Abstract
Motivation: In functional genomics, it is frequently useful to correlate expression levels of genes to identify transcription factor binding sites (TFBS) via the presence of common sequence motifs. The underlying assumption is that co-expressed genes are more likely to contain shared TFBS and, thus, TFBS can be identified computationally. Indeed, gene pairs with a very high expression correlation show a significant excess of shared binding sites in yeast. We have tested this assumption in a more complex organism, Drosophila melanogaster, by using experimentally determined TFBS and microarray expression data. We have also examined the reverse relationship between the expression correlation and the extent of TFBS sharing.
Results: Pairs of genes with shared TFBS show, on average, a higher degree of co-expression than those with no common TFBS in Drosophila. However, the reverse does not hold true: gene pairs with high expression correlations do not share significantly larger numbers of TFBS. Exception to this observation exists when comparing expression of genes from the earliest stages of embryonic development. Interestingly, semantic similarity between gene annotations (Biological Process) is much better associated with TFBS sharing, as compared to the expression correlation. We discuss these results in light of reverse engineering approaches to computationally predict regulatory sequences by using comparative genomics.
Contact: amarcoca@asu.edu
1 INTRODUCTION
Gene expression is typically mediated by transcription factors, which bind to specific DNA sequences (transcription factor binding sites, TFBS). The same transcription factor frequently binds regulatory regions of different genes, inducing their coordinated expression (Davidson, 2001; Latchman, 2005). Therefore, genes containing the same TFBS are often co-expressed. Because the experimental identification of TFBS is difficult, investigators frequently use shared sequence motifs in co-expressed genes to identify putative TFBS (reviews in Chua et al., 2004; Hannenhalli, 2008; Wasserman and Sandelin, 2004). This reverse engineering approach uses the co-expression of genes as a tool to predict regulatory sequences, which has been employed in yeasts, plants and animals (e.g. Cheng and Li, 2008; Dai et al., 2007; De Bleser et al., 2007; Fogel et al., 2004; Perco et al., 2005; Tavazoie et al., 1999).
Are genes with high expression correlation more likely to share TFBS, as compared to those with low expression correlation? This question has been addressed in Saccharomyces cerevisiae for which both extensive microarray expression data and experimentally verified TFBS exist (Yu et al., 2003; Allocco et al., 2004; Yeung et al., 2004). In this single-celled organism, only gene pairs with a very high degree of co-expression share significantly larger number of TFBS (Allocco et al., 2004). While these results point to limitations of the use of the reverse engineering approach, they do encourage the use of shared sequence motifs as putative TFBS for some pairs of genes (Allocco et al., 2004).
The regulation of gene expression in animals is far more complex than that in unicellular organisms such as yeast or prokaryotes. At the whole-organism level, the existence of compartmentalization and multiple cell types leads to enhanced complexity in animal regulatory networks. Thus, the experimental characterization of regulatory sequences in multicellular organisms is more difficult, and the use of computational tools to predict putative TFBS more valuable. However, the prediction of regulatory sequences is expected to be generally less successful in animals than in unicellular organisms (see also Tompa et al., 2005; Vavouri and Elgar, 2005). The extent of such limitations in the context of high-throughput microarray expression data remains unknown for animals. Here, we use a compilation of experimentally-verified TFBS and extensive microarray experiment results available for D.melanogaster in efforts to (i) quantify the average degree of co-expression for genes containing common TFBS, and (ii) estimate the relative propensity of co-expressed genes to share common TFBS in a multicellular organism.
2 METHODS
We extracted all experimentally verified TFBS data from the REDfly 2.0 database (Halfon et al., 2008), which contains associations between transcription factors and their target genes from systematic curation of DNase I footprint experiments from multiple developmental stages in Drosophila (ranging from antero-posterior to mesoderm and imaginal disk patterning). We retrieved the expression patterns for the target Drosophila genes in the dataset from microarrays compiled by Spellman and Rubin (2002). The co-expression level is measured as the Pearson correlation coefficient of their expression values in different stages/experiments. Expression levels in these experiments were measured for the whole organisms.
