Abstract
Objective
To determine changes in chondrocyte transcription of a range of anabolic, catabolic and signaling genes following simultaneous treatment of cartilage with IGF-1 and ramp-and-hold mechanical compression, and compare with effects on biosynthesis.
Methods
Explant disks of bovine calf cartilage were slowly compressed (unconfined) over 3-min to their 1mm cut-thickness (0%-compression) or to 50%-compression with or without 300 ng/ml IGF-1. Expression of 24 genes involved in cartilage homeostasis was measured using qPCR at 2, 8, 24, 32, 48 hours after compression ± IGF-1. Clustering analysis was used to identify groups of co-expressed genes to further elucidate mechanistic pathways.
Results
IGF-1 alone stimulated gene expression of aggrecan and collagen II, but simultaneous 24-hour compression suppressed this effect. Compression alone upregulated expression of MMP-3, MMP-13, ADAMTS-5 and TGF-β, an effect not reversed by simultaneous IGF-1 treatment. Temporal changes in expression following IGF-1 treatment were generally slower than that following compression. Clustering analysis revealed five distinct groups within which the pairings, TIMP-3 and ADAMTS-5, MMP-1 and IGF-2, and IGF-1 and Collagen II, were all robustly co-expressed, suggesting inherent regulation and feedback in chondrocyte gene expression. While aggrecan synthesis was transcriptionally regulated by IGF-1, inhibition of aggrecan synthesis by sustained compression appeared post-transcriptionally regulated.
Conclusion
Sustained compression markedly altered the effects of IGF-1 on expression of genes involved in cartilage homeostasis, while IGF-1 was largely unable to moderate the transcriptional effects of compression alone. The demonstrated co-expressed gene pairings suggest a balance of anabolic and catabolic activity following simultaneous mechanical and growth factor stimuli.
Keywords: IGF-1, Chondrocyte Gene Expression, Mechanical Compression, Growth Factor treatment, mechanobiology
Introduction
Insulin-like growth factor-1 (IGF-1, M.W. 7.6 kDa, pI = 8.5) is a potent anabolic factor capable of endocrine and paracrine/autocrine signaling. While IGF-1 is primarily produced in the liver and transported throughout the body via the blood stream, studies have shown that chondrocytes produce this important growth factor, and that IGF-1 can stimulate ECM biosynthesis by chondrocytes in native cartilage and tissue engineered constructs. The use of growth factors as therapeutics to reverse or inhibit cartilage degradation has been an important focus in cartilage research. The avascular and alymphatic nature of cartilage suggests the need for local delivery, which is complicated since diffusion rates and penetrance into tissue are affected by joint motion and cartilage structure[1-4].
Previous studies with free swelling explants in vitro have delineated certain effects of exogenous IGF-1 on chondrocyte protein biosynthesis and gene transcription. IGF-1 can stimulate chondrocyte synthesis of ECM in a dose dependent manner[5-10]. While culture conditions have varied, (e.g., explants, cell-seeded gels, cell monolayers), chondrocyte biosynthesis levels increased from 1.5- [8] to 2-6 fold over[7,10-12] control levels. IGF-1 can also inhibit production of specific proteins such as MMP-13[13]. At the gene expression level, type II collagen was significantly up-regulated by IGF-1[7,14-16]. Aggrecan transcripts showed no[7,8] or slight up-regulation by 48 hours of IGF-1 treatment[16], though moderate up-regulation (130%) after 1-3 weeks[14]. The transcription factor, Sox-9, also showed no significant response to IGF-1[15]. Interestingly, IGF-1 transcript levels peaked at 24 hours after exogenous IGF-1 treatment, suggesting an autocrine response[5].
