Abstract
Systems pharmacology is an emerging area of pharmacology which utilizes network analysis of drug action as one of its approaches. By considering drug actions and side effects in the context of the regulatory networks within which the drug targets and disease gene products function, network analysis promises to greatly increase our knowledge of the mechanisms underlying the multiple actions of drugs. Systems pharmacology can provide new approaches for drug discovery for complex diseases. The integrated approach used in systems pharmacology can allow for drug action to be considered in the context of the whole genome. Network-based studies are becoming an increasingly important tool in understanding the relationships between drug action and disease susceptibility genes. This review discusses how analysis of biological networks has contributed to the genesis of systems pharmacology and how these studies have improved global understanding of drug targets, suggested new targets and approaches for therapeutics, and provided a deeper understanding of the effects of drugs. Taken together, these types of analyses can lead to new therapeutic options while improving the safety and efficacy of existing medications.
Contact: ravi.iyengar@mssm.edu
1 INTRODUCTION
Translational medical sciences aim to convert our understanding of biological mechanisms into effective ways of treating and preventing diseases. Such understanding has enabled us to move from invasive treatment approaches to therapies that can be delivered orally. For example, the use of surgery to treat peptic ulcers has greatly declined as use of Histamine-2 receptor blockers and proton pump inhibitors gained popularity (Towfigh et al., 2002). As our understanding of complex diseases has grown, it is becoming increasingly feasible to design orally delivered drugs for many pathophysiologies. Drug discovery has also been enabled by combinatorial chemistry that allows for the facile synthesis of many classes and variants of compounds that have the potential to be drugs. High-throughput screening technology facilitates the use to these compound libraries to identify leads that can be further developed into useful therapeutic agents. As technological limitations in drug discovery are reduced, there still exist challenges in identifying appropriate targets for complex diseases. This challenge arises from our incomplete knowledge of the functions of biological systems including those involved in human physiology and disease. New experimental technologies are generating massive amounts of data that characterize complex biological systems. Extracting knowledge from these large datasets requires both bioinformatics and extensive computational analysis. There are many types of computations that aid in knowledge extraction. It seems clear that such computational analysis will be a part of all biomedical sciences.
Systems pharmacology is an emerging field that uses both experiments and computation to develop an understanding of drug action across multiple scales of complexity ranging from molecular and cellular levels to tissue and organism levels. By integrating multi-faceted approaches, systems pharmacology can provide mechanistic understanding of both the therapeutic and adverse effects of drugs. This includes understanding of how drugs act in different tissues and cell types, as well as the issues of multiple actions within a single cell type due to the presence of several interacting pathways. Such studies are important from a translational perspective because they help identify new drug targets, predict adverse events and can improve the safety and efficacy of existing drugs. These goals have become increasingly important in the current practice of medicine as many therapeutic challenges deal with complex diseases such as cancers, psychiatric disorders and the metabolic syndrome.
In previous decades, there has been considerable success in the development of targeted therapies for diseases with single targets, such as Fabry's disease. Additionally, there are many successful ways to treat some complex diseases such as hypertension and inflammatory diseases like arthritis. However, such treatments have been developed empirically and it is not entirely clear why certain drugs are effective in certain patients. Systems pharmacology, if successful, should provide for a general understanding of drug action in individual patients. This includes the balance between the therapeutic action of a medication and the unintended adverse effects.
A general understanding of drug action requires a systems level view rooted in the human genome. Implicit in such understanding of drug action is also the knowledge of how complex diseases originate in the context of the whole genome of an individual. This type of understanding will come from various sources of data such as physiological, biochemical and genomic parameters. Integrating these datasets requires an array of computational approaches. One particularly valuable approach is the use of network analysis of cellular systems. In this review, we describe current network-based approaches for a global understanding of drug action.