In addition to REDFLY TFBS, we analyzed data for six transcription factors (bcd, kni, Kr, hb, cad, gt) from Li et al. (2008), which enabled us to examine correlations of expression and TFBS sharing in Drosophila early development. We used TFBS with a false discovery rate ≤1%, which were filtered using two additional criteria: the target gene should be expressed during early development; and, if two antibodies were used in the detection, the TFBS should be reported in both experiments. Expression patterns for these TFBS were obtained from Tomancak et al. (2002), which captured temporal expression during early development. For these data, we calculated the average expression value for the three experimental replicates in each time window. Alternative analyses using values individually did not change our results.
In further analyses, we also used GO annotation similarity to determine expression correlation instead of microarrays. In order to quantify the GO annotation similarity, we calculated the maximum Resnik semantic similarity between genes (Lord et al., 2003; Resnik, 1999). The calculations were done with the GOSim package (Fröhlich et al., 2007) under the R environment (http://www.r-project.org/). Analyses using other different measures yielded similar results. In all cases we calculated confidence intervals from 1000 randomly permuted datasets or from standard errors of the mean, as applicable.
3 RESULTS
In an analysis of 2485 gene pairs (71 genes from REDFly), genes sharing at least one TFBS showed a significantly higher expression correlation than those with no common TFBS (Fig. 1, P < <0.01). The average expression correlation is higher in magnitude for genes sharing at least two TFBS as compared to only one TFBS, although the difference was not statistically significant.
Fig. 1.
Number of common TFBS and expression correlation. Height of the boxes represents the average Pearson correlation values for gene pairs sharing different numbers of transcription factor binding sites. The lines over the boxes represent 95% confidence intervals (1.96 times the standard error of the mean). Significance of the differences was assessed by using Mantel tests (Sokal and Rohlf, 1995). Results are shown for absolute, only positive and only negative expression correlation values.
Separate examination of these trends for gene pairs with negative and positive expression correlations showed that genes sharing at least one TFBS had, on average, a 10% more positive expression correlation than those not sharing TFBS (Fig. 1, P < <0.01). We also observed this trend for negative expression correlation, although the differences between categories were much smaller than in the case of positive correlation (Fig. 1).
In these negative correlated gene pairs, the observation that genes sharing TFBS have higher negative co-expression may be taken as an indication of the presence of dual regulators (i.e. transcription factors acting both as activators and repressors). A dual regulator would activate one gene of the pair while repressing the other at the same time. This would produce a negative (complementary) expression correlation between the two genes. Indeed, dual regulators seem to play an important role during Drosophila development (Papatsenko and Levine, 2008). In our data, 25% TFBS (22 out of 88) were common to genes with negative expression correlation. The list of potentially dual regulators contained segmentation genes (bcd, kni, hb, Kr) and homeotic genes (abd-A, Antp, Ubx), which are known to act simultaneously as repressors and activators during fly development (Lawrence, 1992).
Trends for absolute, positive and negative correlations between D.melanogaster and S.cerevisiae were similar (Fig. 2), with the exception that all differences were statistically highly significant in yeast analysis presumably due to much larger sample sizes. However, the fraction of negatively co-expressed genes in yeast is much lower than the fraction in Drosophila (Fig. 2C), which could suggests a lower frequency of dual regulators in yeast.
Fig. 2.
Number of common TFBS and co-expression correlation comparing Drosophila and yeast. The plot compares the average and 95% confidence intervals for yeast and Drosophila co-expression correlation values for genes bound by none, more than one or more than two common TFBS. Confidence intervals in yeast are negligible. Dashed line indicates the 1:1 ratio. Different panels show: (A) absolute; (B) only positive and (C) only negative co-expression correlation values.
