Abstract
Summary: Mapping of next-generation sequencing data derived from RNA samples (RNAseq) presents different genome mapping challenges than data derived from DNA. For example, tags that cross exon-junction boundaries will often not map to a reference genome, and the strand specificity of the data needs to be retained. Here we present RNA-MATE, a computational pipeline based on a recursive mapping strategy for placing strand specific RNAseq data onto a reference genome. Maximizing the mappable tags can provide significant savings in the cost of sequencing experiments. This pipeline provides an automatic and integrated way to align color-space sequencing data, collate this information and generate files for examining gene-expression data in a genomic context.
Availability: Executables, source code, and exon-junction libraries are available from http://grimmond.imb.uq.edu.au/RNA-MATE/
Contact: n.cloonan@imb.uq.edu.au
Supplementary information: Supplementary data are available at Bioinformatics Online.
High-throughput sequencing technologies can generate hundreds of millions of short tags (typically only 25–50 nt) from a single experiment, and this capacity is enabling a new wave of genomic research. Not only is this technology being used for genomic sequencing, re-sequencing and epigenomic applications, but several groups have recently applied this technology to sequence RNA for gene-expression studies (RNAseq).
Conceptually, RNAseq protocols are simple. Single-stranded RNA molecules are captured between two sequencing adaptors, either through serial ligation (Lister et al., 2008) or through random-primed PCR (Cloonan et al., 2008), yielding strand specific information. More commonly, RNAseq data is generated from sheared, double-stranded cDNA libraries (Mortazavi et al., 2008; Nagalakshmi et al., 2008; Sultan et al., 2008; Wilhelm et al., 2008), however this approach loses strand information, which can not always be assumed from annotations (since loci can produce antisense transcripts, and loci on different strands can overlap).
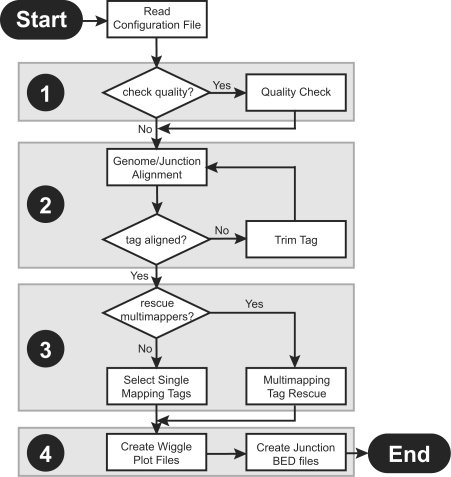
Some of the benefits of using RNAseq over microarrays to study gene-expression data include: (i) the potentially unlimited dynamic range of expression; (ii) the greater sensitivity of the sequencing data; (iii) the improved ability to discriminate regions of high sequence identity; and (iv) the ability to profile transcription without prior assumptions of which genomic regions are expressed. However, for mammalian genomes, there are technical challenges associated with mapping and counting short-tag sequences. Firstly, mammalian transcripts are non-contiguous due to the splicing of introns from the pre-mRNA, therefore a proportion of tags (those that cross exon-exon boundaries) will not map directly to the genome. The presence of genome wide repeats and other repetitive sequence in the mouse and human genomes means that a sizeable proportion of short tags can not be placed uniquely (Faulkner et al., 2008). Finally, depending on the specific library preparation protocol, a proportion of fragments may be shorter than the full length of the tag sequenced. Such tags will contain adaptor sequence that prevents mapping to the genome. Here we present a computational pipeline to map RNAseq data. RNA-Mapping and Alignment Tools for Expression (RNA-MATE) generates both tag counting and genome-browser visualization of genomic and exon-junction mapping results, addressing the issues above (Fig. 1).
Fig. 1.
The RNA-MATE recursive mapping pipeline. The pipeline consists of four major components. (1) Tags are (optionally) filtered based on the quality values for each basecall. (2) The alignment module attempts to align tags first to the genome, and then to a library of known exon-junction sequences. If a tag fails to align, then the tag is truncated, and the process is repeated. (3) The optional tag rescue module uses information derived from both single- and multi-mapping tags to uniquely place multi-mapping tags. (4) Finally, UCSC genome browser compatible wiggle plots and BED files are generated.