2 WHY USE NETWORK ANALYSIS IN SYSTEMS BIOMEDICINE?
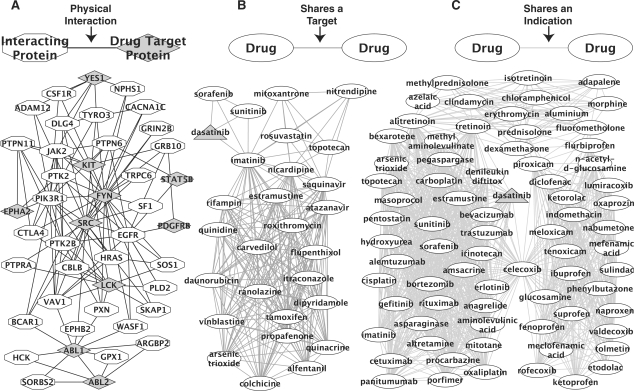
Network approaches in biology have proven to be useful for organizing high-dimensional biological datasets and extracting meaningful information. A network is a way of representing datasets emphasizing the relationships between nodes. A graph is constructed where these nodes, which represent genes, proteins, small molecules or any other entity capable of interacting in the system being modeled, are connected by edges, which represent the nature of the interaction. Nodes and edges can have various attributes and annotations. Depending on the nature of the study, interactions can be experimentally determined physical and chemical interactions, genetic regulatory interactions, higher order relationships such as co-expression or some other shared property linking the nodes. Edges represent these interactions between the nodes and, when the information is available, edges can have directions, weights and other attributes that provide information about the hierarchy of effects. Different relationships between nodes which can be used to study drug are depicted in Figure 1. These three examples feature dasatinib, a tyrosine kinase inhibitor used to treat chronic mylogenous leukemia. A protein interaction-based approach shown in Figure 1A shows the relationship between the targets of dasatinib and their interacting proteins. A network connecting drugs based on shared targets is shown in Figure 1B. This shows dasatinib connected to other tyrosine kinase inhibitors which are also connected to drugs which interact with efflux pumps such as the multi-drug resistant transporters that pump drugs out of cells. Figure 1C shows a different network which connects drugs based on shared indications. Dasatinib falls in a cluster of antineoplastic agents. This cluster is connected to anti-inflammatory agents through celecoxib which has both indications. Both these clusters have drugs which share indications with topical acne treatments. These examples demonstrate how different network views of drug relationships can highlight different aspects of the same drug.
Fig. 1.
Different relationships between nodes in networks used in systems pharmacology. In systems pharmacology, networks can be used to understand the relationship between drugs, their targets and diseases. Nodes can be genes, proteins, small molecules, drugs, diseases or any other biological entity capable of interacting in the system of interactions being modeled. Edges can be directed or undirected, weighted or not weighted and can represent direct physical interactions, activation, inhibition, coregulation or any other relationship between the nodes. Using networks centered on dasatinib, a tyrosine kinase inhibitor used to treat chronic mylogenous leukemia as an example, depicted above are small sub-networks demonstrating different types of nodes, represented by shaded shapes and edges, represented by lines. (A) Nodes are proteins connected by physical interactions found in literature. Diamond shaped nodes are annotated as targets of dasatinib and interacting proteins shown as octagons physically interact with at least two of the drug targets. (B) Oval nodes are drugs connected by shared targets within two steps of dasatinib, shown in a triangle. Dasatinib connects directly to other tyrosine kinase inhibitors. Imatinib also connects to clusters of drugs which interact with the ABCB1 and ABCG2 drug efflux pumps. (C) Oval nodes are drugs connected by sharing a therapeutic indication as described by the Anatomical Therapeutic Chemcial Classification System (ATC) third level codes. Within two steps of dasatinib shown in a triangle, the network forms three connected clusters. The cluster on the left of anti-neoplastic agents is connected to the cluster of anti-inflammatory agents on the right through celecoxib which has both indications. Both these clusters have connections to drugs above them in which are topical acne treatments. Drug targets and indication codes were taken from Drugbank (Wishart et al., 2008) and the protein interaction network was taken from Genes2Networks (Berger et al., 2007). These examples demonstrate different ways that one can study different aspects of the same drug using different types of networks.