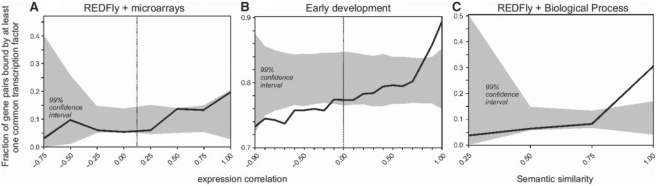
Next, we examined the reverse relationship between gene expression correlation and the number of shared TFBS. This is important for evaluating the predictive use of co-expression in detecting TFBS. In particular, we tested whether gene pairs with high (either positive or negative) correlation show an increased probability of sharing TFBS. For positively correlated genes, the number of shared TFBS is 0.20 per pair on average, which is more than two times that observed for negatively correlated genes (0.09). The plot of the proportion of gene pairs sharing TFBS as a function of the expression correlation revealed a mild positive trend (Fig. 3A), with positively correlated genes showing slightly higher propensity of sharing TFBS than the negatively correlated genes. Gene pairs with higher expression correlation shared more TFBS, but the relationship is not statistically significant. Even for genes with expression correlations ≥0.75, only one in five gene pairs shares at least one TFBS (Fig. 3A). Therefore, the use of expression correlation to find genes via comparative genomics of shared motifs may not be very productive.
Fig. 3.
Relationship between degree of co-expression or semantic similarity, and fraction of common TFBS. The fraction of gene pairs sharing at least one common TFBS (y-axis) is plotted for intervals of co-expression levels in (A) and (B), and semantic similarity in (C) (x-axis). A 99% confidence interval for random expectation is represented in gray. Positive and negative correlation values are separated by a dashed vertical line. (A) Comparison between REDFLY 2.0 reported TFBS and Spellman and Rubin (2002) compilation of microarray expression data. (B) Comparison between early development dataset of TFBS generated by Li et al. (2008) and Tomancak et al. (2002) microarray dataset. (C) Fraction of gene pairs sharing at least one common TFBS in REDFLY 2.0 for different intervals of semantic similarity.
In contrast to D.melanogaster, about 40% of yeast gene pairs with expression correlation above 0.75 shared at least one TFBS and 100% of the genes with expression correlation ≥0.90 shared at least one TFBS (Allocco et al., 2004). In Drosophila, gene pairs with an expression correlation ≥0.90 share a TFBS only 24% of the time. This is a rather large difference, which prompted us to examine whether a small number of TFBS available for Drosophila analysis, as compared to yeast (∼10 times), adversely impacted our investigations. In order to test this possibility, we analyzed randomly selected subsets of the yeast TFBS data, such that the random yeast subsets had the same number of transcription factors as those present in our Drosophila dataset. The yeast subsets still showed a significant enrichment for highly co-expressed genes in 80% of the analyses, which suggests that the smaller number of TFBS available in Drosophila is not expected to obscure the relationship between expression correlation and the number of genes with common TFBS.
Next, we compared the temporal microarray results with those observed in the analysis of adult tissues by using the FlyAtlas data (Chintapalli et al., 2007), which contain genes expressed in 26 tissues. We calculated the proportion of tissues in which genes sharing TFBS were co-expressed and the proportion of co-expressed genes not sharing TFBS. Genes sharing TFBS are co-expressed, on average, in 55% of the analyzed tissues, which was similar to the proportion of co-expressed genes not sharing TFBS (52%). Thus, the expression correlations based on the proportion of tissues in which genes are co-expressed will not be effective as a tool for finding TFBS. On the other hand, genes were enriched for common TFBS in specific tissues. For instance, genes expressed in the brain, share many more common TFBS than expected by chance (2.9 times). For fat body this ratio is 3.1, and for the heart organ as high as 4.7.
