Abstract
A brief history of human genetics and genomics is provided, comparing recent progress in those fields with that in pharmacogenetics and pharmacogenomics, which are subsets of genetics and genomics, respectively. Sequencing of the entire human genome, the mapping of common haplotypes of single-nucleotide polymorphisms (SNPs), and cost-effective genotyping technologies leading to genome-wide association (GWA) studies—have combined convincingly in the past several years to demonstrate the requirements needed to separate true associations from the plethora of false positives. While research in human genetics has moved from monogenic to oligogenic to complex diseases, its pharmacogenetics branch has followed, usually a few years behind. The continuous discoveries, even today, of new surprises about our genome cause us to question reviews declaring that “personalized medicine is almost here” or that “individualized drug therapy will soon be a reality.” As summarized herein, numerous reasons exist to show that an “unequivocal genotype” or even an “unequivocal phenotype” is virtually impossible to achieve in current limited-size studies of human populations. This problem (of insufficiently stringent criteria) leads to a decrease in statistical power and, consequently, equivocal interpretation of most genotype-phenotype association studies. It remains unclear whether personalized medicine or individualized drug therapy will ever be achievable by means of DNA testing alone.
Keywords: Binary traits, Complex disease, CYP2C19 gene, CYP2D6 gene, Cystic fibrosis, dbGAP, ENCODE, Extreme discordant phenotype design and analysis, Gaucher disease, GAIN, Genetic Association Information Network, Oligogenic diseases
Introduction
As a subset of genetics, pharmacogenetic studies have generally followed a few years behind the more fundamental studies of human genetics. Perhaps one exception to this is what some claim to be the “first pharmacogenetic observation.” In the village of Croton (southern Italy) about 510 B.C., Pythagoras was credited as the first to point out “dangers in eating the fava bean.” Favism is now known to be caused by glucose-6-phosphate dehydrogenase (G6PD) deficiency, an X-linked trait as prevalent as one in three males in southern Italy and Sardinia (Nebert et al., 1999). Whereas the complete absence of G6PD is incompatible with life, a deficiency of 80–95% of the G6PD enzyme activity can lead to fava bean-induced hemolytic anemia. The hemolytic crisis usually presents as severe abdominal pain and cramping, dangers against which Pythagoras had warned.
As we proceed through the history of genetics and genomics (especially the past decade), it will become obvious that the genome is exceedingly complex—far more so than had previously been appreciated. It therefore comes as no surprise that our final section concludes that “individualized drug therapy” is impossible now, or in the foreseeable future. Many claim that it is, and that simply identifying a single nucleotide change and modifying therapy accordingly, represents “individualized drug therapy.” Unfortunately, the genome is much more complex than this.
From Beginning of Human Genetics to Start of Molecular Genetics
Resurrecting the concepts of Gregory Mendel who had used garden peas in the 1860s, Sir Archibald Garrod in the early 1900s described several clinical “inborn errors of metabolism.” Albinism, alkaptonuria, cystinuria and pentosuria were explained as four diseases—inherited as autosomal recessive traits—a pattern of inheritance similar to that of the white color in Mendel's garden pea flower. Garrod is credited with pioneering the field of inborn errors of metabolism and, subsequently, ushered in the era of human genetics. The underlying tenet, from the 1900s into the 1980s, was “one gene, one disease,” which then evolved into “one wild-type healthy allele, and one disease allele.” Two parents who are heterozygous for the disease allele have a one-in-four chance of producing a child having both disease alleles and, hence, inheriting the unwanted disorder.
During the next several decades, many additional examples of traits inherited in an autosomal recessive manner were described (e.g., maple syrup urine disease, phenylketonuria, cystic fibrosis, Gaucher disease, congenital adrenal hyperplasia) in which the disorder requires two mutant alleles for expression and, hence, typically skips a generation. Autosomal dominant traits were also detailed (e.g., Huntington disease, achondroplastic dwarfism, Marfan syndrome, neurofibromatosis, hereditary spherocytosis) in which the disorder typically appears in each generation. Traits transmitted as X-linked recessives were reported (e.g., red-green colorblindness, hemophilia) in which the female carrier has a one-in-two chance of passing the defective allele to her male offspring. X-linked dominant traits were described (e.g., incontinentia pigmenti, Coffin-Lowry syndrome) in which the affected female carrier has a 50% chance of passing the defective allele to her offspring or the affected male has a 100% chance of passing the defective allele to his daughters.
In the Red × White cross of snapdragons, Lorenz in the 1880s noted that the F1 progeny produced flower petals that were pink, and the F2 generation displayed a 1 red:2 pink:1 white distribution (i.e. contrary to the recessive or dominant inheritance above, the phenotype directly reflects the genotype); Lorenz described this trait as a form of additive (codominant; gene-dose) inheritance of these two colors. Indeed, human enzyme subunits (e.g., lactate dehydrogenase) could be separated on starch electrophoresis gels in the 1960s (Vesell, 1965), showing this 1:2:1 ratio of subunits; this was a demonstration of additive inheritance. Human genetic examples of codominant inheritance have been described from the 1940s to 1980s (e.g., ABO blood groups, breast cancer, familial hypercholesterolemia, psoriasis); however, later it became clear that many of these disorders actually reflect complex diseases, i.e., dozens or hundreds of genes (polygenic) combined with additional modifying effects such as penetrance, incidence of the disease allele in a population, and the contribution of environmental factors.
Pharmacogenetics During this Same Period
Larry Snyder (1932) is often recognized as ushering in the era of pharmacogenetics (Table 1). He showed that a difference in response to a chemical (inability to taste phenylthiourea) was inherited as an autosomal recessive trait—similar to what Garrod had reported with albinism and the other monogenic disorders three decades earlier. In 1957, Arno Motulsky proposed that “inheritance might explain why many individuals differ in drug efficacy and in experiencing adverse drug reactions” (Motulsky, 1957). Shortly thereafter, Friedrich Vogel first coined the term “pharmacogenetics” and defined it as the “study of the role of genetics in drug response” (Vogel, 1959). Table 1 lists some notable advances in the field of pharmacogenetics; in general, these watershed studies focused on a “simple” gene-drug interaction, inherited as a high-penetrance predominantly monogenic (hPpM) trait; this terminology is discussed next. Studies in the 1960s compared plasma drug half-lives in monozygotic and dizygotic twin pairs (Vesell and Page, 1968); these findings also demonstrated the importance of heritability in plasma drug half-lives, but rather than revealing the effects of a single gene, this trait of plasma half-life probably represents a larger, unknown number of genes.
Table 1.
Notable advances between 1932 and 1995 in shaping the field of pharmacogenetics.a
| Observation | Key Reference |
|---|---|
| Phenylthiourea (PTU) nontaster traitb | (Snyder, 1932) |
| Glucose-6-phosphate dehydrogenase (G6PD) deficiency | (Alving et al., 1956) |
| N-acetylation (NAT) polymorphism | (Knight et al., 1959; Evans et al., 1960) |
| Federation Proceedings of the Am. Soc. Exp. Biologists (FASEB): two symposia gave high visibility to the field. | (Kalow, 1965; O'Reilly and Aggeler, 1965; Armaly and Becker, 1965; Wigle, 1965; La Du, 1965; Eichelbaum, 1984; Spielberg, 1984; Vesell, 1984; Weber, 1984; La Du and Eckerson, 1984; Vesell and Penno, 1984; Weinshilboum, 1984a; Kalow, 1984a; Weinshilboum, 1984b; Kalow, 1984b) |
| Genetic variation in ethanol metabolism; mutation in aldehyde dehydrogenase (ALDH) gene | (Von Wartburg et al., 1964) |
| The aryl hydrocarbon receptor and [Ahr] gene battery | (Nebert et al., 1975) |
| Debrisoquine/sparteine oxidation polymorphism (CYP2D6 gene) | (Mahgoub et al., 1977; Eichelbaum et al., 1979) |
| Thiopurine methyltransferase (TPMT) gene polymorphism | (Weinshilboum and Sladek, 1980) |
| Identification of the human GSTM1 null allele; GSTT1 null allele later | (Seidegård et al., 1986) |
| Molecular cloning of the CYP2D6 gene and variant alleles | (Gonzalez et al., 1988) |
| Molecular cloning of the NAT2 gene and variant alleles | (Blum et al., 1990) |
| Molecular cloning of the CYP2C19 gene and variant alleles | (Goldstein and de Morais, 1994) |
| Molecular cloning of the TPMT gene and variant alleles | (Krynetski et al., 1995) |
Detailed in (Nebert, 1997); please see this Opinion Article to find additional discussion and other relevant studies and references. Just as monogenic disorders in human genetics are proposed to involve genes encoding “moonlighting” enzymes having more than one function (Sriram et al., 2005), so have all xenobiotic-metabolizing enzymes (XMEs) been proposed to carry out endogenous functions—in addition to functions of metabolizing a drug or environmental toxicant (Nebert, 1991;Nebert and Dalton, 2006).
The taste receptor gene TAS2R1 was identified 70 years later (Kim et al., 2003) as responsible for the PTU nontaster trait. Contrary to the XME and XRT genes, such a receptor polymorphism more likely is inherited as a monogenic human disease than as an hPpM pharmacogenetic disorder.
Just as human genetics had been promoting the idea of “one healthy allele, one disease allele,” the field of pharmacogenetics endorsed the concept of “one normal (metabolism or receptor) allele, one abnormal allele.” This notion was finally dispelled, once and for all, during the mid to late 1980s.
Effect of Molecular Biology on Human Genetics
In the 1970s, Victor McKusick stored all of his human genetics data on 3″ × 5″ index cards in one small file box. Because advances in molecular biology caused an explosion of knowledge during the 1970s and early 1980s, combined with the beginning of the internet, McKusick decided that the amount of expanding human genetics data required an online web site; thus began the Online Mendelian Inheritance in Man (OMIM)—summarizing all recent information, plus making it readily available to the clinical geneticist (McKusick, 2005).
Then came the realization that “monogenic” diseases do not reflect only one “disease allele.” Phenylketonuria (PKU) was known to be an autosomal recessive disorder caused by phenylalanine hydroxylase deficiency. The PAH gene was first cloned (Woo et al., 1983), and then a GT > AT transition defect at the 5′ splice donor site of intron 12 was demonstrated in a PKU carrier individual (Marvit et al., 1987). At first, this discovery was declared widely to be the disease allele for PKU. Within a few months, however, a second mutation (this one changing an amino acid) was reported, and by the time of a 1990 review (Cotton, 1990), this unexpected allelic heterogeneity (at that time, a total of 18 mutations) in the PAH gene was addressed.
Today, on the PKU database web site (PAHdb Knowledgebase), there are worldwide reports of more than 500 mutations in and near the PAH gene responsible for the disease; degrees of severity and time-of-onset are controlled by mutations in an unknown number of modifier genes, and by genes encoding other enzymes in the phenylalanine pathway that influence the severity of the disease (Scriver, 2007). Additional genes are responsible for affecting the age-at-onset for Huntington disease (HD) (Metzger et al., 2006). Hence, PKU and HD are not strictly monogenic diseases, but are affected to some degree by other genes, as discussed further below.
The same story can be told for cystic fibrosis and the CFTR (transmembrane conductance regulator) gene, which now has more than 1,000 variant alleles (almost all are point mutations or deletions of anywhere from one to 84 bp). The pattern is also true for Gaucher disease and the GBA (glucosylceramide β-glucosidase gene) gene, which now has more than 200 mutations in and near the gene; even subsets (with distinct phenotypes) of the syndrome exist (Grace et al., 2007). The same applies for many other monogenic human disorders: all monogenic disorders appear to have innumerable variant alleles (allelic heterogeneity). These disorders are also influenced, at least to some small degree, by additional genes.
Contributions of Molecular Biology to Pharmacogenetics
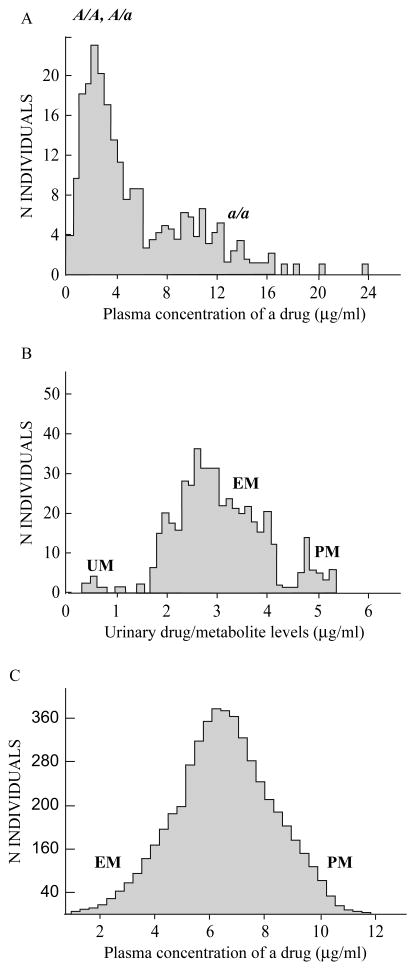
Several early clinical pharmacology studies had demonstrated bimodal distributions of subjects' plasma drug levels, following administration of a standard drug dosage (Fig. 1A); this distribution, combined with the fact that two “poor-metabolizer” (PM) parents produced only PM children, led to the conclusion that the PM trait is autosomal recessively inherited. Bimodal distributions in patterns of urinary drug metabolite elimination (Fig. 1B) were reported; in this case the “extensive-metabolizer” (EM) phenotype can be said to be inherited as an autosomal dominant trait. In other clinical pharmacology studies, unimodal polymorphisms were described (Fig. 1C) —indicating either additive inheritance but much more likely polygenic inheritance, also termed multiplex phenotype (i.e., contribution of numerous genes plus environmental factors).
Figure 1.