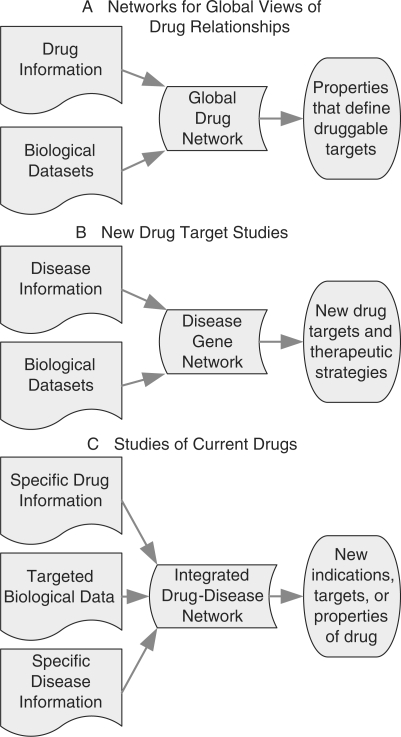
Network data structures are amenable to many sophisticated forms of computational analysis which can uncover important, non-obvious, properties of nodes and the relationships between them. Networks allow integration of diverse sources of experimental data and biological knowledge into a framework which provides new insight into the systems. These approaches can combine genome scale datasets with information about specific genes and proteins. In recent years, studies of metabolic networks, gene regulatory networks, protein–protein interaction networks and other biological networks have provided insight into the origins of overall cellular behaviors and evolutionary design principles, as well as more focused fields of study concerning specific cell biological processes or diseases. From these analyses, one can devise experimentally testable hypotheses ranging from prediction of novel functions of specific genes to genome scale properties of human cellular networks. In a similar manner, analysis of networks for pharmacologic studies has the promise of allowing for the identification of new drug targets for many diseases, better understanding of what makes a good drug target, improved ability to predict effective drug combinations and drug adverse events. These studies contribute to shifting the paradigm of drug action from a relatively simple cascade of signaling events downstream of a target to a coordinated response to multiple perturbations of the cellular network. As illustrated by Figure 2, network studies in systems pharmacology can be divided into three main categories based on the type and scale of data being studied and the type of information sought: networks for global views of drug relationships, new drug target studies and studies of current drugs. These categories are discussed below.
Fig. 2.
Types of network studies in systems pharmacology. Network studies in systems pharmacology can be grouped into three broad categories. (A) Global drug network studies that incorporate information about many types of drugs and biological datasets such as protein–protein interaction data can generate network properties of drug targets. These properties give information about historical drug development trends and can suggest properties of what makes a target druggable. (B) Disease specific network studies use information about a specific disease to identify potential new drug targets and therapeutic strategies. (C) Studies that integrate information about specific diseases and drugs can identify new indications for drugs, unknown targets of drugs, and other potentially interesting properties of the drugs.
3 NETWORKS FOR GLOBAL VIEWS OF DRUG RELATIONSHIPS: WHAT MAKES A GOOD DRUG TARGET?
Networks provide global views of the relationships between nodes and thus allow for network properties to be calculated based on a node's connections and relationship to the rest of the network. For example, each node has a degree, the number of nodes that it interacts with. Studies of many biological networks, often protein–protein interaction or gene regulatory networks, have focused on the degree distribution of the global network and found that there are often hubs, nodes which connect to relatively many other nodes (Barabasi and Albert 1999; Jeong et al., 2000). These are often important for the functioning of multiple biological processes (Jeong et al., 2001). Similarly, several different measures of node centrality have been studied to assign value to a node based on its relative importance in a network (Jeong et al., 2001; Jovelin and Phillips, 2009; Vinogradov, 2009; Wang et al., 2008). Other studies have looked at network motifs, small sub-graphs of three to five nodes which have patterns of connectivity overrepresented in the network (Milo et al., 2002). These have shed light onto the information processing capabilities of the network and suggested mechanisms of network evolution (Barabasi and Albert 1999; Ma'ayan et al., 2005; Muller et al., 2008; Wuchty et al., 2003). Networks connecting drug targets based on the chemical similarity of their ligands have been shown to have also important topological attributes distinct from the attributes of networks based on structural similarity of the proteins (Hert et al., 2008). Properties such as these are interesting as they represent non-obvious, but intrinsically important, attributes of a gene, protein or set of proteins, that arise from its interactions and position in the cellular network topology. Applying studies such as these to drugs and targets of drugs can allow for more rapid identification of some of these non-obvious network properties that define potentially good drug targets, proteins which can interact with a drug to provide a therapeutic response.