Chondrocytes are highly responsive to mechanical compression at both protein and gene transcript levels. Recent studies of cartilage deformation in vivo using combined dual fluoroscopic and MR imaging technique with healthy human subjects showed that within 20s of steady, full body-weight loading, contact compressive strains within ankle cartilage increased to 24-38%, and static creep equilibrium was reached by ~50s[17]. This transient-to-static deformation in vivo provides an important benchmark for the ranges of stresses, strains and strain rates applied to cartilage specimens in vitro in studies of chondrocyte mechanotransduction. Static compression of cartilage explants[9,18], chondrocyte-seeded alginate and type I collagen gels[19,20] decreased synthesis of collagen and proteoglycans in a dose-dependent manner. In explants, this inhibition occurred within 1-2 hours of load application[21,22]. In contrast, static compression caused a transient increase in aggrecan and type II collagen transcripts for 1-4 hours post-compression, followed by decreased expression to control levels[18,20,23,24]. Fitzgerald[24] applied a ~3 min ramp compression to cartilage explants and found time-dependent gene expression patterns during 24-hours of subsequent static compression that depended on intracellular calcium and cyclic AMP for a range of ECM molecules, proteinases, TIMPs, cytokines, growth and transcription factors.
Relevant to the present study, Bonassar[9] found a 2-3 fold increase in proteoglycan and protein synthesis when cartilage explants were treated with 300 ng/ml IGF-1 at 0%-compression (cut thickness) for 48 hour. Under 50% static compression alone, biosynthesis decreased by 50% compared to 0%-compressed controls. However, when 50%-compressed explants were treated with IGF-1, biosynthesis rates significantly increased back to levels comparable to non-compressed, non-IGF-1-treated controls. Thus, compression diminished but did not eliminate the effects of IGF-1, and IGF-1 could up-regulate synthesis rates in statically compressed explants. Plumb[25] reported similar trends using elderly human cartilage. Taken together, these findings have suggested that chondrocyte biosynthetic response to simultaneous mechanical and biochemical stimuli can occur through separate cellular pathways. However, the corresponding changes in gene expression in response to the physiologically relevant combinations of growth factor (IGF-1) treatment and compression have not been studied, to our knowledge. Thus, our objectives were (1) to elucidate and compare the changes in chondrocyte transcription of a range of anabolic, catabolic and signaling genes following stimulation by compression alone, IGF-1 alone, or combined treatment with IGF-1 and sustained compression in cartilage explants, and (2) to test the hypothesis that IGF-1 treatment combined with sustained compression causes co-expression of gene groupings indicative of attempts by chondrocytes to maintain homeostasis.
Methods
Cartilage Harvest, Compression, and Growth Factor Treatment
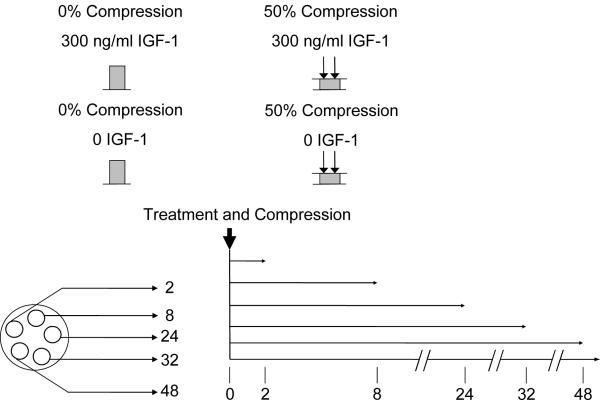
Cartilage-bone plugs were harvested from the patellofemoral grooves of 1-2 week old calves (Research 87, Hopkinton, MA). Cartilage disks (1mm-thick X 3mm-diameter) were cored and punched from the middle zone as described previously[24] and equilibrated for two days under free-swelling conditions in serum-free medium (high-glucose Dulbecco's modified essential medium with 10 mM Hepes Buffer, 0.1 mM nonessential amino acids, 20 μg/ml ascorbate, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B) (Sigma, St. Louis). Five anatomically matched disks were separated for each time point (Fig. 1), and placed in polysulfone loading chambers[24]. Each time point included four separate conditions: 8 disks each were allocated to 0%-compression (uniaxial unconfined compression to 1-mm cut-thickness from free swelling), 50%-compression, 0%-compression + 300 ng/ml IGF-1, and 50%-compression + 300 ng/ml IGF-1, the concentration found previously to be the lowest that maximally stimulated similar free-swelling calf cartilage explants[9].