General examples of phenotypes in pharmacogenetic studies. A, slow metabolism inherited as a predominantly autosomal recessive trait (a allele). B, efficient metabolism inherited as a predominantly autosomal dominant trait. C, gradient of slow-to-rapid metabolism involving multiple genes and showing a unimodal distribution possibly inherited as a codominant, gene-dose or additive trait; this could also reflect a complex disorder. The amount of “scatter”, typical in studies such as those depicted in parts A, B and C, most likely represents contributions from modifier genes, gene-gene interactions, and/or environmental factors. Another interesting example (not shown) is the CYP2D6 polymorphism in which the poor-metabolizer (PM), intermediate-metabolizer (IM), efficient-metabolizer (EM), and ultra-rapid-metabolizer (UM) phenotypes can sometimes be distinguished as four distinct peaks (Ingelman-Sundberg, 2005).
Molecular cloning and identification of defective alleles were then reported for the CYP2D6, NAT2, CYP2C19 and TPMT genes between 1988 and 1995 (Table 1). Dozens of other purportedly “monogenic” pharmacogenetic disorders followed in the scientific literature.
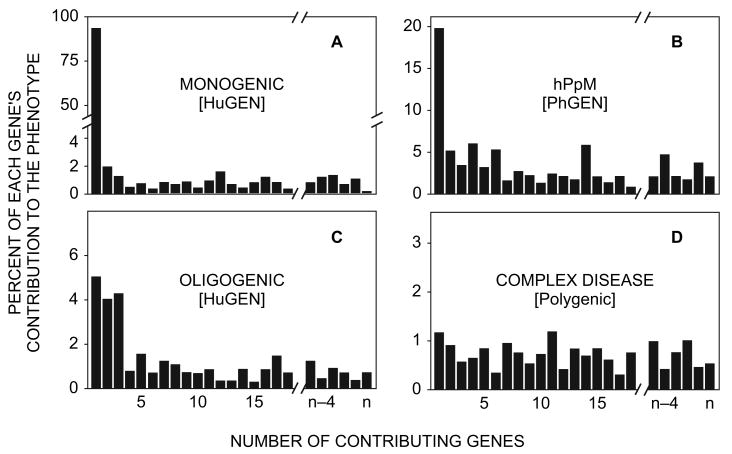
From previous reviews (Nebert et al., 2003; Nebert and Vesell, 2004; Nebert, 2005), it seems that those few hPpM genes that do make a substantial difference in metabolism of various drug substrates (e.g., CYP2D6, NAT2, TPMT, CYP2C19) might contribute perhaps 15% to 20% to the (EM or PM) phenotype (Fig. 2B), compared with a contribution of 90% or more to the single gene responsible for a monogenic disease (Fig. 2A). For example, one large retrospective study that correlated thiopurine-related adverse drug reactions with the TPMT genotype by DNA-testing (van Aken et al., 2003) noted that 78% of adverse drug reactions were not associated with the TPMT gene polymorphism, but rather attributable to factors other than mutations in this gene.
Figure 2.
Theoretical scheme showing the number of genes and extent of their contribution to the trait. A monogenic human disease (A) and an hPpM pharmacogenetic disorder (B) are compared with an oligogenic (C) and a complex disease (D). The gene responsible for a monogenic human disease contributes much more to expression of that disease (A) than does an hPpM gene responsible for expression of a pharmacogenetic disorder (B). If we use the same measure for genetic “contribution”, the allelic architecture (number of risk loci, inheritance mode, allele frequency, etc.) of complex diseases should be determined by both evolution and selective pressures. Thus, the dissimilar patterns in A versus B (and in fact in all four panels) represent different roles during human evolution and environmental adaptations (see text). HuGEN, human genetics; PhGEN, pharmacogenetics.
One might conclude that perhaps the TPMT polymorphism itself contributes about 22% to adverse drug reactions. It should be noted that TPMT and paraoxonase-1 are two enzymes that can be phenotyped by red-blood-cell assays, both of which have been demonstrated to be orders of magnitude more predictive than their respective DNA (genotype) assays (Nebert et al., 2003). The major gene's percent contribution to a “monogenic” pharmacogenetic trait (such as the CYP2D6, TPMT, NAT2 or CYP2C19 polymorphisms) is therefore considerably less (Fig. 2B) than that for a monogenic human genetic trait (Fig. 2A).
When as shown in Figs. 1A and 1B, the plasma drug concentration (or urinary metabolite level) is predominantly determined by a genetic variant, the “genetic contribution” of that gene is 15–20% to the overall variance of the quantitative trait (i.e., level of drug or metabolite). If the total population is divided into “PM” and “EM” phenotypes by using specific threshold criteria, however, the “contribution” of that gene to the binary trait is 80% or even higher. Since the biology of drug or metabolite levels seems to be the same as a human monogenic trait, a geneticist might call this phenomenon “artificial”: in other words, perhaps the disparity is generated by using a different measure of “genetic contribution,” especially if the measure based on dichotomous “PM” versus “EM” traits is relevant to where the threshold lies.
What is the fundamental difference between a monogenic human disease (Fig. 2A) and an hPpM pharmacogenetic disorder (Fig. 2B)? Most likely, the difference lies primarily in the redundancy function of the gene products: drugs are notoriously metabolized by a myriad of different enzymes, each enzyme encoded by a different gene. Thus, if PKU were equally caused by defects in any of six enzymes and transporters, instead of just phenylalanine hydroxylase encoded by the one PAH gene, then mutations in only the PAH gene might contribute 15–20% (rather than >90%) to the overall trait of phenylalanine hydroxylase deficiency. We may still treat this hypothetical PKU as “monogenic,” but with “locus heterogeneity”; because we could divide this hypothetical PKU into six subtypes, each of them is caused by a distinct genetic defect.
A disease is therefore classified as monogenic if the cause of the disorder is necessarily and sufficiently determined by mutation(s) in a single gene (allelic heterogeneity) —regardless of whether age-at-onset or severity is influenced by multiple genetic or environmental factors. In pharmacogenetic studies, drug dosage and overlapping substrate specificity are two of many factors (Nebert et al., 2003; Nebert and Vesell, 2004; Nebert, 2005) contributing to a drug response measured by quantitative (continuous) variables. Hence, it is preferable to call pharmacogenetic traits “hPpM”, even in instances (Figs. 1A and 1B) where a bimodal distribution suggests that the trait is controlled predominantly by a single gene.
Alternatively, the difference between a single gene's contribution to a monogenic human disease (>90% in Fig. 2A) and a pharmacogenetic trait (15–20% in Fig. 2B) may simply be due to different choices in how “contribution to the trait” is measured (Risch, 2000). The difference is at least partially due to the definition of a dichotomous disease versus a quantitative pharmacogenetic trait. The physician is more interested in differentiating between “affected” and “unaffected” individuals—this is called diagnosis (recognition of a disease or condition by its outward signs and symptoms). However, a pharmacologist needs to evaluate the drug response or the reverse effect on a quantitative scale—which is, by its very nature, influenced by multiple genetic and environmental factors.
Effects of the Human Genome Project
Starting in October 1990 and continuing to advance even today, the Human Genome Project (HGP) has provided huge amounts of new (and often totally unexpected) knowledge, which has been highly beneficial to human genetics and genomics—as well as to pharmacogenetics and pharmacogenomics. The Environmental Genome Project (EGP), initiated in October 1997, was a natural spin-off; clearly, information from the EGP overlaps and complements that in the HGP.
In the late 1990s, what had been termed “nucleotide substitutions” by (yeast, worm and fly) geneticists for several decades became known as single-nucleotide polymorphisms (SNPs). “SNP fever” then began. Hundreds of publications sought a statistically significant (P <0.05) association between one or a few SNPs and a multiplex phenotype—complex diseases such as hypertension, Alzheimer disease, asthma or schizophrenia. Not far behind, clinical pharmacologists also sought statistically significant (P <0.05) associations between one or a few SNPs and a multiplex phenotype—response of schizophrenia to clozapine; and effects of a new drug on asthma, a new drug on diabetes or heart disease, or a new drug on treating a type of cancer. Such publications were often followed by several reports refuting the original conclusion. This dilemma has been discussed in detail (Nebert and Vesell, 2004).
Various investigators suggested reasons for discrepancies between genotype-phenotype association studies; these included: (a) publication bias (small studies are prone to large variation in risk estimates, and only selected strong positive results are reported); (b) weak genetic effects and lack of power, i.e., small sample size is a frequent problem and can result in insufficient power to detect minor effect genetic variants; (c) population stratification; (d) heterogeneity in classification of phenotypes across studies (i.e., lack of an unequivocal phenotype); (e) different linkage disequilibrium (LD) patterns between populations or different SNP markers between studies; (f) gene-gene or gene-environment interactions, modifier genes, or other major genes epistatic to the allele under study that differ between populations; (g) regional population ethnic differences; and (h) another functional variant chromosomally linked to the allele under study (Ioannidis et al., 2001; Hirschhorn et al., 2002; Ioannidis, 2003; Ioannidis et al., 2003). In fact, in 2002, it was noted that, out of 166 putative genotype-phenotype association studies that had been visited three or more times, only six have been consistently replicated (Hirschhorn et al., 2002). Further requirements were suggested to make such studies more reproducible; these included: need for more accurate diagnoses of disease and reproducibility in other ethnic populations (i.e., unequivocal phenotype), need for haplotype information or a much greater SNP density per DNA segment under study, and larger numbers of subjects. Very recently, standards for genetic association studies were proposed—including the initial study, replicability, and publication (Chanock et al., 2007).
Many shortcomings of such studies were described earlier (Nebert et al., 2003; Nebert and Vesell, 2004). The term unequivocal phenotype was introduced, defined as “the assignment of a trait by scientific investigators without any room for error.” Factors such as overlapping drug-substrate specificity, renal clearance, error in clinical diagnosis of a disease, phenocopy, age and gender, diet, cigarette smoking, disease state, and noncompliance of patients taking a drug—all contribute to the lack of an unequivocal phenotype. More than 18 factors that can cause an equivocal phenotype have previously been listed (Nebert and Vesell, 2004).
The term unequivocal genotype was also introduced, defined as “the assignment of a genotype by scientific investigators without any room for error.” Factors such as allelic heterogeneity, locus (non-allelic) heterogeneity, large number of genes contributing to a trait, genocopy, synonymous mutations that unpredictably alter the trait, and ethnic differences in allelic frequencies of any gene—all contribute to the lack of an unequivocal genotype. More than three dozen factors that can cause an equivocal genotype have previously been listed (Nebert and Vesell, 2004; Nebert, 2005). Clearly, if one compared 200 cases with 200 controls in a genotype-phenotype association study that included five or ten cases and five or ten controls having equivocal phenotypes and/or equivocal genotypes, the amount of “noise” would easily conceal any real risk factors with moderate effect, and the statistical power would be seriously limited. If the genetic risk factor has a sufficiently “large-effect” impact, one would still be fortunate to detect it; however, this is not always the case.
New discoveries about our amazing human genome—since the previous reviews (Nebert et al., 2003; Nebert and Vesell, 2004; Nebert, 2005) —are listed in Table 2; these phenomena further contribute to the problem of our being unable to assign unequivocal genotypes. Copy-number variations (CNVs) encompassing neighboring genes (Table 2) came as a surprise. “Systems biology” i.e., gene-gene interactions (Gandhi et al., 2006) can easily complicate interpretation of a SNP in one of the genes within a complex pathway.
Table 2.
Unexpected phenomena found in the human genome, contributing to additional reasons for the difficulty in not being able to achieve an unequivocal genotype.a
| Phenomenon | Reference(s) |
|---|---|
| Synonymous SNPs inducing exon-skipping, changes in splicing | (Chao et al., 2001; Cartegni et al., 2002) |
| Synonymous SNPs affecting tRNA pool size | (Polesskaya and Sokolov, 2002) |
| Modulation of gene expression by loci located cis of that gene | (Mekus et al., 2003) |
| Synonymous SNPs affecting membrane attachment and degradation pathway of the protein | (Goldman et al., 2004) |
| Population stratification | (Campbell et al., 2005) |
| Genotype of one gene affects basal and inducible expression of a second gene (in trans) | (Lamba et al., 2006) |
| Copy-number variations (CNVs) | (Freeman et al., 2006; Locke et al., 2006; McEwen et al., 2006; Redon et al., 2006; Aitman et al., 2006; Long and Miano, 2007; Kikuta et al., 2007; Wong et al., 2007; Cooper et al., 2007; Stranger et al., 2007a) |
| Mutation affects bidirectional long-range intergenic transcription in cis | (Takeda et al., 2006; Khaitovich et al., 2006) |
| Enhancer on one chromosome controls gene on another chromosome | (Lomvardas et al., 2006) |
| Locus control regions: long-range gene activation | (Ho et al., 2006; Berrozpe et al., 2006) |
| MicroRNAs, small interfering RNAs, and repeat-associated small interfering RNAs | (Vagin et al., 2006; Chen and Rajewsky, 2006; Levine et al., 2007) |
| Crosstalk between RNA-editing and RNA-interference | (Nishikura, 2006) |
| Genetic architecture is modular and multifunctional | (Khaja et al., 2006; Kapranov et al., 2007) |
| Heritability of alternative splicing | (Kwan et al., 2007) |
| Transcripts of unknown function that encode 11-amino-acid peptides | (Galindo et al., 2007; Gingeras, 2007) |
| Splicing efficiency between individuals | (Hull et al., 2007) |
| RNA regulons controlling posttranscriptional gene-expression networks | (Keene, 2007) |
| Ancient population bottlenecks affect phenotypic, ethnic variation and genome architecture | (Manica et al., 2007; Spielman et al., 2007; Nomura et al., 2007; Gherman et al., 2007) |
| Proteins involved in mRNA translation | (Scheper et al., 2007) |
| Transcriptional factories in nucleus | (Akhtar and Gasser, 2007) |
| Ultraconserved elements of unknown function | (Katzman et al., 2007) |
| First sequencing effort to compare all alleles on both chromosomes | (Levy et al., 2007) |
| Interindividual regulatory variation | (Stranger et al., 2007b) |
These items are provided here, in addition to approximately three dozen others listed in previous reviews (Nebert and Vesell, 2004; Nebert, 2005).