Several studies have looked at network properties of drug targets. Ma'ayan et al. constructed a bipartite network connecting drug targets and drugs and subsequently analyzed the targets in the context of a global protein–protein interaction network (Ma'ayan et al., 2007). This initial work showed that drug targets are not randomly distributed throughout the cellular interaction network. For example, drug-targeted proteins frequently have annotated localization to the cellular membrane. This is reasonable as this localization provides easier access to the target for the drug and demonstrates that importance in a biological process is not the only property that is required for a good drug target. Yildirim et al. constructed a similar bipartite network of drugs and targets, but used it to generate projections (Yildirim et al., 2007). In one projection, nodes are drugs and are connected if they share a common target. Analysis of this network as drugs are added in order of development demonstrated that most new drugs interact with previously targeted cellular components and there are relatively few drugs entering the market with novel targets. In the other projection, nodes are the drug targets, the cellular components which interact with a drug. These are connected by edges if they are affected by a common drug. Overlaying this with a protein–protein interaction network allowed several observations of network properties of drug targets. For example, drug targets tend to have a higher degree than other nodes in the protein–protein interaction network. This means that drug targets, on average, participate in more interactions than other cellular components despite not tending to be essential for viability as defined by mouse knockout studies. Nacher et al. also constructed a bipartite graph, but theirs connected drugs to their therapeutic indications (Nacher and Schwartz, 2008). The authors made projections of this graph to make a network of drugs connected if they are used for similar indications and a network of diseases connected if they are treated with the same drugs. From this, the authors found diseases clustered based on their treatments as well as treatments clustered based on the diseases. Various measures of network centrality were used to calculate the topological importance of the drugs in the network and found that certain drugs, with multiple targets, are used to treat distinct diseases in different parts of the network. Another type of interaction network involving drugs was studied by Yeh et al. (2006). In this study, a network of antibiotics was constructed based on experiments where edges connected antibiotics that either worked synergistically or antagonized each other in inhibiting the growth of Escherichea coli. This network allowed drugs to be grouped into classes that were either mutually synergistic or antagonistic and showed that drugs with similar mechanisms could be grouped into classes using this network approach.
From these analyses, one can begin to formulate a set of network criteria that define a good drug target and allow for selection of new drug targets from the network. For example, measures of network centrality can relate to the importance of a node and thus ability to disrupt a biological process of interest. However, since too much disruption of a cellular network can lead to undesirable effects, other properties of a node can also suggest whether or not a protein may serve as a good drug target. Hwang et al. propose an approach for identifying ‘bridging nodes’ as potential drug targets (Hwang et al., 2008b). These node are not core regulators of important pathways and thus not likely to have major global effects and drug side effects when targeted. However, they are situated in the network topology in such that their disruption would prevent information flow between modules of interest and thus specifically target the disease process effectively. Given that nodes in networks can thus be identified as potential drug targets, ideally networks for specific diseases must be constructed to find targets useful in treatment of these diseases.
4 NEW DRUG TARGETS: IDENTIFICATION AND TARGETING STRATEGIES
In addition to global cellular network studies, network analysis has proven useful for more focused studies investigating a particular disease or pathophysiology. It has been shown that proteins which interact with each other are frequently involved in a common biological process (Luo et al., 2007). Similarly, interacting proteins can be involved in a disease process (Goh et al., 2007; Ozgur et al., 2008). Approaches based on this idea have led to several candidate gene prediction tools which use the relative location in a network and node network properties to suggest proteins topologically related to a set of disease genes (Berger et al., 2007; Chen et al., 2009; Kohler et al., 2008). From a pharmacological point of view, any new candidate disease gene, which is suspected of playing a role in a pathophysiological process, can also be considered a candidate drug target for modifying that disease process. To this end, specific networks around several disease processes have been constructed. These include asthma, schizophrenia, cardiovascular disease, various cancers and various infectious diseases (Camargo et al., 2007; Chu and Chen, 2008; Durmus et al., 2009; Fatumo et al., 2009; Hwang et al., 2008a; Hyduke et al., 2007; Lim et al., 2006; Raman and Chandra, 2008; Tanaka et al., 2008). Whether these are constructed by careful literature review, generated from high-throughput experiments or computationally extracted from a larger network, these disease centered networks provide insights into the specific disease processes and therefore can suggest novel drug targets.