The overall lack of relationship between the extent of co-expression and the number of shared TFBS may be related to the greater diversity of tissues and cell types resulting from the complex developmental pathways in higher metazoans. This attribute is less severe in the earliest stages of Drosophila embryogenesis. In order to assess the relationships between co-expression and shared TFBS in the early development, we took advantage of a recently published genome-wide experimental detection of TFBS for six transcription factors acting during early development (Li et al., 2008). The expression correlation values were measured for the temporal expression pattern during the first 6 h of development (Tomancak et al., 2002). We examined the fraction of shared TFBS as a function of the degree of co-expression, and found that gene pairs with expression correlations ≥0.80 were statistically enriched for common TFBS (Fig. 3B). Genes showing substantial negative correlation had a smaller fraction of common TFBS. The positive association between co-expression and shared TFBS is likely due to two primary factors. First, there are no differentiated tissues at these early stages. Second, the embryonic chromatin is not organized in heterochromatic structures prior to the third hour of development which, potentially, makes all genomic DNA accessible to the action of transcription factors (Lu et al., 1998; Vlassova et al., 1991). These two factors make the early Drosophila embryo to exhibit higher similarities with those observed in yeast. Therefore, expression correlation is expected to be more useful to find TFBS during early development.
Next, we examined whether genes involved in the same biological processes show an overabundance of shared TFBS. Annotations in the Biological Process category in Gene Ontology (GO; Ashburner et al., 2000) reflects our knowledge on tissues and cell types in which genes are found to be co-expressed. This information is more comprehensive (and of different type) than that of selected adult organs in FlyAtlas, and covers embryonic tissues as well. We calculated an index that reflects the degree of similarity in the GO annotation of two gene products (Resnik similarity measure; Lord et al., 2003). This index ranges from 0 to 1 (from completely unrelated annotations to identical annotation for genes). The average level of annotation similarity between gene pairs was higher for genes sharing more TFBS (0.68 versus 0.56). In reverse, gene pairs with very similar GO annotation were more highly enriched in common TFBS (Fig. 3C). The percentage of gene pairs sharing TFBS, when the similarity in the annotation was over 0.75, was ∼30% and significantly higher than expected.
These results indicate that the spatial patterns of expression captured in gene annotations have a better association with the number of shared TFBS as compared to the expression correlation obtained from Drosophila microarray experiments. In contrast to Biological Process, analyses using the other two GO categories (Molecular Function and Cellular Component) yielded no significant association with the number of TFBS shared (data not shown). Therefore, Biological Process semantic similarity could be a useful tool for finding genes with common TFBS.
We also examined the relationships between the microarray-based gene expression and semantic similarity. The overall correlation coefficient was rather small (R2 ≈ 0.1), however, gene pairs sharing TFBS with expression correlation >0.75 showed high semantic similarities. On the other hand, pairs of genes sharing TFBS with a semantic similarity >0.75 had a lower average expression correlation. Therefore, many gene pairs with high semantic similarity have low expression correlations. For example, slbo and btl are co-activated during cell migration processes (Murphy et al., 1995), and have semantic similarity value of 0.77. But, the co-expression correlation is <0.01 in the microarray experiments. The expression of rho and sim provide another example, they are both expressed in the neuroectoderm (Martín-Bermudo et al., 1995; Nibu et al., 1998) and have a semantic similarity of 0.94. However, the co-expression correlation is only 0.21. Many such gene pairs are significantly enriched for common TFBS.
4 DISCUSSION
Computational genomics studies often predict TFBS assuming that co-expressed genes are more likely to share binding sites than those not co-expressed (Chua et al., 2004; Hannenhalli, 2008; Wasserman and Sandelin, 2004). However, one can begin to assess the underlying principles of these studies only by comparing experimentally verified TFBS and expression patterns. Here we addressed this question in one model animal, D.melanogaster, and compared the results to those observed in the yeast. We find that genes bound by the same TFBS have expression correlations greater than those not sharing TFBS. Nevertheless, unlike yeast, gene pairs with a high degree of co-expression in DNA microarray experiments are not statistically more likely to share a TFBS (except in the early development). We obtained similar results by using other microarray experiments (Arbeitman et al., 2002; Manak et al., 2006; Tomancak et al., 2002).