Short transcripts of unknown function, often located in gene-desert regions (Galindo et al., 2007; Gingeras, 2007), are among the latest unexpected findings. Of 300 women from high-risk families for breast cancer who had received negative BRCA1 and BRCA2 test results from Myriad Genetics (Salt Lake City, Utah), 12% of these patients were discovered to carry gene rearrangements not included in Myriad's vast array of DNA tests and therefore should have been designated to have positive test results (Walsh et al., 2006). From very recent sequencing of the diploid genome from a single individual (Levy et al., 2007), the 22 pairs of autosomes differed by 0.5% (!!)—suggesting far more genetic heterogeneity than previously expected. Problems with ever attaining, with near certainty, an unequivocal genotype (Table 2, plus [Nebert et al., 2003; Nebert and Vesell, 2004; Nebert, 2005]) have therefore led to major changes in our thinking (Sebat, 2007).
Oligogenic Diseases
The first oligogenic disorder to be described was one form of retinitis pigmentosa, shown to be due to strict digenic inheritance, requiring the presence of heterozygous mutations in the RDS and ROMI genes (Kajiwara et al., 1994). Katsanis and coworkers (2001) first coined the term “tri-allelic inheritance.” This concept of oligogenic traits led to the elegant phrase “The land between Mendelian and multifactorial inheritance” (Burghes et al., 2001). It was suggested that, if complex diseases include too many genes to examine effectively in genotype-phenotype association studies and “monogenic” diseases are too simple, then perhaps oligogenic diseases should be considered. Oligogenics studies might provide some understanding of the interactions among just a few genes and how these contribute to a particular trait.
Table 3 lists nine so-called oligogenic diseases, plus one review (Ming and Muenke, 2002) that enumerates eleven additional, and another review (Badano and Katsanis, 2002) that specifies 30 additional, oligogenic traits, totaling approximately 50 oligogenic disorders. Some diseases listed as “oligogenic”, however, are questionable, notably: BRCA1 and APC mutations causing breast cancer, SOD1 and CNTF mutations causing familial amyotrophic lateral sclerosis, CDKN2A and MC1R mutations causing melanoma, MYOC and CYP1B1 mutations responsible for autosomal dominant congenital glaucoma (buphthalmos), and APP and TGFB1 mutations causing Alzheimer disease. Many geneticists would agree that each of these disorders is more likely a complex disease (involving dozens, if not hundreds, of contributing genes, combined with an unknown number of environmental factors). Are these disorders complex diseases or oligogenic diseases? It is presently unclear, since the dividing line between these two is blurred and, hence, difficult to define.
Table 3.
Partial list of oligogenic diseases (in which two, or a small number of, genes contribute in a major way to the phenotype).a
| Phenotype | Genes | Reference(s) |
|---|---|---|
| Bardet-Biedl syndrome “tri-allelic inheritance” | BBS2, BBS6 | (Katsanis et al., 2001) |
| Digenic inheritance (a review of 11 syndromes) | (2 genes each) | (Ming and Muenke, 2002) |
| Oligogenic inheritance (a review of 30 syndromes) | (2–4 genes each) | (Badano and Katsanis, 2002) |
| Polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy | DAP12, TREM2 | (Paloneva et al., 2002) |
| Hirschsprung disease | (mutations in 8 genes) | (Carrasquillo et al., 2002) |
| Refsum disease | PHYH, PEX7 | (van den Brink et al., 2003; Jansen et al., 2004) |
| Bardet-Biedl syndrome | BBS1, BBS2, BBS6 | (Beales et al., 2003) |
| Cortisone reductase deficiency | HSD11B1, H6PDH | (Draper et al., 2003) |
| Holoprosencephaly | SHH, ZIC2, SIX3, TGIF | (Dubourg et al., 2004) |
| Polycystic ovary syndrome | HSD11B1, H6PDH | (San Millan et al., 2005) |
| Restless legs syndrome | BTBD9, MEIS1, MAP2K5, LBXCOR1 | (Mignot, 2007; Winkelmann et al., 2007) |
No new publications mentioning “tri-allelic digenic inheritance” have appeared since the 2005 paper. The term “oligogenic” has been used in several hundred publications, but the term now seems to be utilized quite loosely.
To summarize, Fig. 2 shows the possible number of genes, and to what percent of the phenotype they might contribute, for human monogenic diseases (Fig. 2A), compared with oligogenic disorders (Fig. 2C). For PKU or cystic fibrosis, respectively, for example, mutations in the PAH and CFTR genes probably contribute 90% or more to the disease; the remaining 5–10% would be contributed by modifier genes (Fig. 2A). For what appears to be bona fide oligogenic disorders, such as Bardet-Biedl syndrome or Hirschsprung disease, we suggest that mutations in two or three genes might each contribute 3% to 6%; we propose that the remaining 94% to 97% would be contributed by modifier genes (Fig. 2C).
Do some pharmacogenetic disorders fall under the oligogenic classification? None has been specifically defined to date, but some may be identified and characterized. On the other hand, since oligogenic disorders and complex diseases represent a continuum, perhaps oligogenics is no longer a useful term or concept.
Polygenic Diseases
From many studies of numerous generations and large families, the contribution of a single variant to a complex disease trait is now believed to be usually between 1% and 5% (in terms of the heritability of liability). There have been many claims of an optimistic identification of a major gene contributing significantly (15% to 35%) to a complex disease, followed by the inability to replicate or corroborate the initial study; examples include: (a) DYX1C1 gene associated with dyslexia (Taipale et al., 2003), but these initial findings have not been replicated by six other association analyses (Scerri et al., 2004; Marino et al., 2005; Meng et al., 2005; Bellini et al., 2005; Schumacher et al., 2007; Anthoni et al., 2007) using independent samples of predominantly European origin; (b) PDE4D gene and vascular stroke (Gretarsdottir et al., 2003), but several follow-up studies (Reneland et al., 2005; Rosand et al., 2006) have not yet resolved the validity of the original association; (c) BMP2 gene and osteoporosis (Styrkarsdottir et al., 2003), yet current evidence (Medici et al., 2006) remains inconclusive about the precise association of BMP2 gene variants with osteoporosis; and (d) ITPR3 gene and type-1 diabetes (Roach et al., 2006), but whereas the P value is highly significant (10−6) this study has not been replicated, and the observed signal near the ITPR3 gene is probably generated from an adjacent MHC region (moreover, this association was not corroborated by a recent GWA study (The Wellcome Trust Case Control Consortium, 2007)).
Additional convincing proof for the moderate contribution (1% to 5%) of any single gene to a complex trait comes from the analysis of a large population of heterogeneous-stock mice, descended from eight inbred progenitor strains; 101 complex traits in 1,904 progeny derived from 50 generations of interbreeding were analyzed (Valdar et al., 2006). These data indicated that most quantitative trait loci (QTL) are able to explain only 1% to 5% of phenotypic variation. In addition, genotype-environment interactions are important, and experimental covariates show substantial interaction with genetic variation. These complexities, familiar to quantitative geneticists for years, pose substantial challenges into the analysis of complex traits, to the determination of how much each individual major gene might contribute to that trait, and at what level the contribution to a trait can be detected.
Epigenetics and Epigenomics
Epigenetics and epigenomics are among the many reasons for our lack of achieving an unequivocal genotype (Nebert and Vesell, 2004). DNA methylation patterns, covalent modifications of histones and chromatin, and RNA interference represent examples of epigenetics—so-called non-Mendelian inheritance of a trait located somewhere between “genotype” and “phenotype.” There has been a recent explosion in this field (Eckhardt et al., 2006; Brena et al., 2006; Martin and Zhang, 2007). Some epigenetic variants can be inherited by the offspring (or the offspring's offspring), indicating the existence of a mechanism for biological heredity not based on DNA sequence. For example, cigarette smoking by a grandmother increases the risk of asthma in the granddaughter (Li et al., 2005).
Famine at the time a male is entering puberty sends an unknown transgenerational message to his grandson, resulting in a 4-fold less risk of diabetes type-2; famine in the same population, at the time a female's oocytes are forming in utero, increases the risk of obesity and diabetes 4-fold in that baby's granddaughter (Pembrey, 2002). In utero exposure of the mother to an environment rich in microbial compounds (farm and barn stables) protects against the development of atopic sensitization and enhances the innate immune system in school-age children (Ege et al., 2006).
A brief exposure of endocrine disruptors, vinclozolin (anti-androgenic compound) or methoxychlor (estrogenic compound), to a pregnant rat during the period of gonadal sex development in utero produces an adult phenotype in the F1 to F4 generations of decreased spermatogenesis and greater risk male infertility (Anway et al., 2005). Recent work from mouse models, human monozygotic twin studies, and large-scale epigenetic profiling suggests that epigenetically determined phenotypes and epigenetic inheritance are far more common than previously appreciated (Rakyan and Beck, 2006).
It is now well known that epigenetic changes are involved in various human diseases including cancer and asthma, as well as during normal development (Feinberg, 2007). There are currently no known human examples of pharmacogenetic disorders that are caused by, or affected by, epigenetic events. Drug tolerance (to the anesthetic, benzyl alcohol) in the fly has very recently been shown to be caused by epigenetic histone modification and transcriptional induction of slo, an ion-channel gene (Wang et al., 2007). Epigenetic effects increase with each passing decade of life, however, due to constant bombardment of environmental stimuli (which would include drugs, as well as chemical or metal toxicants). Within the next several years, we expect to see examples of epigenetic-mediated effects on gene-drug and (perhaps especially) on gene-environment interactions.
Common SNPs in Candidate Genes
There are many clinicians, clinical pharmacologists, wet-bench pharmacologists and toxicologists, biochemists, epidemiologists—and even molecular biologists and scientists who sequence DNA or generate transgenic and other knockout mouse lines—who have not had formal training in human genetics. Yet, there exist many publications written by such investigators—in which genotype-phenotype association studies are considered “statistically significant” (P <0.05) and, by implication, biologically meaningful. Table 4 lists only 20 such examples. If an “association” exists between a SNP in some gene and a multiplex phenotype (such as risk of: lung cancer, myocardial infarction, asthma, schizophrenia, bipolar disorder, atherosclerosis, renal stones, gallstones, multiple sclerosis) or efficacy in response to one or another drug, and the P value is <0.05 but far from <10−6, of what clinical or therapeutic value is this to the patient, physician, or basic scientist?
Table 4.
Partial list of recent publications in which one or very few SNPs “are associated with” a complex disease, also termed multiplex phenotype (P-values between <0.05 and 0.001).a
| Phenotype | Gene | P value(s) | Reference |
|---|---|---|---|
| Lung cancer and smoking | NQO1 | 0.048 | (Rosvold et al., 1995) |
| Induced CYP1A2 metabolic activity | CYP1A2 | <0.05 | (Nakajima et al., 1999) |
| Risk of colorectal cancer | SULT1A1 | 0.009 | (Bamber et al., 2001) |
| Myocardial infarction | GCLM | <0.001 | (Nakamura et al., 2002) |
| Ethnic differences in risk of heart failure | ADRA2C, ADRB1 | <0.001, 0.004 | (Small et al., 2002) |
| Successful weight reduction by sibutramine | GNB3 | 0.031, 0.004 | (Hauner et al., 2003) |
| Vulnerability to illegal drug abuse | HTR2B | 0.0335 | (Lin et al., 2004) |
| Caffeine metabolism | CYP1A2 | 0.036, 0.038 | (Chen et al., 2005) |
| Risk of young onset, late onset Parkinson disease | NAT2 | 0.003 | (Chaudhary et al., 2005) |
| Coffee intake and risk of myocardial infarction | CYP1A2 | 0.04 | (Cornelis et al., 2006) |
| Smoking and obesity in prostate, lung, colorectal and ovarian cancer patients | DRD2 | 0.02, 0.007 | (Morton et al., 2006) |
| Antidepressant efficacy in depression patients | GNB3 | 0.02, 0.03 | (Wilkie et al., 2007) |
| Risk of aspirin-intolerant asthma | PTGER2, PTGER3, PTGIR, TBXA2R | 0.023, 0.038 | (Kim et al., 2007) |
| Gefitinib responsiveness in non-small-cell lung cancer | EGFR | 0.014, 0.029 | (Han et al., 2007) |
| Risk of atherosclerosis | CYP2J2 | 0.036 | (Lee et al., 2007) |
| Risk of toxic liver injury | ABCC2 | 0.04 | (Choi et al., 2007) |
| Risk of primary lung cancer | ERCC1 | 0.034 | (Ma et al., 2007) |
| Induction of extrapyramidal symptoms by antipsychotics | RGS2 | 0.003, 0.009 | (Greenbaum et al., 2007) |
| Fracture risk (bone mass) after estrogen treatment | P2RX7 | <0.01, 0.02 | (Ohlendorff et al., 2007) |
| Glatiramer acetate therapy for multiple sclerosis | TRB@ locus | 0.049 | (Grossman et al., 2007) |
This list is limited to 20 examples; many hundreds of additional references could be cited. Presumably, most of these findings represent false positives (see text).