Often disease or pathogen-related networks are amenable to further analysis which identifies the specific nodes and have network properties of druggable targets. Singh et al. examined the metabolic network of E.histolytica, a pathogen causing amoebiasis (Singh et al., 2007). They performed choke point analysis, an approach which identifies enzymes that either uniquely produce or consume a given metabolite. Sufficient inhibition of these enzymes would either cause a lethal inability to produce an essential metabolite or toxic accumulation of another metabolite in the pathogenic organism. While without detailed kinetic information a true rate limiting step can not be identified, this analysis suggests that network topology alone is enough to propose reasonable drug targets. Sridhar et al. have suggested several other approaches to identifying drug targets in metabolic networks with a focus on minimizing potential side effects (Sridhar et al., 2007, 2008). They developed an algorithm for identifying a set of enzymes, which when inhibited blocks production of a desired set of target metabolites while minimizing effects on other metabolites. Similar approaches have been applied by Ruths et al. on biological signaling networks (Ruths et al., 2006). By formulating the constrained downstream problem and suggesting an algorithmic solution, they identify all the proteins affected by targeting a specified node in the network. This allows one to find nodes that inhibit a specified biological process while leaving others generally intact which extends to a drug having a desired effect while decreasing the risk of undesired side effects.
When a particular potential target is identified in a biological network, there are often multiple different approaches to targeting it therapeutically. For example, a drug binding a specific conformation of a protein may be more effective than a drug which binds a different conformation. While a purely interaction-based network does not contain the information to make this distinction, incorporation of rate constants and other kinetic information can allow more detailed analysis. Stites et al. performed dynamical model analysis of the Ras signaling network in cancer and wild-type cells (Stites et al., 2007). Their ordinary differential equation model was then used to compare the effect of hypothetical drugs which inhibits the GTP-bound Ras, GDP-bound Ras or both forms of Ras. The results of this demonstrated that only the hypothetical drug which preferentially binds GDP-bound Ras would have the desired effect of decreasing Ras effector activity in cancer cells more than in the healthy wild-type network.
Another promising area in network-based prediction of drug targets is the ability to predict combinations of targets, or protein complexes, which will prove to be most efficacious and safe when targeted together. Since networks emphasize the relationships between proteins, they can suggest pairs or groups of drug targets that can work well together to treat a disease. Ruths et al. formulate this as the minimum knockout problem where they identify the set of nodes that are required to be removed from a signaling network in order to disconnect a set of signaling molecules from their downstream effectors (Ruths et al., 2006). Similarly, Dasika et al. formulate a solution to the Min-Interference problem (Dasika et al., 2006). This approach also allows them to identify the minimal sets of nodes needed to be disrupted in order to block an undesired process, but it also aims to preserve desired ones. These approaches will allow combinations of drugs against these targets to be developed where neither drug alone would block a process, but together the drugs would work. The global understanding of diseases and drug effects based on network relationships between drug targets can also allow repurposing of existing drugs with known targets for use in combination therapies for different diseases. Such reuse of existing drugs has the advantage that their individual safety has already been established. So, combination therapy using previously approved drugs could lead to safer and more efficient treatments. Such an ability to better understand and predict synergistic effects of drug combinations is one of the more powerful goals of systems pharmacology.
5 CURRENT DRUGS: IMPROVED UNDERSTANDING OF THEIR USE AND EFFECTS
Biological network analyses can also contribute to understanding the effects of clinically used pharmaceuticals. Various approaches can identify previously unknown targets of the drug, pathways affected by the drug and pharmacogenomic factors affecting the usage of the drug. These can in turn be used to explain off-target effects, adverse events or suggest additional indications or contraindications for the usage of a drug.
While many drugs have known therapeutic targets, many drugs that are currently used work through unknown mechanisms. Furthermore, even drugs with a known target often have ‘off-target’ effects. These are effects, often undesirable, of a drug which can not be explained through its interaction with its primary targets. For example, many drugs can cause cardiac arrhythmias by blocking potassium channels in the heart even though this was not the primary target or indication for these drugs (Hoffmann and Warner, 2006). Network studies of drugs have allowed identification of some of these secondary targets of drugs. Campillos et al. constructed a network of drugs connected if they shared both a degree of structural similarity and similar side effect profiles (Campillos et al., 2008). By identifying pairs of drugs in this network that have distinct targets, the authors were able to assign the targets of one drug to the drugs it was connected to and subsequently validate the binding of the drug to its predicted secondary target. In another network study with similar goals performed by Iorio et al., a network of drugs was connected based on similarity of gene expression profiles after treatment of cells with drugs (Iorio et al., 2009). They were able to identify regions in this drug network which consisted of medications with similar modes of action. Another approach to identifying drug modes of action using expression datasets was described by Xing et al. (Xing and Gardner, 2006). They used a large diverse set of gene expression datasets to create a network model of the regulator influences between genes. Using microarray data from test treatments with a drug, their network model can distinguish the targets of the treatment from changes downstream of these targets.