Depending on the downstream applications of the mapped data, the quality of individual tags may need to be assessed before inclusion in the mapping dataset. To accommodate this, there is an optional tag quality module, which assesses the tags by the number of basecalls with PHRED scores of <10. If this option is disabled, all tags are passed to the alignment module. Alignment of the short tags to a reference is done using mapreads (http://solidsoftwaretools.com/gf/project/mapreads/), an algorithm that maps color-space data. Tags are mapped to the reference genome, and then against a library of known exon-junctions (although this default behavior can be modified). Tags that fail to map are chopped to user-defined lengths, and the genomic mapping is restarted. In this way, tags that have adaptor sequence, or poor quality ends are recovered at their longest length. Although this strategy uses more CPU time, it will typically yield between 1.6 and 3 times the mappable tags, providing significant cost savings. The number of mismatches between the reference and tag is user defined, and when unique genomic mappings are selected, only the mappings at the highest level of stringency are retained. Advice on mapping parameters is included in the documentation.
For most downstream applications, tags are only informative if they can be placed uniquely within a genome. The proportion of transcriptome tags that align to multiple places within a genome will vary depending on the length of the tags, the genome, and the expression in the individual sample, however this is typically between 13–38% for mammalian libraries (Cloonan et al., 2008; Mortazavi et al., 2008). Strategies to rescue ambiguous sequences have recently been applied to high-throughput sequencing data, and we have refined our previously described algorithm (Faulkner et al., 2008) to work efficiently with large data sets. For every multi-mapping tag, the algorithm considers all tags that map near to each of the possible locations of the tag (within a user-specified window) to determine the most likely mapping position of the tag. Where a tag can not be unambiguously assigned, a fractional weighting to the relevant positions is assigned. Between 40 and 60% of multi-mapping tags can be assigned a single position with ≥60% likelihood, depending on the relative sequence coverage (Supplementary Table 1). The recommended window size for shotgun sequencing is 10 nt (Cloonan et al., 2008), though for disparate data types currently available this can vary. For instance, Cap Analysis of Gene Expression (CAGE) tags are rescued using a window of 100 nt, a size previously shown to optimize mammalian promoter detection (Carninci et al., 2006).
Finally, UCSC genome browser compatible wiggle plots for genome mapped data, and BED files for exon-junction mapped data are generated automatically from the collated results. The wiggle plots are strand-specific, single-nucleotide resolution coverage plots, and directly represent the number of times an individual nucleotide has been seen in the sequencing data. BED files depict hits to junction sequences, and graphically display exon combinatorics. In addition, plots containing only start sites of tags are included to facilitate tag-counting applications. Plots generated from particularly deep sequencing may be difficult to upload directly to the UCSC genome browser, and a post-processing script to filter wiggle plots to aid visualization in this way is provided.
The assignment of tags to genes is facilitated Galaxy (Giardine et al., 2005), and a tutorial is provided in the user manual. This allows the comparison of RNAseq data to microarray data, or the visualization and analysis using tools developed for microarrays. In this step, particular care needs to be taken to ensure that different RNAseq protocols are processed with the strand of capture in mind. For example, serial-ligation approaches will generate sequences from the sense strand, relative to the annotated gene, whereas the random-primed strand-specific protocols will generate tags mapping to the anti-sense strand.
This pipeline described here is written in Perl, and makes use of a PBS queue manager, however it can be configured to use LSF or SGE (detailed in the documentation). Due to the long computational times required for recursive-mapping, implementation on machines without access to a cluster is not recommended, but possible. Future releases will support alternative alignment algorithms, SNP calling, and an easy to use web based GUI. All source code, instructions, testing data, and additional scripts are available from http://grimmond.imb.uq.edu.au/RNA-MATE/.
Supplementary Material
ACKNOWLEDGEMENTS
The authors greatly appreciate the advice and sequencing data given to us by Ehsan Nourbakhsh and Brooke Gardiner, and the help with LSF and SGE queues given to us by Xuanzhong Li and Till Bayer.
Funding: National Health and Medical Research Council [455857 to S.M.G. 456140, 631701]; the Australian Research Council [DP0988754]; the Australian Stem Cell Centre [to G.J.F, G.K. and D.F.T.]; and the University of Queensland [to N.C. and Q.X.].
Conflict of Interest: none declared.
REFERENCES
- Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Meth. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, et al. A rescue strategy for multimapping short sequence tags refines surveys of transcriptional activity by CAGE. Genomics. 2008;91:281–288. doi: 10.1016/j.ygeno.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008:1160342. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.