Figure 1.
A schematic of the four conditions measured. 5 disks were cored for each time point and matched for time. IGF-1 treatment and static compression were applied at time 0, and disks were flash frozen at 2, 8, 24, 32, and 48 hours.
At time zero, all chambers were slowly compressed in a ramp-and-hold fashion to 50% strain over a 3-minute period to avoid injurious effects of high strain rates. Disks were maintained at 50%-compression for 2,8,24,32, and 48 hours for each of the four conditions (Fig. 1). The disks exhibited a modest increase in stress during application of slow compression, followed by a slow (5-10 min) stress relaxation associated with the poroelastic properties of cartilage. Thus, the five time points were chosen to capture the kinetics of gene expression associated with both the initial transient and final equilibrium components of compression[24]. At each time point, disks were promptly removed, flash frozen in liquid nitrogen, and stored at -80° C. This entire experiment, using 160 disks from 1 animal (2 joints), was repeated 3 more times (4 animals total.)
RNA Extraction and Quantization, Primer Design, and qPCR
The 8 frozen disks from each time point/condition were pulverized in a liquid nitrogen cooled apparatus to prevent RNA degradation. RNA extraction and quantitation were performed as previously described[26]. Forward and reverse primers for 24 relevant genes (Table 1) were designed and qPCR was performed as described[24]. Measured threshold values (Ct) were converted to RNA copy number according to previously calculated standard curves.
Table 1.
24 Cartilage relevant genes. Primers were designed Primer3 software (www.genome.wi.mit.edu/cgi-bin/primer/primer3www.cgi). Standard dilutions were used to calculate relative mRNA copy number.
| Matrix Molecules | Proteases | Protease inhibitors | Cytokines | Growth Factors | Transcription Factors | Stress Activated Genes | Housekeeping Gene |
|---|---|---|---|---|---|---|---|
| Type II Collagen | MMP1 | Timp-1 | TNF-a | IGF-1 | c-Jun | HSP90 | 18s |
| Aggrecan | MMP3 | Timp-2 | IL-1 | IGF-2 | c-Fos | Txnip | |
| Link Protein | MMP13 | Timp-3 | IL-4 | TGF-B | Sox-9 | ||
| Fibronectin | ADAMTS-5 | IL-6 |
Data Normalization and Statistical Analyses
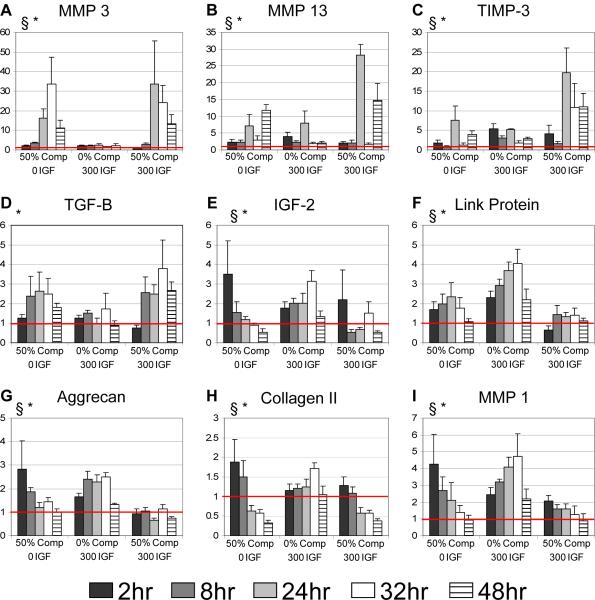
Under each loading condition and time point, each gene RNA copy number was normalized to the 18s housekeeping gene from that same condition and time point[24,26]. Expression levels due to experimental error, as defined by 5σ from mean, were removed. Mixed-effect modeling: The effects of compression and IGF-1 treatment on expression of 23 genes at each time point were evaluated using linear mixed-effects models (R software). The mixed-effect framework allows explicit incorporation of correlations among observations in an experimental design via random effects, which leads to more efficient estimates[27]. Independent crossed-random effects for animal, compression, IGF-1, time, and interactions were considered. Likelihood ratio tests were used to assess the contribution of the random effects to the corresponding model goodness of fit for each gene. Random effects that