Another way drug targets can be linked together into a network involves a chemoinformatic approach. Keiser et al. developed a method of scoring the similarity between the sets of ligands for different receptor. The authors use this score to construct a network of receptors connected together if they bind structurally similar ligands (Keiser et al., 2007). This analysis showed that many biologically related drug targets clustered together by ligand similarity even though the targets themselves have minimal sequence similarity. Using this approach, the authors made specific predictions such as suggesting that methadone interacts with Muscarinic-3 receptors and experimentally validated the predictions. This type of analysis based on the ability of different targets to bind the same ligand allowed for the identification of off-targets for certain drugs.
Drug action is not only related to the targets of the drug, but can also be affected by variations in metabolic enzymes, transporter proteins and downstream effects of drug action. The field of pharmacogenomics identifies genetic variations that can change drug effects. Network analyses can contribute to identification of such pharmacogenes, genes which modulate the response to a drug. Hansen et al. constructed a classifier which considered each gene in the context of a gene–gene–drug interaction network (Hansen et al., 2009). Physical protein–protein interactions were used to approximate functional modules. The physical and genetic interaction of genes with drugs, along with the structural and indication similarities between drugs, were used to rank genes based on their likelihood of affecting the pharmacology of a drug of interest. This allowed prediction of known and novel pharmacogenes.
In addition to identifying unknown targets of drugs and pharmacogenes, network-based approaches can suggest potential alternative uses of drugs. Macpherson et al. (2009) developed software to examine networks in context of various annotations. The authors tested this software by studying a network consisting of interactions between human proteins and HIV infections as well as drugs and their targets. Analysis of this network suggested possible mechanisms by which several different types of drugs, ranging from antineoplastic and immunosuppressive drugs to statins, might be useful in treating HIV-1 infections. Qu et al. (2009) developed a technique to integrate information about drugs, treatments and diseases into a Disease–Drug Correlation Ontology. By querying the complex network structure surrounding the disease Systemic Lupus Erythematosus, they predicted which drugs might modify the course of the disease. Their top prediction, the selective estrogen receptor modulator Tamoxifen, is an experimentally studied, promising candidate for the treatment of Lupus. Another study, by Bromberg et al. (2008) investigated the transcription factors activated downstream of cannabinoid receptor 1. A network was constructed between this receptor, which is a known target of certain analgesics, antiemetics, as well as drugs of abuse and downstream activated transcription factors involved in neuronal differentiation. This network allowed key regulators of the pathway to be elucidated and suggested additional targets which might synergize with effects of cannabinoid agonists to have potential therapeutic effects such as a role in neuroregeneration.
Systems pharmacology studies are changing the way we think about drug actions as shown in Figure 3. Whereas originally medications were thought to hit a specific target and modulate effects through pathways downstream of that target, we now know that many drugs hit multiple targets, each of which exist within a complex network. The effects of the drug, both therapeutic actions and adverse events, are therefore a result of perturbation of the complex network landscape. This was suggested in a study by Xie et al. (2009) where structural homology between protein drug binding sites was used to predict potential off targets of Cholesteryl Ester Transfer Protein Inhibitors. Incorporating these predicted off-targets into a network of metabolic, signaling and gene regulatory pathways explained the different side-effect profiles of drugs in this class. Variations from person to person in the cellular networks due to environmental factors and genetic variation will likely affect the therapeutic response to medications, required dosing and susceptibility to adverse events and side effects. Furthermore, drugs can have both acute effects happening immediately as the drug interacts with its targets, and later effects as downstream signaling adjusts transcriptional regulation and leads to network rewiring resulting in delayed physiological action.
Fig. 3.
Changing views of drug actions. Network studies in systems pharmacology are changing the way we think about the actions of drugs. (A) The classic view of drug action has a drug that interacts with a therapeutic target which transduces its signal through its effector pathway to mediate a therapeutic effect and certain side effects. Additional off-targets can mediate effects through distinct effector pathways to lead to other side effects and other adverse events. (B) In a systems pharmacology view of drug action, a drug interacts with multiple primary and secondary targets. These targets exist within a complex network which can mediate the response to the drugs leading to both the therapeutic and adverse effects.