For example, several reports showed a statistically significant (P <0.05) association between CYP1A2 metabolic activity (Nakajima et al., 1999), rate of caffeine metabolism (Chen et al., 2005), or risk of myocardial infarction from drinking coffee (Cornelis et al., 2006) with a particular SNP in or near the CYP1A2 gene. It is well known (Eaton et al., 1995; Nebert et al., 1999) that hepatic CYP1A2 activity differs among individuals by more than 60-fold, for reasons not yet established. A recent study examining 16 tagSNPs across a 39.6-kb region of the entire CYP1A1_1A2 locus in 280 individuals (from very-high, to very-low, CYP1A2 activity) concluded that it is impossible at the present time to devise a DNA test to detect high-CYP1A2 versus low-CYP1A2 individuals (Jiang et al., 2006). These three examples listed above (and in Table 4) therefore should be regarded as “inconclusive,” “under-powered,” and in need of further replication before they can be regarded as informative studies.
Fig. 2 reveals small contributions to the phenotype under study from an unknown, but considerable, number n (dozens or hundreds), of genes—especially through the middle and to the right side of all four panels. Hence, a study or two in Table 4 might have identified a gene in the middle or to the right side of all four panels in Fig. 2. Such “P-values of <0.05 but not <10−4 studies” (e.g., Table 4), however, most likely represent a false positive. Occasionally but rarely likely, such studies might have identified a modifier gene with an extremely low contribution (<1%) to the trait under study; one can never be certain which explanation of such studies is correct.
Candidate Gene SNPs and LD Studies Helped by the HapMap
The goal of the International HapMap Project (Gibbs and [The International HapMap Consortium], 2003), completed between 2003 and 2006, has been to determine common patterns of DNA sequence variation in the human genome and to make this information freely available to all scientists. DNA samples studied in this first phase included: Yoruba in Ibadan, Nigeria (YRI), Japanese in Tokyo, Japan (JPT), Han Chinese in Beijing, China (CHB), and Centre d'Etude du Polymorphisme Humain (CEPH) samples from Utah, having Caucasian ancestry from northern and western Europe (CEU). The final data set of phase I of the HapMap project was released in mid-2005 (Release #16c.1), which contains genotype data for more than 1 million SNPs from the 269 subjects (The International HapMap Consortium, 2005). Phase II of the project increased the density of genotyped SNPs to one SNP per kilobase; the most recent full data set of combined phase I and phase II HapMap project was released in January 2007 (Release #21a), which contains genotype data of almost 4 million non-redundant SNPs and a total number of 7 million genotyped SNPs (http://www.hapmap.org).
It has been estimated (Kruglyak and Nickerson, 2001;McVean et al., 2005) that there are 10 million to 11 million such variants in the human genome having a minor allele frequency (MAF) of >1% in the human population, and about 7 million SNPs with MAF >5%. Recent estimates based on the HapMap phase II data (unpublished data) indicate that the number of common SNPs (MAF >5%) is around 10 million in the YRI population, i.e., the oldest population being examined.
There also have been discussions about extending HapMap studies to include the other two major geographically isolated human subgroups, Oceanian and Amerindian. One study reports the haplotype structure across 12 Mb of DNA sequence in 927 individuals representing 52 populations (Conrad et al., 2006). Another study examines dense SNP data uniformly across samples of the four HapMap populations and eleven other populations (de Bakker et al., 2006). These two studies provide convincing reassurance that detailed SNP discovery in these two additional human subgroups might be unnecessary.
One major insight from the HapMap data is that the most important determinant of tagSNP “portability” (i.e., how a tagSNP in one population efficiently captures the LD in other populations) is the level of genetic linkage disequilibrium (LD) in the population to be “tagged”: those with high LD (e.g. Oceanian, Amerindian) tend to be tagged well, whereas those with low LD (e.g., Africa) tend to be difficult to tag—regardless of their proximity to the tag-source population (Need and Goldstein, 2006). This is because Africans have been on the planet longer than Oceanians or Amerindians, and thus the LD blocks in Africans are much smaller in length. Because it is not that expensive to include a small number of extra tagSNPs, it now seems preferable to use a universal set of SNPs, perhaps including a specific supplement for tagging in populations having substantial African ancestry. From HapMap data, the current general rule for GWA studies, discussed below, is to screen the genome at a density of one tagSNP per 5 kb, except for Africans where the density should be one tagSNP per 2.5 kb.
Using LD blocks and common SNPs from candidate genes, or later HapMap data and its haplotype information, a number of statistically very solid genotype-phenotype association studies have been carried out during the past several years. Table 5 lists several of the truly replicable studies. All these examples have been confirmed by extensive replication in other labs and in other ethnic populations. Although smaller P-values usually provide greater support for a true association, there is no universal threshold that can be applied under all circumstances. Generally, P-values are not comparable between different studies. Therefore, instead of setting a stringent significant level (i.e., P-value <10−6), replication studies in different populations are the most important requirement for establishing a positive association.
Table 5.
Exemplary studies—involving LD and not a GWA study—wherein one or a few variants in a candidate gene is associated with a complex disease.a
| Phenotype | Gene(s) | P value | Reference |
|---|---|---|---|
| Risk of Alzheimer disease | APOE | <0.00001 | (Saunders et al., 1993) |
| Risk of deep vein thrombosis | F5 | <10−15 | (Bertina et al., 1994) |
| Risk of type-2 diabetes | PPARG | 0.002 | (Altshuler et al., 2000) |
| Risk of Crohn disease | NOD2 | 2×10−5; 6×10−6 | (Hugot et al., 2001) |
| Type-1 diabetes | PTPN22 | 6.0×10−4 | (Bottini et al., 2004) |
| Rheumatoid arthritis | PTPN22 | 6.6×10−4; 5.6×10−8 | (Begovich et al., 2004) |
| Type-2 diabetes | TCF7L2 | 2. 1×10−9 | (Grant et al., 2006) |
These examples have all been confirmed by repeated replications in other labs and in other ethnic groups. Generally, P-values are not comparable between different studies; hence, instead of setting a stringent level of significance (i.e. P-value <10−6), replication studies in different populations and ethnic groups are preferred in establishing a true positive association.
Again, whereas human genetic disorders have used these data to their advantage, pharmacogenetic studies have lagged behind. Perhaps the closest gene-drug example (using haplotype information, LD blocks, and common SNPs from a candidate gene) would be identification of a particular UGT1A1 haplotype associated with a greater risk of toxic response to irinotecan in Japanese cancer patients (Sai et al., 2004); although a significant advance, however, this study fails to attain the criteria for “individualized drug therapy.” In other words, some patients with that haplotype do not show the toxic response, while other patients not having that haplotype do show the toxic response.
Encode
Another ambitious undertaking has been the Encyclopedia of DNA Elements (ENCODE) Pilot Project (2004–2007), which comprises several hundred investigators located in at least 80 institutes worldwide (The ENCODE Project Consortium, 2004). The plan was to select about 1% of the human genome, to study this in great detail, and to map a large variety of sequences: genes (protein-coding and noncoding exons), promoters, enhancers, repressor/silencer sequences, origin-of-replication and termination sites, conserved sequences, RNA transcripts, transcription-factor binding sites, methylation sites, DNase I hypersensitivity sites, and chromatin modifications. The consortium produced more than 200 data sets, representing >400 million data points, 200 Mb of comparative sequences (e.g., human genome versus that of chimpanzee, mouse or pufferfish), and guidelines for rapid release of all data (http://genome.ucsc.edu/ENCODE).
Some breakthroughs from ENCODE include discoveries (or a more complete understanding) of: extensive overlap of gene transcripts and many non-protein coding regions; complex networks of transcripts, which challenge the concept of “lone transcription units”; many new transcription start-sites, with an arrangement of far more complex regulatory sequences and binding of transcription factors than heretofore imagined; regulation of genes in cis that can be governed by motifs millions of bases away; regulation of genes in trans that can be controlled by factors on other chromosomes; interactions between chromatin structure, regulation of transcription and replication; and finally, an unbelievable 60% of all DNA to be under evolutionary constraint (The ENCODE Project Consortium, 2007). Some of these ENCODE breakthroughs, concerning identification of new genomic complexities, are listed in Table 2 among the additional problems in ever being able to achieve an unequivocal genotype.
These amazingly complex findings by the ENCODE Project have thrown into disarray previous concepts of “what constitutes a gene” (Gerstein et al., 2007). Seven years ago, it was clear that “the gene unit” should include everything upstream including the 5′-most regulatory element and everything downstream including the 3′-most regulatory element. Accordingly, some genes will overlap other genes, some genes will be located on the other DNA strand of an existing gene, and some genes will be completely inside another gene (Nebert, 2000a).
Therefore, the definition of “a gene” has been: “A segment of DNA, including all regulatory sequences for that DNA, which encodes a functional product—whether it is a protein, small peptide, or one of the many classes of regulatory RNA.” The proposed definition this year is: “A union of genomic sequences encoding a coherent set of potentially overlapping functional products”. This definition sidesteps complexities of regulation and transcription (Gerstein et al., 2007). Such discussions of “what is a gene” should also remind pharmacologists of the complexities facing us in ever achieving “individualized drug therapy” concerning one or more of these “genes.”
The next goal of ENCODE is, eventually, to perform all these analyses on the other 99% of the human genome. Has the ENCODE Pilot Project, however, shown that it is possible to build an encyclopedia of sequence elements correlated with biochemical functions? Will there be predictive value in knowing, for each region of the genome, all entries into the encyclopedia and the state of the chromatin? How can these data be applied to human disease? Given that 60% of the DNA is evolutionarily constrained, do more elements need to be mapped experimentally, to account for the remaining 40%? Future plans of the scale-up of the ENCODE Project are indeed staggering (Weinstock, 2007).
Genome-Wide Association (GWA) Studies
With the development and availability of ultra-high-volume genotyping platforms—on the order of 100,000 to 1 million genotypes per sample—at a manageable cost, a GWA workshop convened in April 2005 to compare notes on planned as well as ongoing studies, to discuss statistical methods and study design, and to debate alternative technologies (Thomas et al., 2005). Using the millions of SNPs catalogued by the HapMap Project, and having determined haplotypes across diverse populations so that subsets of highly informative tagSNPs could be identified, GWA scans (involving hundreds of thousands of markers and thousands of subjects) have now become financially feasible and a real possibility, just as Risch and Merikangas (1996) had prophesied a decade earlier.
In this highly cited 2-page paper, the authors question whether genetic studies of complex diseases (because of the persistent lack of replicability) had reached their limits (Risch and Merikangas, 1996). Methods such as linkage studies that had been successful in finding major genes (far left bars of panels in Fig. 2) have limited power to detect genes of modest effect (all bars to the right of the far left bars of each panel in Fig. 2) or lower penetrance; thus, a different method of association study—that tested all candidate genes—would have greater power, even if this meant testing every gene in the genome.
Using a disease susceptibility locus with two alleles, A and a, with population frequencies of p and q = 1–p, there are three possible genotypes: A/A, A/a and a/a. The authors defined genotypic relative risk (GRR) as “the increased chance that an individual with a particular genotype has the disease”. Starting with a number of assumptions [including probability of the A allele transmission from a heterozygous parent to child, number of human genes in the genome, number of diallelic polymorphisms per gene, a LOD score (logarithm of the odds ratio for linkage) of 3.0 for a linkage test with pairs of affected siblings, type I error probability a of ∼10−4, and a linkage study of 500 genomic markers], Risch and Merikangas (1996) estimated that this significance level would provide a probability of >95% for no false positives. For 1 million alleles to be tested, the equivalent false positive rate in 1 million independent association tests was then extrapolated to a significance level a = 5 × 10−8.
Much discussion, (Clark et al., 2005; Eberle et al., 2006; Gibbs and Singleton, 2006; Eberle et al., 2007; Chanock et al., 2007), has ensued since this publication. For example, the original estimate (Risch and Merikangas, 1996) of number of genes in the genome was 100,000; several other assumptions could be off by a factor of 10, but the bottom line is that P-values in GWA studies should preferably reach at least 10−6, if not 10−7, in order to establish true statistical, as well as biological, significance.
The first ever GWA study was probably published in 2002, in which an association between the LTA gene and myocardial infarction was developed by typing 92,788 gene-based SNP markers (Ozaki et al., 2002). Another pioneering GWA study (Klein et al., 2005) reported an association between the CFH gene and age-related macular degeneration by typing 116,204 SNPs. These early reports and subsequent GWA studies have recently been reviewed (Kingsmore et al., 2007). Table 6 lists two dozen representative GWA studies published between the end of 2006 and October 2007, with P-values ranging from 6.7 × 10−6 to 2 × 10−76. These robust GWA studies appear to call into question and further undermine the true informative value of many of the presumed “P <0.05” false-positive studies such as those listed in Table 4.
Table 6.
