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows CJ, et al. Structural elucidation and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanosine oxidation by transition metals. Environ Health Perspect. 2002;110(Suppl5):713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson PT, et al. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 8.Parker JB, et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. Pt 2. [DOI] [PubMed] [Google Scholar]
- 11.Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yavin E, Stemp EDA, O'Shea VL, David SS, Barton JK. Electron trap for DNA-bound repair enzymes: A strategy for DNA-mediated signaling. Proc Natl Acad Sci USA. 2006;103:3610–3614. doi: 10.1073/pnas.0600239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fok P-W, Chou T. Accelerated search kinetics mediated by redox reactions of DNA repair enzymes. Biophys J. 2009;96:3949–3958. doi: 10.1016/j.bpj.2009.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 15.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. Sequence and stacking dependence of 8-oxoG oxidation: Comparison of one-electron vs. singlet oxygen mechanisms. J Am Chem Soc. 1999;121:9423–9428. [Google Scholar]
- 16.Lin J-C, Singh RRP, Cox DL. Theoretical study of DNA damage recognition via electron transfer from the [4Fe-4S] complex of MutY. Biophys J. 2008;95:3259–3268. doi: 10.1529/biophysj.108.132183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misiaszek R, Uvaydov Y, Crean C, Geacintov NE, Shafirovich V. Combination reactions of superoxide with 8-oxo-7,8-dihydroguanine radicals in DNA: Kinetics and end products. J Biol Chem. 2005;280:6293–6300. doi: 10.1074/jbc.M412253200. [DOI] [PubMed] [Google Scholar]
- 18.Markus TZ, et al. Electronic structure of DNA—Unique properties of 8-oxoguanosine. J Am Chem Soc. 2009;131:89–95. doi: 10.1021/ja804177j. [DOI] [PubMed] [Google Scholar]