6 PERSPECTIVE
Network approaches allow biomedical researchers to rapidly organize current knowledge by integrating different types of large datasets. Applying such approaches to problems in pharmacology can allow systems level descriptions of drug action, rapid identification of novel therapeutic strategies and potentially safer and more effective drugs development and prescription. Network analysis provides scientists with a unified approach to understanding complex diseases which can not be understood in terms of a single target or single pathway model. Furthermore, it allows for the prediction and explanation of unexpected effects of medications and suggests factors influencing drug efficacy and safety. Currently, network approaches in systems pharmacology are in their infancy. As more data is collected and generated, more detailed networks will be generated. These will incorporate not only additional drug target relationships, gene expression relationships and physical interaction data, but also additional data about target structure, signaling kinetics and protein localization. Each type of information incorporated into these networks will allow new predictions and understanding of different aspect of pharmaceuticals.
The future of systems pharmacology will require more than additional analysis of existing and updated datasets. New experimental approaches to study drug action broadly at the biochemical as well as physiological levels are needed. Current datasets used to build networks need to be greatly improved with the addition of data specifying tissue expression levels of proteins, localization information, as well as directionality and strength of interactions. Quantitative experimental techniques, such a surface plasmon resonance-based technologies, will facilitate these studies. Such quantitative measurements are currently done with one or a few targets at a time. A new wave of technology development is needed to move quantitative drug–target analysis into high-throughput formats. Moreover, in order to become a truly translational field, the results of network studies need to find a path to clinical relevance. This could be in the form of utilizing systems analysis of drug effects to enhance post market drug surveillance data to better understand and predict drug adverse events. In the end, the true impact of network analyses will be measured by how information gleaned through systems pharmacology analyses will lead to improved drug discovery, safer and more effective drug prescriptions, and ultimately lead to safe, cost effective and effectual patient care.
ACKNOWLEDGEMENTS
We thank Avi Ma'ayan for relevant discussions and Simon Hardy, Aislyn Wist, and the anonymous reviewers for comments.
Funding: National Institutes of Health (grants DK038761); Systems Biology Center New York (grant P50GM07558). Pharmacological Sciences Training (grant GM062754 to S.B.).
Conflict of Interest: none declared.
REFERENCES
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Berger SI, et al. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg KD, et al. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- Campillos M, et al. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LH, Chen BS. Construction of a cancer-perturbed protein-protein interaction network for discovery of apoptosis drug targets. BMC Syst. Biol. 2008;2:56. doi: 10.1186/1752-0509-2-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasika MS, et al. A computational framework for the topological analysis and targeted disruption of signal transduction networks. Biophys J. 2006;91:382–398. doi: 10.1529/biophysj.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmus Tekir S, et al. Drug targets for tumorigenesis: insights from structural analysis of EGFR signaling network. J. Biomed. Inform. 2009;42:228–236. doi: 10.1016/j.jbi.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Fatumo S, et al. Estimating novel potential drug targets of Plasmodium falciparum by analysing the metabolic network of knock-out strains in silico. Infect. Genet. Evol. 2009;9:351–358. doi: 10.1016/j.meegid.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Goh KI, et al. The human disease network. Proc. Natl Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen N, et al. Generating genome-scale candidate gene lists for pharmacogenomics. Clin. Pharmacol. Ther. 2009;86:183–189. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hert J, et al. Quantifying the relationships among drug classes. J. Chem. Inf. Model. 2008;48:755–765. doi: 10.1021/ci8000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann P, Warner B. Are hERG channel inhibition and QT interval prolongation all there is in drug-induced torsadogenesis? A review of emerging trends. J. Pharmacol. Toxicol. Methods. 2006;53:87–105. doi: 10.1016/j.vascn.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hwang S, et al. A protein interaction network associated with asthma. J. Theor. Biol. 2008a;252:722–731. doi: 10.1016/j.jtbi.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Hwang WC, et al. Identification of information flow-modulating drug targets: a novel bridging paradigm for drug discovery. Clin. Pharmacol. Ther. 2008b;84:563–572. doi: 10.1038/clpt.2008.129. [DOI] [PubMed] [Google Scholar]