Many Drosophila microarrays deal with whole organism gene expression level at different time periods (Arbeitman et al., 2002; Manak et al., 2006; Stolc et al., 2004; Tomancak et al., 2002). Given that genes can be transcribed in different tissues simultaneously in multicellular organisms, it is reasonable to expect that the spatial expression patterns of gene products will be more informative than temporal patterns in order to detect TFBS. This may be the reason why there is no statistical enrichment of TFBS in even highly co-expressed genes (Fig. 3A). Of course, other causes might explain the partial uncoupling between microarray (temporal) expression patterns and regulation by common transcription factors: including differential affinity to binding sites (see Discussion in Pankratz and Jackle, 1993); or regulation by miRNA (Sempere et al., 2003; Sokol et al., 2008). The presence of indirect regulatory interactions after early development might also obscure the association between co-expression and TFBS.
Our results suggest that most collections of DNA microarray data in Drosophila may not be useful to predict TFBS, unless we study the action of transcription factors during very early development. These findings are consistent with the success in mammals of reverse engineering approaches to infer regulatory sequences when microarrays measure expression levels in different tissues, that is, spatial expression, instead of quantifying global transcript levels in the whole organism (e.g. De Bleser et al., 2007; Perco et al., 2005). The use of cell cultures in mammals has been also useful to detect TFBS involved in cell-cycle control (e.g. De Bleser et al., 2007; Elkon et al., 2003). Further analyses in Drosophila cell lines could be of interest.
We could not establish a quantitative relationship between the number of tissues in which two genes are co-expressed and TFBS sharing. However, exploration of individual tissues reveals that genes co-expressed in the same organ are indeed enriched for shared TFBS. The fact that genes with similar Biological Process annotations are statistically enriched for common TFBS further supports the above conclusion and suggests that the correlations of spatial patterns of gene expression will provide invaluable for finding TFBS via comparative analysis in multicellular eukaryotes. In particular, correlations in spatial patterns of gene expression captured in high-throughput in situ RNA hybridization techniques in Drosophila may be useful in identifying TFBS using a comparative approach (Tomancak et al., 2002; Lécuyer et al., 2007).
ACKNOWLEDGEMENTS
The authors thank Fabia Battistuzzi and two anonymous reviewers for useful comments on this manuscript.
Funding: National Institutes of Health grant (to S.K.).
Conflict of Interest: none declared.
REFERENCES
- Allocco DJ, et al. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics. 2004;5:18. doi: 10.1186/1471-2105-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, et al. Gene expression during the life cycle ofDrosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Li L. Systematic identification of cell cycle regulated transcription factors from microarray time series data. BMC Genomics. 2008;9:116. doi: 10.1186/1471-2164-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, et al. Using FlyAtlas to identify betterDrosophila melanogastermodels of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Chua G, et al. Transcriptional networks: reverse-engineering gene regulation on a global scale. Curr. Opin. Microbiol. 2004;7:638–646. doi: 10.1016/j.mib.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Dai X, et al. A new systematic computational approach to predicting target genes of transcription factors. Nucleic Acids Res. 2007;35:4433–4440. doi: 10.1093/nar/gkm454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic Press; 2001. [Google Scholar]
- De Bleser P, et al. A distance difference matrix approach to identifying transcription factors that regulate differential gene expression. Genome Biol. 2007;8:R83. doi: 10.1186/gb-2007-8-5-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, et al. Discovery of sequence motifs related to coexpression of genes using evolutionary computation. Nucleic Acids Res. 2004;32:3826–6835. doi: 10.1093/nar/gkh713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich H, et al. GOSim—an R-package for computation of information theoretic GO similarities between terms and gene products. BMC Bioinformatics. 2007;8:166. doi: 10.1186/1471-2105-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon MS, et al. REDfly 2.0: an integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res. 2008;36:D594–D598. doi: 10.1093/nar/gkm876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S. Eukaryotic transcription factor binding sites—modeling and integrative search methods. Bioinformatics. 2008;24:1325–1331. doi: 10.1093/bioinformatics/btn198. [DOI] [PubMed] [Google Scholar]