contributed most to the models' goodness of fit across all genes were selected for the final mixed-effects models. The significance of fixed effects for each gene was also evaluated by likelihood ratio test. Restricted maximum likelihood (REML) was used to estimate the parameters of the fixed effects and the variances of the random effects in the final models for each gene. To represent the time course of gene expression graphically (e.g., Fig. 2), the conditions 0%-compression + IGF-1, 50%-compression, and 50%-compression + IGF-1 were normalized to 0%-compression levels. Thus, gene expression values below or above 1 represent a decrease or increase in expression, respectively, compared to 0%-compression. We note that all mixed effects model analysis was performed on log-transformed data that were not normalized to the 0%-compression, 0-IGF-1 controls and thereby accounts for any variation of control data across time points.
Figure 2.
Gene expression of proteinases, growth factors, and ECM molecules. 8 cartilage disks were pooled for each time point for each experiment. All genes were normalized to 18s and plotted relative to 0%-compression 0 IGF-1. Significance of the fixed effects from the mixed model were calculated. § denotes a significant effect of IGF-1 on gene expression; * denotes a significant effect of compression on gene expression. (p-values < 0.05) Mean ± SE (n=4 animals)
Clustering and Principle Component Analyses
In addition to measuring changes in the expression magnitude of each gene, we further explored patterns of co-expression using principle component analysis (PCA, a tool that reveals internal structure of the data in a way which best explains the variance in the data), and clustering analysis performed on the 3 normalized conditions (0%-compression + IGF-1, 50%-compression, and 50%-compression + IGF-1) and the 5 time points (2,8,24,32,48 hours) over 23 genes. This resulted in a 3×5×23 matrix which was standardized by expression amplitude[24]. After normalization of the data using PCA, the 15-dimensional space was reordered according to greatest dimensional variance, in which the first three detentions (principle components) represent 80% of the variance in the data[28,29]. Once the 15 principle components had been calculated, a k-means clustering algorithm was applied to cluster the components into k groups. The average and variance of each projected coordinate group was calculated to give the group centroid. Centroid vectors were formed by combining the three main principal components weighted by their projected centroid coordinate. Uniqueness of each group's expression patterns was evaluated by student's T-test.
Results
The expression of 24 genes (Table 1) measured at each of the five time points was compared to controls at 0%-compression with no added IGF-1. To more easily focus on specific pathways of interest, selected results for specific genes are reported in Fig. 2; the complete results for all genes are given in Appendix A (Supplemental Material). The significance of the fixed effects of IGF-1, 50% compression, time, and interaction effects for each gene are presented in Table 2.
Table 2.