- Hyduke DR, et al. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl Acad. Sci. USA. 2007;104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F, et al. Identifying network of drug mode of action by gene expression profiling. J. Comput. Biol. 2009;16:241–251. doi: 10.1089/cmb.2008.10TT. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jovelin R, Phillips PC. Evolutionary rates and centrality in the yeast gene regulatory network. Genome Biol. 2009;10:R35. doi: 10.1186/gb-2009-10-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- Kohler S, et al. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Luo F, et al. Modular organization of protein interaction networks. Bioinformatics. 2007;23:207–214. doi: 10.1093/bioinformatics/btl562. [DOI] [PubMed] [Google Scholar]
- Ma'ayan A, et al. Formation of regulatory patterns during signal propagation in a Mammalian cellular network. Science. 2005;309:1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A, et al. Network analysis of FDA approved drugs and their targets. Mt. Sinai. J. Med. 2007;74:27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson JI, et al. JNets: Exploring networks by integrating annotation. BMC Bioinformatics. 2009;10:95. doi: 10.1186/1471-2105-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Muller M, et al. Network topology determines dynamics of the mammalian MAPK1,2 signaling network: bifan motif regulation of C-Raf and B-Raf isoforms by FGFR and MC1R. FASEB J. 2008;22:1393–1403. doi: 10.1096/fj.07-9100com. [DOI] [PubMed] [Google Scholar]
- Nacher JC, Schwartz JM. A global view of drug-therapy interactions. BMC Pharmacol. 2008;8:5. doi: 10.1186/1471-2210-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur A, et al. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics. 2008;24:i277–i285. doi: 10.1093/bioinformatics/btn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XA, et al. Inferring novel disease indications for known drugs by semantically linking drug action and disease mechanism relationships. BMC Bioinformatics. 2009;10(Suppl. 5):S4. doi: 10.1186/1471-2105-10-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K, Chandra N. Mycobacterium tuberculosis interactome analysis unravels potential pathways to drug resistance. BMC Microbiol. 2008;8:234. doi: 10.1186/1471-2180-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruths DA, et al. Hypothesis generation in signaling networks. J. Comput. Biol. 2006;13:1546–1557. doi: 10.1089/cmb.2006.13.1546. [DOI] [PubMed] [Google Scholar]
- Singh S, et al. Choke point analysis of metabolic pathways in E.histolytica: A computational approach for drug target identification. Bioinformation. 2007;2:68–72. doi: 10.6026/97320630002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar P, et al. An iterative algorithm for metabolic network-based drug target identification. Pac. Symp. Biocomput. 2007;12:88–99. [PubMed] [Google Scholar]
- Sridhar P, et al. Mining metabolic networks for optimal drug targets. Pac. Symp. Biocomput. 2008;13:291–302. [PubMed] [Google Scholar]
- Stites EC, et al. Network analysis of oncogenic Ras activation in cancer. Science. 2007;318:463–467. doi: 10.1126/science.1144642. [DOI] [PubMed] [Google Scholar]
- Tanaka T, et al. Pharmacogenomics of cardiovascular pharmacology: pharmacogenomic network of cardiovascular disease models. J. Pharmacol. Sci. 2008;107:8–14. doi: 10.1254/jphs.08r03fm. [DOI] [PubMed] [Google Scholar]
- Towfigh S, et al. Outcomes from peptic ulcer surgery have not benefited from advances in medical therapy. Am. Surg. 2002;68:385–389. [PubMed] [Google Scholar]
- Vinogradov AE. Global versus local centrality in evolution of yeast protein network. J. Mol. Evol. 2009;68:192–196. doi: 10.1007/s00239-008-9185-2. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Betweenness centrality in a weighted network. Phys Rev. E Stat. Nonlin. Soft Matter Phys. 2008;77(Pt 2):046105. doi: 10.1103/PhysRevE.77.046105. [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S, et al. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nat. Genet. 2003;35:176–179. doi: 10.1038/ng1242. [DOI] [PubMed] [Google Scholar]
- Xie L, et al. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of cetp inhibitors. PLoS Comput. Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Gardner TS. The mode-of-action by network identification (MNI) algorithm: a network biology approach for molecular target identification. Nat. Protoc. 2006;1:2551–2554. doi: 10.1038/nprot.2006.300. [DOI] [PubMed] [Google Scholar]
- Yeh P, et al. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- Yildirim MA, et al. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]