- Latchman DS. Gene Regulation: A Eukaryotic Perspective. New York: Taylor & Francis; 2005. [Google Scholar]
- Lawrence PA. The Making of a Fly: The Genetics of Animal Design. Boston: Blackwell Scientific; 1992. [Google Scholar]
- Lécuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Li X, et al. Transcription factors bind thousands of active and inactive regions in theDrosophilablastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord PW, et al. Semantic similarity measures as tools for exploring the Gene Ontology. Pacific Symp. Biocomput. 2003;8:601–612. doi: 10.1142/9789812776303_0056. [DOI] [PubMed] [Google Scholar]
- Lu B, Ma J, Eissenberg J. Developmental regulation of heterochromatin-mediated gene silencing inDrosophila. Development. 1998;125:2223–2234. doi: 10.1242/dev.125.12.2223. [DOI] [PubMed] [Google Scholar]
- Manak , et al. Biological function of unnanotated transcription during the early development ofDrosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Martin-Bermudo MD, et al. Neurogenic genes control gene expression at the transcriptional level in early neurogenesis and in mesectoderm specification. Development. 1995;121:219–224. doi: 10.1242/dev.121.1.219. [DOI] [PubMed] [Google Scholar]
- Murphy AM, et al. The breathless FGF receptor homolog, a downstream target ofDrosophilaC/EBP in the developmental control of cell migration. Development. 1995;121:2255–2263. doi: 10.1242/dev.121.8.2255. [DOI] [PubMed] [Google Scholar]
- Nibu Y, et al. dCtBP mediates transcriptional repression by knirps, Kruppel and snail in theDrosophilaembryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz MJ, Jackle H. The Development of Drosophila Melanogaster. New York: Cold Spring Harbor Laboratory Press; 1993. Blastoderm segmentation; pp. 505–509. [Google Scholar]
- Papatsenko D, LevineM.S. Dual regulation by the Hunchback gradient in theDrosophilaembryo. Proc. Natl Acad. Sci. USA. 2008;105:2901–2906. doi: 10.1073/pnas.0711941105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perco P, et al. Detection of coregulation in differential gene expression profiles. Bio. Systems. 2005;82:235–247. doi: 10.1016/j.biosystems.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Resnik P. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J. Artificial Intell. Res. 1999;11:95–130. [Google Scholar]
- Sempere LF, et al. Temporal regulation of microRNA expression inDrosophila melanogastermediated by hormonal signals andbroad-complexgene activity. Dev. Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. New York: Freeman; 1995. [Google Scholar]
- Sokol NS, et al. Drosophilalet-7microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in theDrosophilagenome. J. Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc , et al. A gene expression map for the euchromatic genome ofDrosophila melanogaster. Science. 2004;306:665–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Tavazoie S, et al. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Tomancak P, et al. Systematic determination of patterns of gene expression duringDrosophilaembryogenesis. Genome Biol. 2002;3:R88. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Elgar G. Prediction of cis-regulatory elements using binding site matrices—the successes, the failures and the reasons for both. Curr. Opin. Genet. Develop. 2005;15:395–402. doi: 10.1016/j.gde.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Vlassova IE, et al. Constitutive heterochromatin in early embryogenesis ofDrosophila melanogaster. Mol. Gen. Genet. 1991;229:316–318. doi: 10.1007/BF00272172. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Yeung KY, et al. From co-expression to co-regulation: how many microarray experiments do we need? Genome Biol. 2004;5:R48. doi: 10.1186/gb-2004-5-7-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, et al. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 2003;19:422–427. doi: 10.1016/S0168-9525(03)00175-6. [DOI] [PubMed] [Google Scholar]