Partial list (limited to two dozen examples) of recent GWA studies in which one or a few SNPs are associated with a complex disease or multiplex phenotype (note highly significant P-values of <1 × 1(10−6).
| Phenotype | Gene(s) | P-value | Reference |
|---|---|---|---|
| Risk of inflammatory bowel disease | IL23R | 1.60 × 10−9 to 3.36 × 10−13 | (Duerr et al.,2006) |
| Risk of type-2 diabetes | Four loci plus TCF7L2 | 3.2 × 10−17 | (Sladek et al.,2007) |
| Risk of breast cancer | Nine previous genes plus CASP8 | 1.1 × 10−7 | (Cox et al., 2007) |
| Susceptibility to Crohn disease | IL23R, CARD15, PTGER4 a | l0−7 to 10−9 | (Libioulle et al.,2007) |
| Risk of prostate cancer | Chr 8q24 (two independent loci) | 1.41 × 10−11; 6.62 × 10−10 | (Yeager et al.,2007) |
| Risk of prostate cancer | Chr 8q24 (two independent loci) | 1.4 × 10−10; 1.6 ×10−14 | (Gudmundsson et al., 2007a) |
| Association with body mass index and predisposition to obesity | FTO | 3 × 10−26 | (Frayling et al., 2007) |
| Susceptibility to Crohn disease | IL23R, CARD15, ATG16L1, PHOX2B, NCF4, FAM92B, and intergenic region on 10q21.1b | < 10−10 | (Rioux et al., 2007) |
| Bipolar disorder, coronary artery disease, Crohn disease, hypertension, rheumatoid arthritis, type-1 diabetes and type-2 diabetes | 24 independent association signals | <5 × 10−7 | (The Wellcome Trust Case Control Consortium, 2007) |
| Association with coronary heart disease | Region with no annotated genes, near CDKN2A & CDKN2B | 6.7 × 10−6 | (McPherson et al., 2007) |
| Risk of myocardial infarction | Region with no annotated geneb, near CDKN2A & CDKN2B | 1.4 × l0−7 to 1.2 × 10−20 | (Helgadottir et al.,2007) |
| Susceptibility of breast cancer | FGFR2, LSP1 & 3 other loci | 3 × 10−9 to 2 × l0−76 | (Easton et al., 2007) |
| Risk of childhood asthma | Several sites in cis with ORMDL3 expression | <10−12; <10−22 | (Moffatt et al.,2007) |
| Risk of sporadic postmenopausal breast cancer | FGFR2 | 1.1 × 10−10 | (Hunter et al., 2007) |
| Risk of celiac disease | KIAA1109-TENR-IL2-IL21 linkage disequilibrium block | 2.0 × 10−7; 1.3 × 10−14 | (van Heel et al., 2007) |
| Association with obesity-related traits | FTO, PFKP | l.l × 10−7 to 3.4 × 10−8 | (Scuteri et al., 2007) |
| Major determinants for host control of HIV-1 | HCP5 | 9.36 × 10−12 | (Fellay et al., 2007) |
| Risk of type-1 diabetes | KIAA0350, INS, PTPN22, COL1A2, LPHN2 | 7.53 × 10−8 to 1.11 × 10−12 | (Hakonarson et al., 2007) |
| Coronary artery disease | Chr 9p21.3 | 1.80 × 10−14 | (Samani et al., 2007) |
| Protection against type-2 diabetes | TCF2 | 4.7 × 10−9 | (Gudmundsson et al., 2007b) |
| Susceptibility to colorectal cancer | Chr 8q24.21 | 1.27 × 10−14; 3.16 × 10−11 | (Tomlinson et al., 2007; Zanke et al., 2007) |
| Risk of type-2 diabetes | WFS1 | 1.4 & 3.4 × 10−7 | (Sandhu et al.,2007) |
| Susceptibility to gallstone disease | ABCG8 | 1.4 × 10−14 | (Buch et al., 2007) |
| Association with type-1 diabetes | IL2RA, RBM17 | 1.92 × 10−28 | (Lowe et al., 2007) |
Interestingly, the segment of DNA that regulates the PTGER4 gene lies within a 1.25-Mb gene-desert region at 5p l3.1.
Another example of a SNP associated with a complex disease that lies within a large gene-desert region, perhaps explained by the short open-reading-frame transcripts encoding 11-amino-acid peptides [see Table 2].
It should be recognized that multiple parameters will affect the power, the reported P-values of putative findings, and the confidence interval (CI) of effect-size estimators for any genotype-phenotype association study—whether a human disease or a pharmacogenetic disorder. Statistical power improves, of course, with larger numbers of cases and controls. If the MAF increases, usually fewer subjects are needed in the study group and the level of detectable contribution by a gene to a phenotype will be smaller. If the MAF is low, greater numbers per group will be required and the level of detectable contribution by a gene to a phenotype will be higher.
The above statements may not, however, necessarily hold. For example, when the “contribution” (in genetic terms, the “effect size”) of the risk variant is measured by heritability, the influence of MAF on the statistical power of an association study is not significant. If the effect size is measured by odds ratio (OR) or genotypic relative risk (GRR), however, the influence of MAF will be tremendous, especially for MAF <0.05. In addition to the MAF and prevalence of the disorder, the inheritance model of the risk allele (“multiplicative,” “dominant,” or “recessive”) can also influence the power of an association study.
It must be emphasized that GWA studies are not always error-free. For example, a GWA study of risk of Parkinson disease identified a number of SNPs having a high association with the phenotype (Maraganore et al., 2005); subsequently, four independent investigative teams (Myers, 2006) could not corroborate any of these data, concluding that even the LRRK2 gene that had been identified by Maraganore and coworkers (Maraganore et al., 2005) is likely to be a false positive. Some of the same reasons for these discrepancies were discussed above in the early SNP-fever publications: population stratification differences between cases and controls are a primary source of false positive associations; there is substantial evidence in Parkinson disease for low-penetrance genes; the original study (Maraganore et al., 2005) would have benefited greatly by replicability in a second ethnic group; finally, the etiology of Parkinson disease is likely to be a complex interaction between multiple genes and numerous environmental factors (Tanner et al., 1999; Myers, 2006).
Future Directions: Personalized Medicine and Individualized Drug Therapy?
Perhaps the method by which most investigators evaluate the outcomes of a complex disease is too simplistic: non-afflicted versus afflicted, or zero versus one. Such binary traits fail to reflect the QTL nature of most attributes which could be immediately related to disease causation. Dividing a large population into more valuable subsets can enhance the statistical power of a study, while reducing the number of individuals studied. Dividing a large population into subsets such as “highly sensitive” versus “highly resistant,” has been described as the extreme discordant phenotype (EDP) method of design and analysis (Nebert, 2000b). The statistical power of the EDP approach has been demonstrated; the EDP approach is best applied to traits that are relatively common, and quantitation of a phenotype (e.g., number of cigarette-pack years, dosage of a drug for how long) clearly improves the statistical power (Zhang et al., 2006). We predict that, in the near future, a similar (EDP, or gradient) approach to GWA studies, instead of the “binary state” assignment, will improve GWA studies—in both human genomics and pharmacogenomics.
Improved statistical analyses will also help future GWA studies. For example, principal components analysis, using hundreds of thousands of markers, can help correct for population stratification (Price et al., 2007). Bayesian inference in case-control epistasis-mapping studies is significantly more powerful than existing approaches and is feasible with many thousands of markers (Zhang and Liu, 2007). The importance of collaboration, by way of (for example) the Genetic Association Information Network (The GAIN Collaborative Research Group, 2007) and the NIH Database of Genome-Wide Association Studies dbGAP (Mailman et al., 2007), will be necessary for the integration of all genotyping and phenotyping data in the near future (West et al., 2006).
One ultimate goal of human genetics and genomics, and now GWA studies, is to provide to the patient of the internist, surgeon or pediatrician personalized medicine in preventing or treating complex diseases. One ultimate goal of pharmacogenetics is to provide the patient of clinical pharmacologists or other physicians with individualized drug therapy. As can be seen from this review—especially the unanticipated, surprising, multiple findings about our genome uncovered during the past several years—it continues to be virtually impossible to assign a patient to an unequivocal phenotype and especially to an unequivocal genotype. This can be firmly stated, although many optimistic reviews to the contrary continue to appear (West et al., 2006; Eichelbaum et al., 2006; Roden et al., 2006; Lesko, 2007; Roses, 2007; Hunter, 2007; Swen et al., 2007). Moreover, most of the March 2007 issue of Clinical Pharmacology and Therapeutics and several articles in the September 2007 issue of the Journal of Clinical Pharmacology are optimistic about “prospects in the near future for individualized therapy based mainly on DNA testing.”
What is the current assessment of gene-drug interactions? How close are we to achieving individualized drug therapy? From all that has been discussed above, it should be clear by now that drawing a conclusion from a study of 200 or 1,000 patients is one thing; drawing a conclusion from a DNA test on an individual patient walking into your office is quite another.
Where are we, today, in our understanding of the role of human genetics and genomics in drug toxicity, efficacy and therapeutic failure? From previous reviews (Nebert et al., 2003; Nebert and Vesell, 2004; Nebert, 2005), and from what has been discussed above (Fig. 2), it is clear that those few hPpM genes that do make a difference in metabolism of various drug substrates (e.g., CYP2D6, NAT2, TPMT, CYP2C19) might contribute perhaps 15% to 20% to the (EM or PM) phenotype (Fig. 2B), whereas a contribution of 90% or more is expected for the gene responsible for a monogenic human disease (Fig. 2A). If we know that dozens or hundreds of additional downstream genes might affect the ultimate outcome of a particular drug, how can we integrate and assemble this knowledge into a diagram or equation?
In discussing possible confounding factors, it is useful to distinguish genetic factors—specific to the drug used—from those influencing the nature of the condition itself. Drug-specific effects include both pharmacokinetics [e.g., drug- or xenobiotic-metabolizing enzymes (XMEs) and XME-related transporters (XRTs)] and pharmacodynamics (e.g., drug targets and target-related proteins, often not involved in disease etiology) of the particular drug (Tate and Goldstein, 2004; Nebert and Dalton, 2006).
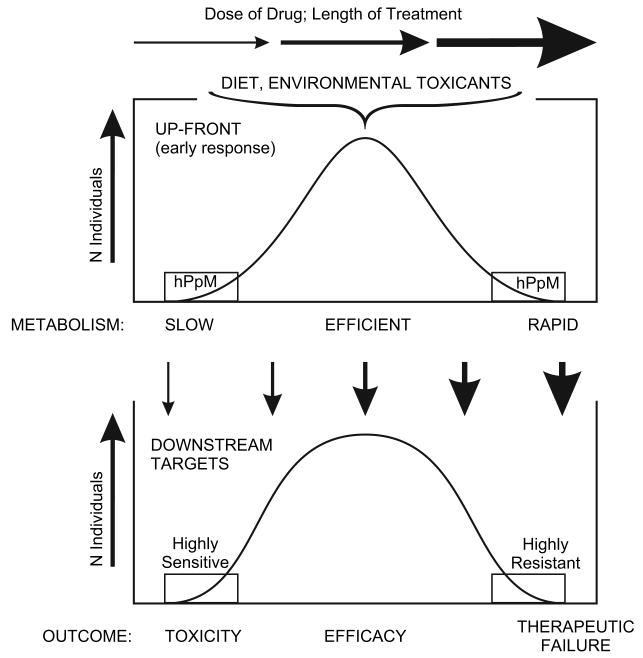
As illustrated in Fig. 3, we propose a 2-tier system of: [a] front-end differences in hPpM gene polymorphisms associated with pharmacokinetics, and [b] downstream genes that will contribute to the phenotype (toxicity, efficacy, or therapeutic failure of a particular drug) associated with pharmacodynamics. Because of hPpM inheritance, the up-front genes can provide a substantial impact on the pharmacokinetics of any incoming drug or combination of drugs. Because the downstream targets are not generally encoded by hPpM genes, the ultimate outcome (toxicity, efficacy, or therapeutic failure) is considerably more complicated, i.e., reflects more closely genes that contribute to a polygenic disease trait (Fig. 2D).
Figure 3.
Scheme proposing an “up-front” (or “early-response”) network of hPpM genes involved in pharmacokinetics, followed by downstream targets involving a network of innumerable low-penetrance genes responsible for pharmacodynamics. This diagram assumes that the parent drug possesses efficacy but that its accumulation can cause toxicity. Some examples of drugs occur wherein the metabolite rather than the parent drug is the active agent.
In clinical pharmacology, most of the up-front gene products are encoded by XME or XRT genes (Nebert and Dalton, 2006); in any given population, when these genes initially respond to a drug, one can expect a gradient—from slow-to-rapid drug metabolism (Fig. 3). The few early-response genes that are truly hPpM (Table 1) might predict, in part, those individuals at the extreme low and extreme high ends of the spectrum (Fig. 3).
Downstream targets include genes encoding receptors, transporters, ion channels, cell-adhesion molecules, signal transduction pathway moieties, chaperones, nucleic acid or protein repair systems, posttranslational-processing, cell cycle and growth factors, and transcription factors. SNPs in any one, or in any combination, of the genes encoding these downstream targets might render an individual “more sensitive” or “more resistant” to a given level of the potentially toxic drug. Again, one would expect to find a gradient of resistance or sensitivity to all the downstream targets combined (Fig. 3). In the case illustrated, we have assumed that the parent drug is the active form and causes the adverse reaction(s). Patients having the genetic predisposition for slowest drug metabolism, due to the up-front early-response network, combined with having downstream targets most sensitive to the parent drug, would be at highest risk for accumulation of that drug leading to toxicity. Subjects having the genetic predisposition for rapid metabolism, due to the up-front early-response network, combined with having downstream targets most resistant to damage by the specific drug, would clear the drug most rapidly and be at greatest risk for therapeutic failure. Those with intermediate metabolism (not too slow, not too fast) in the up-front genes—would provide the optimum amount of drug to the downstream targets, resulting in efficacy. In addition, the dose, accumulation of drug, and length of treatment time might all affect this balance (as well as age, gender, renal clearance, disease, etc.) and, hence, could ultimately lead to drug toxicity or therapeutic failure, after the drug had demonstrated efficacy for some months or years of time (Fig. 3). Fig. 3 shows the effect of a drug in a large population. Only a few DNA tests provide 100% accuracy in monogenic human diseases. To date, no DNA tests provide 100% accuracy in predicting a pharmacogenetic disorder—in any given patient.
In conclusion, we have compared the research advances and development of human genetics and genomics with those of pharmacogenetics and pharmacogenomics, and we can appreciate how many “new surprising findings” continue to appear almost monthly. Hence, it should be readily appreciated as to why personalized medicine, as well as individualized drug therapy, will be most challenging to achieve in the near future.
Acknowledgments
We thank our many colleagues for a careful reading of this manuscript. This work was supported, in part, by NIH P30 ES06096 (D.W.N.).