Significance of the fixed effects (IGF-1, Compression, Time) from the mixed-effects model of gene expression, and for interactions (IGF-1 and Compression; Time and Compression)
| IGF-1 | 50%-Compression | Time | IGF-1 and 50%-Compression | Time and 50%-Compression | |
|---|---|---|---|---|---|
| TGF-B | - | < 0.0001 | < 0.0001 | - | 0.0008 |
| TNF-a | < 0.0001 | < 0.0001 | 0.0419 | 0.0010 | - |
| IL-6 | - | - | - | - | - |
| IL-4 | - | - | - | - | - |
| IL-1 | < 0.0001 | < 0.0001 | - | < 0.0001 | - |
| IGF-2 | < 0.0001 | < 0.0001 | - | 0.0002 | - |
| IGF-1 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0192 | 0.0004 |
| c-Jun | < 0.0001 | < 0.0001 | < 0.0001 | 0.0058 | 0.0001 |
| c-Fos | < 0.0001 | < 0.0001 | < 0.0001 | 0.0002 | 0.0010 |
| Sox-9 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0224 |
| HSP90 | < 0.0001 | < 0.0001 | - | < 0.0001 | - |
| Txnip | < 0.0001 | < 0.0001 | < 0.0001 | 0.0015 | 0.0138 |
| TIMP-3 | 0.0005 | < 0.0001 | < 0.0001 | - | 0.0001 |
| TIMP-2 | 0.0156 | < 0.0001 | < 0.0001 | - | 0.0038 |
| TIMP-1 | - | - | - | - | - |
| ADAMTS-5 | - | 0.0083 | 0.0019 | - | - |
| MMP-13 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0014 |
| MMP-3 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0031 | < 0.0001 |
| MMP-1 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0011 |
| Link | < 0.0001 | < 0.0001 | - | < 0.0001 | - |
| Fibronectin | - | < 0.0001 | < 0.0001 | - | 0.0104 |
| Collagen II | < 0.0001 | < 0.0001 | < 0.0001 | 0.02 | < 0.0001 |
| Aggrecan | < 0.0001 | < 0.0001 | - | < 0.0001 | - |
Effects of compression alone
Consistent with previous studies[24], aggrecan and collagen II were both up-regulated at early times and then decreased to control values (aggrecan) or below controls (collagen II) at later times (Fig. 2G,H). Genes that were consistently up-regulated over time included MMP-3, MMP-13, TGF-β (Fig. 2), and ADAMTS-5, c-Fos, c-Jun, and Sox-9 (Appendix A). Peak expression of MMP-3 by 30-fold occurred at 32 hours (Fig. 2A). ADAMTS-5 and MMP-13 increased over time, peaking at 48 hours by >6-fold and >17-fold, respectively (Fig. 2B, Appendix A). TGF-β was up-regulated 2.5-fold from 8-32 hours (Fig. 2D). Also consistent with previous data[24], c-Fos and c-Jun were up-regulated in response to 50%-compression at all times; c-Fos, c-Jun and Sox-9 up-regulation peaked at 8 hours (23-fold, 30-fold, and 6.8-fold, respectively (Appendix A). Interestingly, IGF-1 was the only gene that was significantly down-regulated by 50% compression (by 50-70%, Appendix A). Significant interaction between 50% compression and time (Table 2) suggested that the temporal evolution of expression of many genes behaved very differently in the presence of compression.
Effects of IGF-1 alone
As expected, treatment with 300 ng/mL IGF-1 under 0%-compression caused up-regulation of anabolic-induced genes, aggrecan and link protein, as well as TIMP-3. Link and aggrecan were up-regulated with time, peaking at 32 hours (4-fold and 2.5-fold respectively), and still above control levels at 48 hours (Fig. 2F,G). These results agree with previous gene expression data showing up-regulation of aggrecan and collagen II by 18 hrs after 100 ng/ml IGF-1 treatment of bovine articular chondrocytes seeded in monolayer[30]. Unexpectedly, collagen II was not as dramatically up-regulated compared to a previous study[30]. IGF-1 also caused up-regulation at three or more time points of the proteases and cytokines, MMP-13, MMP-1, TNF-α, and IL-1β, which are thought to play catabolic/remodeling roles in cartilage. MMP-13, TNF-α, and IL-1β peaked at 24 hours to 8-fold, 2.5-fold, and 2.75-fold, respectively, above no-IGF-1 controls (Fig. 2B, Appendix A). MMP-1 and IGF2 were up-regulated, peaking at 32 hours (4.75-fold and 3-fold, respectively), after which expression returned to control levels (Fig. 2I,E). As with 50% static compression, Sox-9 was up-regulated at 8 hours to a comparable level of 6.3-fold (Appendix A). Finally, TIMP-3 and HSP90 were up-regulated at all time points, and both displayed an initial transient peak in expression (5-fold and 3-fold, respectively), followed by a decrease over time (Fig. 2C, Appendix A). No genes measured were down-regulated by treatment with IGF-1 up to 48 hours.