Abbreviations
- CEPH
Centre d'Etude du Polymorphisme Humain
- CEU
Individuals from Utah, having Caucasian ancestry from northern and western Europe
- CHB
Han Chinese in Beijing, China
- CI
Confidence interval
- CNVs
Copy-number variations
- DNaseI
Endonuclease that digests double- or single-stranded DNA into oligonucleotides or mononucleotides
- EDP
Extreme discordant phenotype
- EGP
Environmental Genome Project
- EM
Extensive-metabolizer
- ENCODE
Encyclopedia of DNA Elements
- G6PD
Glucose-6-phosphate dehydrogenase
- GRR
Genotypic relative risk
- GWA
Genome-wide association
- HGP
Human Genome Project
- hPpM
High-penetrance predominantly monogenic
- HD
Huntington disease
- JPT
Japanese in Tokyo, Japan
- LD
Linkage disequilibrium
- LOD
Logarithm of the odds ratio for linkage
- MAF
Minor allele frequency
- OR
Odds ratio
- PKU
Phenylketonuria
- PM
Poor-metabolizer
- QTLs
Quantitative trait loci
- SNPs
Single-nucleotide polymorphisms
- UM
Ultrarapid metabolizer
- XMEs
Xenobiotic-metabolizing enzymes
- XRTs
XME-related transporters
- YRI
Yoruba in Ibadan, Nigeria
Footnotes
This review is dedicated to the memory of our dear friend and colleague Professor Werner Kalow (1916–2008)
Note added in proof. After the writing of this article was completed in October, Science magazine decided [Pennisi, E., Science 21 Dec 2007; 318: 1842–1843] that the “Scientific Breakthrough of the Year 2007” was the newly appreciated recognition of “Human Genetic Variation,” i.e. that variation in our genome is far greater than most of us could ever have imagined, just a year or two ago.
Contributor Information
Daniel W. Nebert, Division of Human Genetics, Department of Pediatrics & Molecular Developmental Biology, Department of Environmental Health and Center for Environmental Genetics, University of Cincinnati Medical Center, Cincinnati, Ohio, USA
Ge Zhang, Department of Environmental Health, University of Cincinnati Medical Center, Cincinnati, Ohio, USA.
Elliot S. Vesell, Department of Pharmacology, Pennsylvania State University College of Medicine, Hershey, Pennsylvania, USA
References
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, Hodges MD, Bhangal G, Patel SG, Sheehan-Rooney K, Duda M, Cook PR, Evans DJ, Domin J, Flint J, Boyle JJ, Pusey CD, Cook HT. Copy number polymorphism in FCGR3 gene predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARG Pro12Ala polymorphism is associated with decreased risk of Type-2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Alving AS, Carson PE, Flanagan CL, Ickes CE. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science. 1956;124:484–485. doi: 10.1126/science.124.3220.484-a. [DOI] [PubMed] [Google Scholar]
- Anthoni H, Zucchelli M, Matsson H, Muller-Myhsok B, Fransson I, Schumacher J, Massinen S, Onkamo P, Warnke A, Griesemann H, Hoffmann P, Nopola-Hemmi J, Lyytinen H, Schulte-Korne G, Kere J, Nothen MM, Peyrard-Janvid M. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated with dyslexia. Hum Mol Genet. 2007;16:667–677. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaly MF, Becker B. Intraocular pressure response to topical corticosteroids. Fed Proc. 1965;24:1274–1278. [PubMed] [Google Scholar]
- Badano JL, Katsanis N. Beyond Mendel: An evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- Bamber DE, Fryer AA, Strange RC, Elder JB, Deakin M, Rajagopal R, Fawole A, Gilissen RA, Campbell FC, Coughtrie MW. Phenol sulphotransferase SULT1A1*1 genotype is associated with reduced risk of colorectal cancer. Pharmacogenetics. 2001;11:679–685. doi: 10.1097/00008571-200111000-00006. [DOI] [PubMed] [Google Scholar]
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, Lupski JR, Katsanis N. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in Non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SY, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini G, Bravaccio C, Calamoneri F, Donatella CM, Fiorillo P, Gagliano A, Mazzone D, del Giudice EM, Scuccimarra G, Militerni R, Pascotto A. No evidence for association between dyslexia and DYX1C1 functional variants in a group of children and adolescents From Southern Italy. J Mol Neurosci. 2005;27:311–314. doi: 10.1385/jmn:27:3:311. [DOI] [PubMed] [Google Scholar]
- Berrozpe G, Agosti V, Tucker C, Blanpain C, Manova K, Besmer P. A distant upstream locus control region is critical for expression of the Kit Receptor Gene in mast cells. Mol Cell Biol. 2006;26:5850–5860. doi: 10.1128/MCB.01854-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertina RM, Koeleman BP, Koster T, Rosendaal FR, Dirven RJ, de R H, van d V, Reitsma PH. Mutation in blood coagulation factor V associated with resistance to activated Protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- Blum M, Grant DM, McBride W, Heim M, Meyer UA. Human Arylamine N-Acetyltransferase genes: Isolation, chromosomal localization, and functional expression. DNA Cell Biol. 1990;9:193–203. doi: 10.1089/dna.1990.9.193. [DOI] [PubMed] [Google Scholar]
- Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. A functional variant of lymphoid tyrosine phosphatase is associated with Type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- Brena RM, Huang TH, Plass C. Toward a human epigenome. Nat Genet. 2006;38:1359–1360. doi: 10.1038/ng1206-1359. [DOI] [PubMed] [Google Scholar]
- Buch S, Schafmayer C, Volzke H, Becker C, Franke A, von Eller-Eberstein H, Kluck C, Bassmann I, Brosch M, Lammert F, Miquel JF, Nervi F, Wittig M, Rosskopf D, Timm B, Holl C, Seeger M, El Sharawy A, Lu T, Egberts J, Fandrich F, Folsch UR, Krawczak M, Schreiber S, Nurnberg P, Tepel J, Hampe J. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39:995–999. doi: 10.1038/ng2101. [DOI] [PubMed] [Google Scholar]
- Burghes AH, Vaessin HE, de La CA. The land between Mendelian and multifactorial inheritance. Science. 2001;293:2213–2214. doi: 10.1126/science.1065930. [DOI] [PubMed] [Google Scholar]
- Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, Altshuler D, Ardlie KG, Hirschhorn JN. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, McCallion AS, Puffenberger EG, Kashuk CS, Nouri N, Chakravarti A. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet. 2002;32:237–244. doi: 10.1038/ng998. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chao HK, Hsiao KJ, Su TS. A silent mutation induces exon skipping in the phenylalanine hydroxylase gene in phenylketonuria. Hum Genet. 2001;108:14–19. doi: 10.1007/s004390000435. [DOI] [PubMed] [Google Scholar]
- Chaudhary S, Behari M, Dihana M, Swaminath PV, Govindappa ST, Jayaram S, Singh S, Muthane UB, Juyal RC, T BK. Association of N-Acetyl transferase-2 gene polymorphism and slow acetylator phenotype with young onset and late onset Parkinson Disease among Indians. Pharmacogenet Genomics. 2005;15:731–735. doi: 10.1097/01.fpc.0000173485.59430.49. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang L, Zhi L, Zhou G, Wang H, Zhang X, Hao B, Zhu Y, Cheng Z, He F. The -113G>A polymorphism in CYP1A2 affects the caffeine metabolic ratio in a Chinese population. Clin Pharmacol Ther. 2005;78:249–259. doi: 10.1016/j.clpt.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Choi JH, Ahn BM, Yi J, Lee JH, Lee JH, Nam SW, Chon CY, Han KH, Ahn SH, Jang IJ, Cho JY, Suh Y, Cho MO, Lee JE, Kim KH, Lee MG. MRP2 haplotypes confer differential susceptibility to toxic liver injury. Pharmacogenet Genomics. 2007;17:403–415. doi: 10.1097/01.fpc.0000236337.41799.b3. [DOI] [PubMed] [Google Scholar]
- Clark AG, Boerwinkle E, Hixson J, Sing CF. Determinants of the success of whole-genome association testing. Genome Res. 2005;15:1463–1467. doi: 10.1101/gr.4244005. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39:522–529. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- Cornelis MC, El-Sohemy A, Kabagambe EK, Campos H. Coffee, CYP1A2 genotype, and risk of myocardial infarction. JAMA. 2006;295:1135–1141. doi: 10.1001/jama.295.10.1135. [DOI] [PubMed] [Google Scholar]
- Cotton RG. Heterogeneity of phenylketonuria at the clinical, protein and DNA levels. J Inherit Metab Dis. 1990;13:739–750. doi: 10.1007/BF01799577. [DOI] [PubMed] [Google Scholar]
- Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, Lissowska J, Brinton L, Peplonska B, Southey MC, Hopper JL, McCredie MR, Giles GG, Fletcher O, Johnson N, dos SS I, Gibson L, Bojesen SE, Nordestgaard BG, Axelsson CK, Torres D, Hamann U, Justenhoven C, Brauch H, Chang-Claude J, Kropp S, Risch A, Wang-Gohrke S, Schurmann P, Bogdanova N, Dork T, Fagerholm R, Aaltonen K, Blomqvist C, Nevanlinna H, Seal S, Renwick A, Stratton MR, Rahman N, Sangrajrang S, Hughes D, Odefrey F, Brennan P, Spurdle AB, Chenevix-Trench G, Beesley J, Mannermaa A, Hartikainen J, Kataja V, Kosma VM, Couch FJ, Olson JE, Goode EL, Broeks A, Schmidt MK, Hogervorst FB, Van't Veer LJ, Kang D, Yoo KY, Noh DY, Ahn SH, Wedren S, Hall P, Low YL, Liu J, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Sigurdson AJ, Stredrick DL, Alexander BH, Struewing JP, Pharoah PD, Easton DF. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S, Onofrio RC, Lyon HN, Stram DO, Haiman CA, Freedman ML, Zhu X, Cooper R, Groop L, Kolonel LN, Henderson BE, Daly MJ, Hirschhorn JN, Altshuler D. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- Draper N, Walker EA, Bujalska IJ, Tomlinson JW, Chalder SM, Arlt W, Lavery GG, Bedendo O, Ray DW, Laing I, Malunowicz E, White PC, Hewison M, Mason PJ, Connell JM, Shackleton CH, Stewart PM. Mutations in the genes encoding 11β-hydroxysteroid dehydrogenase Type-1 and hexose-6-phosphate dehydrogenase interact to cause cortisone reductase deficiency. Nat Genet. 2003;34:434–439. doi: 10.1038/ng1214. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Lazaro L, Pasquier L, Bendavid C, Blayau M, Le DF, Durou MR, Odent S, David V. Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Hum Mutat. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le ML, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schurmann P, Dork T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den OA, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DL, Gallagher EP, Bammler TK, Kunze KL. Role of cytochrome P450 1A2 in chemical carcinogenesis: Implications for human variability in expression and enzyme activity. Pharmacogenetics. 1995;5:259–274. doi: 10.1097/00008571-199510000-00001. [DOI] [PubMed] [Google Scholar]
- Eberle MA, Pauline NG, Kuhn K, Zhou L, Peiffer DA, Galver L, Viaud-Martinez KA, Taylor C, Gunderson KL, Shen R, Murray SS. Power to detect risk alleles using genome-wide tag-SNP panels. PLoS Genet. 2007;2 doi: 10.1371/journal.pgen.0030170. ePub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle MA, Rieder MJ, Kruglyak L, Nickerson DA. Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet. 2006;2:e142. doi: 10.1371/journal.pgen.0020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ege MJ, Bieli C, Frei R, van Strien RT, Riedler J, Ublagger E, Schram-Bijkerk D, Brunekreef B, van HM, Scheynius A, Pershagen G, Benz MR, Lauener R, von M E, Braun-Fahrlander C. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006;117:817–823. doi: 10.1016/j.jaci.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- Eichelbaum M. Polymorphic drug oxidation in humans. Fed Proc. 1984;43:2298–2302. [PubMed] [Google Scholar]
- Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–137. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- Eichelbaum M, Spannbrucker N, Steincke B, Dengler HJ. Defective N-oxidation of sparteine in man: A new pharmacogenetic defect. Eur J Clin Pharmacol. 1979;16:183–187. doi: 10.1007/BF00562059. [DOI] [PubMed] [Google Scholar]
- Evans DA, Manley KA, McKusick VA. Genetic control of isoniazid metabolism in man. Br Med J. 1960;2:485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagnam A, Cossarizza A, Cozzi-Lepri A, de la Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation: New insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, Post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- Gherman A, Chen PE, Teslovich TM, Stankiewicz P, Withers M, Kashuk CS, Chakravarti A, Lupski JR, Cutler DJ, Katsanis N. Population bottlenecks as a potential major shaping force of human genome architecture. PLoS Genet. 2007;3:e119. doi: 10.1371/journal.pgen.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, Singleton A. Application of genome-wide single nucleotide polymorphism typing: Simple association and beyond. PLoS Genet. 2006;2:e150. doi: 10.1371/journal.pgen.0020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Gingeras TR. Origin of phenotypes: Genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Accumulation of mutant Lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford Progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JA, de Morais SM. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics. 1994;4:285–299. doi: 10.1097/00008571-199412000-00001. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, Gelboin HV, Hardwick JP, Meyer UA. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988;331:442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- Grace ME, Balwani M, Nazarenko I, Prakash-Cheng A, Desnick RJ. Type 1 Gaucher Disease: Null and hypomorphic novel chitotriosidase mutations—implications for diagnosis and therapeutic monitoring. Hum Mutat. 2007;28:866–873. doi: 10.1002/humu.20524. [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of Type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Strous RD, Kanyas K, Merbl Y, Horowitz A, Karni O, Katz E, Kotler M, Olender T, Deshpande SN, Lancet D, Ben-Asher E, Lerer B. Association of the RGS2 gene with extrapyramidal symptoms induced by treatment with antipsychotic medication. Pharmacogenet Genomics. 2007;17:519–528. doi: 10.1097/FPC.0b013e32800ffbb4. [DOI] [PubMed] [Google Scholar]
- Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM, Hawkins M, Gudmundsson G, Gudmundsdottir H, Andrason H, Gudmundsdottir AS, Sigurdardottir M, Chou TT, Nahmias J, Goss S, Sveinbjornsdottir S, Valdimarsson EM, Jakobsson F, Agnarsson U, Gudnason V, Thorgeirsson G, Fingerle J, Gurney M, Gudbjartsson D, Frigge ML, Kong A, Stefansson K, Gulcher JR. The gene encoding phosphodiesterase 4D confers risk of ischemic atroke. Nat Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- Grossman I, Avidan N, Singer C, Goldstaub D, Hayardeny L, Eyal E, Ben-Asher E, Paperna T, Pe'er I, Lancet D, Beckmann JS, Miller A. Pharmacogenetics of glatiramer acetate therapy for multiple sclerosis reveals drug-response markers. Pharmacogenet Genomics. 2007;17:657–666. doi: 10.1097/FPC.0b013e3281299169. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, bers-Akkers MT, Godino-Ivan MJ, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007a;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez BvanBE, van BE, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen TO, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on Chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against Type 2 diabetes. Nat Genet. 2007b;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C. A genome-wide association study identifies KIAA0350 as a Type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- Han SW, Jeon YK, Lee KH, Keam B, Hwang PG, Oh DY, Lee SH, Kim DW, Im SA, Chung DH, Heo DS, Bang YJ, Kim TY. Intron 1 CA dinucleotide repeat polymorphism and mutations of epidermal growth factor receptor and gefitinib responsiveness in non-small-cell lung cancer. Pharmacogenet Genomics. 2007;17:313–319. doi: 10.1097/FPC.0b013e328011abc0. [DOI] [PubMed] [Google Scholar]
- Hauner H, Meier M, Jockel KH, Frey UH, Siffert W. Prediction of successful weight reduction under sibutramine therapy through genotyping of the G-Protein B3 subunit gene (GNB3) C825T polymorphism. Pharmacogenetics. 2003;13:453–459. doi: 10.1097/00008571-200308000-00003. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Liebhaber SA, Cooke NE. Locus control region transcription plays an active role in long-range gene activation. Mol Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Hull J, Campino S, Rowlands K, Chan MS, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, Kwiatkowski D. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ. Sharing the rankings. N Engl J Med. 2007;357:437–439. [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Genetic associations: False or true? Trends Mol Med. 2003;9:135–138. doi: 10.1016/s1471-4914(03)00030-3. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG. Genetic associations in large versus small studies: An empirical assessment. Lancet. 2003;361:567–571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- Jansen GA, Waterham HR, Wanders RJ. Molecular basis of Refsum disease: Sequence variations in phytanoyl-coA hydroxylase (PHYH) and the PTS2 receptor (PEX7) Hum Mutat. 2004;23:209–218. doi: 10.1002/humu.10315. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Dragin N, Jorge-Nebert LF, Martin MV, Guengerich FP, Aklillu E, Ingelman-Sundberg M, Hammons GJ, Lyn-Cook BD, Kadlubar FF, Saldana SN, Sorter M, Vinks AA, Nassr N, von R O, Jin L, Nebert DW. Search for an association between the human CYP1A2 Genotype and CYP1A2 metabolic phenotype. Pharmacogenet Genomics. 2006;16:359–367. doi: 10.1097/01.fpc.0000204994.99429.46. [DOI] [PubMed] [Google Scholar]
- Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked Peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- Kalow W. Contribution of hereditary factors to the response to drugs. Fed Proc. 1965;24:1259–1265. [PubMed] [Google Scholar]
- Kalow W. Pharmacoanthropology: Drug Metabolism. Fed Proc. 1984a;43:2326–2331. [PubMed] [Google Scholar]
- Kalow W. Pharmacoanthropology: Outline, problems, and the nature of case histories. Fed Proc. 1984b;43:2314–2318. [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Kelso J, Franz H, Visagie J, Giger T, Joerchel S, Petzold E, Green RE, Lachmann M, Paabo S. Functionality of intergenic transcription: An evolutionary comparison. PLoS Genet. 2006;2:e171. doi: 10.1371/journal.pgen.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaja R, Zhang J, MacDonald JR, He Y, Joseph-George AM, Wei J, Rafiq MA, Qian C, Shago M, Pantano L, Aburatani H, Jones K, Redon R, Hurles M, Armengol L, Estivill X, Mural RJ, Lee C, Scherer SW, Feuk L. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, Ghislain J, Pezeron G, Mourrain P, Ellingsen S, Oates AC, Thisse C, Thisse B, Foucher I, Adolf B, Geling A, Lenhard B, Becker TS. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim YK, Park HW, Jee YK, Kim SH, Bahn JW, Chang YS, Kim SH, Ye YM, Shin ES, Lee JE, Park HS, Min KU. Association between polymorphisms in prostanoid receptor genes and aspirin-intolerant asthma. Pharmacogenet Genomics. 2007;17:295–304. doi: 10.1097/01.fpc.0000239977.61841.fe. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Lindquist IE, Mudge J, Beavis WD. Genome-wide association studies: progress in identifying genetic biomarkers in common complex diseases. Biomarker Insights. 2007;2:283–292. [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RA, Selin MJ, Harris HW. Genetic factors influencing isoniazid blood levels in humans. Trans Conf Chemother Tuberc. 1959;8:52–56. [Google Scholar]
- Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- Krynetski EY, Schuetz JD, Galpin AJ, Pui CH, Relling MV, Evans WE. A single-point mutation leading to loss of catalytic activity in human thiopurine S-methyltransferase. Proc Natl Acad Sci USA. 1995;92:949–953. doi: 10.1073/pnas.92.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Serre D, Zuzan H, Clark TA, Schweitzer A, Staples MK, Wang H, Blume JE, Hudson TJ, Sladek R, Majewski J. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Du BN. Altered drug response in hereditary disease. Fed Proc. 1965;24:1287–1292. [PubMed] [Google Scholar]
- La Du BN, Eckerson HW. The polymorphic paraoxonase/arylesterase isozymes of human serum. Fed Proc. 1984;43:2338–2341. [PubMed] [Google Scholar]
- Lamba J, Strom S, Venkataramanan R, Thummel KE, Lin YS, Liu W, Cheng C, Lamba V, Watkins PB, Schuetz E. MDR1 genotype is associated with hepatic cytochrome P450 3A4 basal and induction phenotype. Clin Pharmacol Ther. 2006;79:325–338. doi: 10.1016/j.clpt.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Lee CR, North KE, Bray MS, Couper DJ, Heiss G, Zeldin DC. CYP2J2 and CYP2C8 polymorphisms and coronary heart disease risk: The Atherosclerosis Risk in Communities (ARIC) study. Pharmacogenet Genomics. 2007;17:349–358. doi: 10.1097/FPC.0b013e32809913ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesko LJ. Personalized medicine: Elusive dream or imminent reality? Clin Pharmacol Ther. 2007;81:807–816. doi: 10.1038/sj.clpt.6100204. [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de VM, Dixon A, Demarche B, Gut I, Heath S, Foglio M, Liang L, Laukens D, Mni M, Zelenika D, Van GA, Rutgeerts P, Belaiche J, Lathrop M, Georges M. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Walther D, Yu XY, Drgon T, Uhl GR. The human serotonin receptor HTR2B: Coding region polymorphisms and association with vulnerability to illegal drug abuse. Pharmacogenetics. 2004;14:805–811. doi: 10.1097/00008571-200412000-00003. [DOI] [PubMed] [Google Scholar]
- Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest. 2005;127:1232–1241. doi: 10.1378/chest.127.4.1232. [DOI] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, Eichler EE. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Inter-chromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Long X, Miano JM. Remote control of gene expression. J Biol Chem. 2007;282:15941–15945. doi: 10.1074/jbc.R700010200. [DOI] [PubMed] [Google Scholar]
- Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, Bourget K, Plagnol V, Field S, Atkinson M, Clayton DG, Wicker LS, Todd JA. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in Type 1 diabetes. Nat Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- Ma H, Xu L, Yuan J, Shao M, Hu Z, Wang F, Wang Y, Yuan W, Qian J, Wang Y, Xun P, Liu H, Chen W, Yang L, Jin G, Huo X, Chen F, Shugart YY, Jin L, Wei Q, Wu T, Shen H, Huang W, Lu D. Tagging single nucleotide polymorphisms in excision repair cross-complementing Group 1 (ERCC1) and risk of primary lung cancer in a Chinese population. Pharmacogenet Genomics. 2007;17:417–423. doi: 10.1097/01.fpc.0000239975.77088.17. [DOI] [PubMed] [Google Scholar]
- Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A, Amos W, Balloux F, Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–348. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, de A M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson Disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino C, Giorda R, Luisa LM, Vanzin L, Salandi N, Nobile M, Citterio A, Beri S, Crespi V, Battaglia M, Molteni M. A family-based association study does not support DYX1C1 on 15q21.3 as a candidate gene in developmental dyslexia. Eur J Hum Genet. 2005;13:491–499. doi: 10.1038/sj.ejhg.5201356. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Marvit J, DiLella AG, Brayton K, Ledley FD, Robson KJ, Woo SL. GT to AT transition at a splice donor site causes skipping of the preceding exon in phenylketonuria. Nucleic Acids Res. 1987;15:5613–5628. doi: 10.1093/nar/15.14.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen GK, Woolfe A, Goode D, Vavouri T, Callaway H, Elgar G. Ancient duplicated conserved noncoding elements in vertebrates: A genomic and functional analysis. Genome Res. 2006;16:451–465. doi: 10.1101/gr.4143406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA. The Gordon Wilson Lecture: The clinical legacy of Jonathan Hutchinson (1828-1913): Syndromology and dysmorphology meet genomics. Trans Am Clin Climatol Assoc. 2005;116:15–38. [PMC free article] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on Chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Spencer CC, Chaix R. Perspectives on human genetic variation from the HapMap Project. PLoS Genet. 2005;1:e54. doi: 10.1371/journal.pgen.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici M, van Meurs JB, Rivadeneira F, Zhao H, Arp PP, Hofman A, Pols HA, Uitterlinden AG. BMP2 gene polymorphisms and osteoporosis: The Rotterdam Study. J Bone Miner Res. 2006;21:845–854. doi: 10.1359/jbmr.060306. [DOI] [PubMed] [Google Scholar]
- Mekus F, Laabs U, Veeze H, Tummler B. Genes in the vicinity of CFTR modulate the cystic fibrosis phenotype in highly concordant or discordant F508del homozygous sib pairs. Hum Genet. 2003;112:1–11. doi: 10.1007/s00439-002-0839-7. [DOI] [PubMed] [Google Scholar]
- Meng H, Hager K, Held M, Page GP, Olson RK, Pennington BF, DeFries JC, Smith SD, Gruen JR. TDT-association analysis of EKN1 and dyslexia in a Colorado twin cohort. Hum Genet. 2005;118:87–90. doi: 10.1007/s00439-005-0017-9. [DOI] [PubMed] [Google Scholar]
- Metzger S, Bauer P, Tomiuk J, Laccone F, Didonato S, Gellera C, Mariotti C, Lange HW, Weirich-Schwaiger H, Wenning GK, Seppi K, Melegh B, Havasi V, Baliko L, Wieczorek S, Zaremba J, Hoffman-Zacharska D, Sulek A, Basak AN, Soydan E, Zidovska J, Kebrdlova V, Pandolfo M, Ribai P, Kadasi L, Kvasnicova M, Weber BH, Kreuz F, Dose M, Stuhrmann M, Riess O. Genetic analysis of candidate genes modifying the age-at-onset in Huntington's disease. Hum Genet. 2006;120:285–292. doi: 10.1007/s00439-006-0221-2. [DOI] [PubMed] [Google Scholar]
- Mignot E. A step forward for restless legs syndrome. Nat Genet. 2007;39:938–939. doi: 10.1038/ng0807-938. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: Digenic diseases and holoprosencephaly. Am J Hum Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- Morton LM, Wang SS, Bergen AW, Chatterjee N, Kvale P, Welch R, Yeager M, Hayes RB, Chanock SJ, Caporaso NE. DRD2 Genetic variation in relation to smoking and obesity in the prostate, lung, colorectal, and ovarian cancer screening trial. Pharmacogenet Genomics. 2006;16:901–910. doi: 10.1097/01.fpc.0000230417.20468.d0. [DOI] [PubMed] [Google Scholar]
- Motulsky AG. Drug reactions enzymes, and biochemical genetics. J Am Med Assoc. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- Myers RH. Considerations for genomewide association studies in Parkinson Disease. Am J Hum Genet. 2006;78:1081–1082. doi: 10.1086/504730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Yokoi T, Mizutani M, Kinoshita M, Funayama M, Kamataki T. Genetic polymorphism in the 5′-flanking region of human CYP1A2 gene: Effect on the CYP1A2 inducibility in humans. J Biochem (Tokyo) 1999;125:803–808. doi: 10.1093/oxfordjournals.jbchem.a022352. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Kugiyama K, Sugiyama S, Miyamoto S, Koide S, Fukushima H, Honda O, Yoshimura M, Ogawa H. Polymorphism in the 5′-flanking region of human glutamate-cysteine ligase modifier subunit gene is associated with myocardial infarction. Circulation. 2002;105:2968–2973. doi: 10.1161/01.cir.0000019739.66514.1e. [DOI] [PubMed] [Google Scholar]
- Nebert DW. Pharmacogenetics: 65 candles on the cake. Pharmacogenetics. 1997;7:435–440. [Google Scholar]
- Nebert DW. Proposed role of drug-metabolizing enzymes: Regulation of steady state levels of the ligands that effect growth, homeostasis, differentiation, and neuroendocrine functions. Mol Endocrinol. 1991;5:1203–1214. doi: 10.1210/mend-5-9-1203. [DOI] [PubMed] [Google Scholar]
- Nebert DW. Extreme discordant phenotype methodology: An intuitive approach to clinical pharmacogenetics. Eur J Pharmacol. 2000b;410:107–120. doi: 10.1016/s0014-2999(00)00809-8. [DOI] [PubMed] [Google Scholar]
- Nebert DW. Inter-individual susceptibility to environmental toxicants—A current assessment. Toxicol Appl Pharmacol. 2005;207:34–42. doi: 10.1016/j.taap.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Nebert DW. Suggestions for the nomenclature of human alleles: Relevance to ecogenetics, pharmacogenetics and molecular epidemiology. Pharmacogenetics. 2000a;10:279–290. doi: 10.1097/00008571-200006000-00001. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Ingelman-Sundberg M, Daly AK. Genetic epidemiology of environmental toxicity and cancer susceptibility: Human allelic polymorphisms in drug-metabolizing enzyme genes, their functional importance, and nomenclature issues. Drug Metab Rev. 1999;31:467–487. doi: 10.1081/dmr-100101931. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Jorge-Nebert L, Vesell ES. Pharmacogenomics and “individualized Drug Therapy”: High expectations and disappointing achievements. Am J Pharmacogenomics. 2003;3:361–370. doi: 10.2165/00129785-200303060-00002. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Robinson JR, Niwa A, Kumaki K, Poland AP. Genetic expression of aryl hydrocarbon hydroxylase activity in the mouse. J Cell Physiol. 1975;85:393–414. doi: 10.1002/jcp.1040850407. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Vesell ES. Advances in pharmacogenomics and individualized drug therapy: exciting challenges that lie ahead. Eur J Pharmacol. 2004;500:267–280. doi: 10.1016/j.ejphar.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Need AC, Goldstein DB. Genome-wide tagging for everyone. Nat Genet. 2006;38:1227–1228. doi: 10.1038/ng1106-1227. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Shigematsu H, Li L, Suzuki M, Takahashi T, Estess P, Siegelman M, Feng Z, Kato H, Marchetti A, Shay JW, Spitz MR, Wistuba II, Minna JD, Gazdar AF. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly RA, Aggeler PM. Coumarin anticoagulant drugs: Hereditary resistance in man. Fed Proc. 1965;24:1266–1273. [PubMed] [Google Scholar]
- Ohlendorff SD, Tofteng CL, Jensen JE, Petersen S, Civitelli R, Fenger M, Abrahamsen B, Hermann AP, Eiken P, Jrgensen NR. Single nucleotide polymorphisms in the P2X7 gene are associated to fracture risk and to effect of estrogen treatment. Pharmacogenet Genomics. 2007;17:555–567. doi: 10.1097/FPC.0b013e3280951625. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjaerg L, Konttinen Y, Peltonen L. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembrey ME. Time to take epigenetic inheritance seriously. Eur J Hum Genet. 2002;10:669–671. doi: 10.1038/sj.ejhg.5200901. [DOI] [PubMed] [Google Scholar]
- Polesskaya OO, Sokolov BP. Differential expression of the “C” and “T” alleles of the 5-HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. J Neurosci Res. 2002;67:812–822. doi: 10.1002/jnr.10173. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2007;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Beck S. Epigenetic variation and inheritance in mammals. Curr Opin Genet Dev. 2006;16:573–577. doi: 10.1016/j.gde.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneland RH, Mah S, Kammerer S, Hoyal CR, Marnellos G, Wilson SG, Sambrook PN, Spector TD, Nelson MR, Braun A. Association between a variation in the phosphodiesterase 4D gene and bone mineral density. BMC Med Genet. 2005;6:9. doi: 10.1186/1471-2350-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetics studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, Sheridan CM, Glusman G, Hood L, Lernmark A, Janer M. Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor-3 as a risk factor for type-1 diabetes in Sweden. Am J Hum Genet. 2006;79:614–627. doi: 10.1086/507876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, Krauss RM, McLeod HL, Ratain MJ, Relling MV, Ring HZ, Shuldiner AR, Weinshilboum RM, Weiss ST. Pharmacogenomics: Challenges and opportunities. Ann Intern Med. 2006;145:749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosand J, Bayley N, Rost N, de Bakker PI. Many hypotheses but no replication for the association between PDE4D and stroke. Nat Genet. 2006;38:1091–1092. doi: 10.1038/ng1006-1091. [DOI] [PubMed] [Google Scholar]
- Roses A. “Personalized medicine: Elusive dream or imminent reality?” A commentary. Clin Pharmacol Ther. 2007;81:801–805. doi: 10.1038/sj.clpt.6100227. [DOI] [PubMed] [Google Scholar]
- Rosvold EA, McGlynn KA, Lustbader ED, Buetow KH. Identification of an NAD(P)H:quinone oxidoreductase polymorphism and its association with lung cancer and smoking. Pharmacogenetics. 1995;5:199–206. doi: 10.1097/00008571-199508000-00003. [DOI] [PubMed] [Google Scholar]
- Sai K, Saeki M, Saito Y, Ozawa S, Katori N, Jinno H, Hasegawa R, Kaniwa N, Sawada J, Komamura K, Ueno K, Kamakura S, Kitakaze M, Kitamura Y, Kamatani N, Minami H, Ohtsu A, Shirao K, Yoshida T, Saijo N. UGT1A1 Haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther. 2004;75:501–515. doi: 10.1016/j.clpt.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan JL, Botella-Carretero JI, varez-Blasco F, Luque-Ramirez M, Sancho J, Moghetti P, Escobar-Morreale HF. A study of the hexose-6-phosphate dehydrogenase gene R453Q and 11β-hydroxysteroid dehydrogenase Type-1 Gene 83557insA polymorphisms in the polycystic ovary syndrome. J Clin Endocrinol Metab. 2005;90:4157–4162. doi: 10.1210/jc.2004-1523. [DOI] [PubMed] [Google Scholar]
- Sandhu MS, Weedon MN, Fawcett KA, Wasson J, Debenham SL, Daly A, Lango H, Frayling TM, Neumann RJ, Sherva R, Blech I, Pharoah PD, Palmer CN, Kimber C, Tavendale R, Morris AD, McCarthy MI, Walker M, Hitman G, Glaser B, Permutt MA, Hattersley AT, Wareham NJ, Barroso I. Common variants in WFS1 confer risk of Type 2 diabetes. Nat Genet. 2007;39:951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele E4 with late-onset familial and sporadic Alzheimer disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Scerri TS, Fisher SE, Francks C, MacPhie IL, Paracchini S, Richardson AJ, Stein JF, Monaco AP. Putative functional alleles of DYX1C1 are not associated with dyslexia susceptibility in a large sample of sibling pairs from the UK. J Med Genet. 2004;41:853–857. doi: 10.1136/jmg.2004.018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van der Knaap MS, Proud CG. Translation matters: Protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Hoffmann P, Schmal C, Schulte-Korne G, Nothen MM. Genetics of dyslexia: The evolving landscape. J Med Genet. 2007;44:289–297. doi: 10.1136/jmg.2006.046516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver CR. The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat. 2007;28:831–845. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J. Major changes in our DNA lead to major changes in our thinking. Nat Genet. 2007;39:S3–S5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- Seidegård J, Pero RW, Miller DG, Beattie EJ. A glutathione transferase in human leukocytes as a marker for the susceptibility to lung cancer. Carcinogenesis. 1986;7:751–753. doi: 10.1093/carcin/7.5.751. [DOI] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for Type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of B1- and A2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- Snyder LH. Studies in human inheritance. IX. The inheritance of taste deficiency in man. Ohio J Sci. 1932;32:436–468. [PubMed] [Google Scholar]
- Spielberg SP. In vitro assessment of pharmacogenetic susceptibility to toxic drug metabolites in humans. Fed Proc. 1984;43:2308–2313. [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramm G, Martinez JA, McCabe ER, Liao JC, Dipple KM. Single-gene disorders: What role could moonlighting enzymes play? Am J Hum Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de G A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007a;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavare S, Deloukas P, Dermitzakis ET. Population genomics of human gene expression. Nat Genet. 2007b;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkarsdottir U, Cazier JB, Kong A, Rolfsson O, Larsen H, Bjarnadottir E, Johannsdottir VD, Sigurdardottir MS, Bagger Y, Christiansen C, Reynisdottir I, Grant SF, Jonasson K, Frigge ML, Gulcher JR, Sigurdsson G, Stefansson K. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol. 2003;1:E69. doi: 10.1371/journal.pbio.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swen JJ, Huizinga TW, Gelderblom H, de Vries EG, Assendelft WJ, Kirchheiner J, Guchelaar HJ. Translating pharmacogenomics: Challenges on the road to the clinic. PLoS Med. 2007;4:e209. doi: 10.1371/journal.pmed.0040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, Muller K, Kaaranen M, Lindsberg PJ, Hannula-Jouppi K, Kere J. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci USA. 2003;100:11553–11558. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Caiment F, Smit M, Hiard S, Tordoir X, Cockett N, Georges M, Charlier C. The Callipyge mutation enhances bidirectional long-range DLK1-GTL2 intergenic transcription in Cis. Proc Natl Acad Sci USA. 2006;103:8119–8124. doi: 10.1073/pnas.0602844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner CM, Ottman R, Goldman SM, Ellenberg J, Chan P, Mayeux R, Langston JW. Parkinson disease in twins: An etiologic study. J Am Med Assn. 1999;281:341–346. doi: 10.1001/jama.281.4.341. [DOI] [PubMed] [Google Scholar]
- Tate SK, Goldstein DB. Will tomorrow's medicines work for everyone? Nat Genet. 2004;36:S34–S42. doi: 10.1038/ng1437. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE Pilot Project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The GAIN Collaborative Research Group. New models of collaboration in genome-wide association studies: the genetic association information network. Nat Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A Haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Haile RW, Duggan D. Recent developments in genomewide association scans: A workshop summary and review. Am J Hum Genet. 2005;77:337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of Tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous-stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- van Aken J, Schmedders M, Feuerstein G, Kollek R. Prospects and limits of pharmacogenetics: The thiopurine methyltransferase (TPMT) experience. Am J Pharmacogenomics. 2003;3:147–155. doi: 10.2165/00129785-200303030-00001. [DOI] [PubMed] [Google Scholar]
- van den Brink DM, Brites P, Haasjes J, Wierzbicki AS, Mitchell J, Lambert-Hamill M, de B J, Jansen GA, Waterham HR, Wanders RJ. Identification of PEX7 as the second gene involved in Refsum disease. Am J Hum Genet. 2003;72:471–477. doi: 10.1086/346093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesell ES. Polymorphism of human lactate dehydrogenase isozymes. Science. 1965;148:1103–1105. doi: 10.1126/science.148.3673.1103. [DOI] [PubMed] [Google Scholar]
- Vesell ES. New directions in pharmacogenetics: Introduction. Fed Proc. 1984;43:2319–2325. [PubMed] [Google Scholar]
- Vesell ES, Page JG. Genetic control of drug levels in man: Antipyrine. Science. 1968;161:72–73. doi: 10.1126/science.161.3836.72. [DOI] [PubMed] [Google Scholar]
- Vesell ES, Penno MB. A new polymorphism of hepatic drug oxidation in humans: Family studies of antipyrine metabolites. Fed Proc. 1984;43:2342–2347. [PubMed] [Google Scholar]
- Vogel F. Moderne probleme der humangenetik. Ergeb Inn Med Kinderheilkd. 1959;12:52–125. [Google Scholar]
- Von Wartburg JP, Bethune JL, Vallee BL. Human liver-alcohol dehydrogenase. Kinetic and physicochemical properties. Biochemistry. 1964;3:1775–1782. doi: 10.1021/bi00899a033. [DOI] [PubMed] [Google Scholar]
- Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- Wang Y, Krishnan HR, Ghezzi A, Yin JCP, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5:e265. doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber WW. Acetylation pharmacogenetics: experimental models for human toxicity. Fed Proc. 1984;43:2332–2337. [PubMed] [Google Scholar]
- Weinshilboum RM. Human pharmacogenetics of methyl conjugation. Fed Proc. 1984a;43:2303–2307. [PubMed] [Google Scholar]
- Weinshilboum RM. Human pharmacogenetics: Introduction. Fed Proc. 1984b;43:2295–2297. [PubMed] [Google Scholar]
- Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: Monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM. ENCODE: More genomic empowerment. Genome Res. 2007;17:667–668. doi: 10.1101/gr.6534207. [DOI] [PubMed] [Google Scholar]
- West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16:559–566. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- Wigle ED. Cardiovascular drugs in muscular subaortic stenosis. Fed Proc. 1965;24:1279–1286. [PubMed] [Google Scholar]
- Wilkie MJ, Smith D, Reid IC, Day RK, Matthews K, Wolf CR, Blackwood D, Smith G. A splice site polymorphism in the G-protein β subunit influences antidepressant efficacy in depression. Pharmacogenet Genomics. 2007;17:207–215. doi: 10.1097/FPC.0b013e32801a3be6. [DOI] [PubMed] [Google Scholar]
- Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, Fulda S, Putz B, Eckstein G, Hauk S, Trenkwalder C, Zimprich A, Stiasny-Kolster K, Oertel W, Bachmann CG, Paulus W, Peglau I, Eisensehr I, Montplaisir J, Turecki G, Rouleau G, Gieger C, Illig T, Wichmann HE, Holsboer F, Muller-Myhsok B, Meitinger T. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SL, Lidsky AS, Guttler F, Chandra T, Robson KJ. Cloned human phenylalanine hydroxylase gene allows prenatal diagnosis and carrier detection of classical phenylketonuria. Nature. 1983;306:151–155. doi: 10.1038/306151a0. [DOI] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O'Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- Zhang G, Nebert DW, Chakraborty R, Jin L. Statistical power of association using the extreme discordant phenotype design. Pharmacogenet Genomics. 2006;16:401–413. doi: 10.1097/01.fpc.0000204995.99429.0f. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet. 2007;38:1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]