Combined Effects of IGF-1 and compression
Simultaneous treatment with 300 ng/mL IGF-1 and 50%-compression caused significant interaction in the expression of 15 of the 23 genes tested (Table 2), including MMP-3, MMP-13 (Fig. 2), and c-Fos, c-Jun, and HSP90 (Appendix A). MMP-3 and MMP-13 were up-regulated to peak values of ~30-fold at 24 hours (Fig. 2A,B), while TGF-β and HSP90 were upregulated to peak levels of ~2-4-fold at 32 hours (Fig. 2D, Appendix A). c-Fos and c-Jun were up-regulated at all time points (Appendix A). TIMP-3 showed a slow increase of expression, peaking at 24 hours to ~20-fold (Fig. 2C). Collagen II, IGF-2, TIMP-2, and Txnip (an oxidative stress mediator) were down-regulated at multiple time points. IGF-2 and Txnip showed a sustained decrease over time, reaching minima of 45% and 50% below controls by 48 hours (Fig. 2E, Appendix A). Collagen II and TIMP-2 were steadily down-regulated to 60% and 90%, respectively, by 48 hours (Fig. 2H, Appendix A).
Expression Trends and Groupings
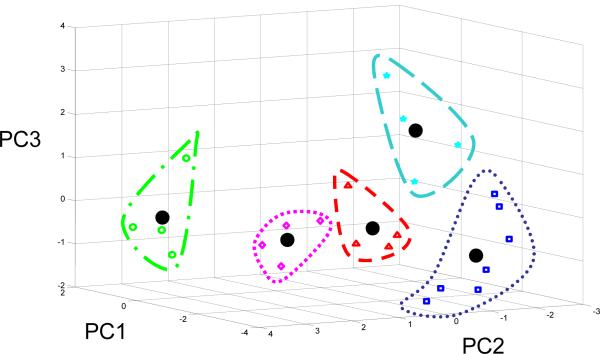
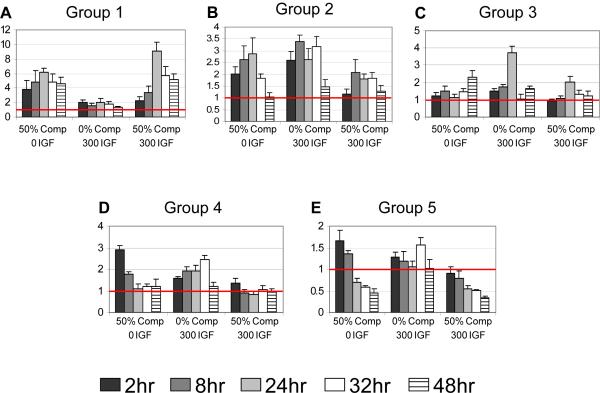
Using the three principle components obtained by PCA, each standardized gene was projected in to the principle component space as visualized in Fig. 3. All 15-dimensions of each gene were used in clustering analyses to ensure that smaller gene variations were represented in the grouping. After dividing the genes into 2-8 clusters and visually comparing their distinctness, 5 groups appeared to be an adequate number (Fig. 3), which was further verified using silhouette graphical scoring[31]. These 5 groups each contained 4-7 genes (Table 3): the mean expression level is represented by a centroid (Fig. 3) and the mean expression profile is shown in Fig. 4. The centroids were found to be significantly separated by taking the Euclidean distance between centroids and calculating gene-to-centroid variance[24] (Table 4).
Figure 3.
Standardized gene expression visualized in principle component space. Principle component 1, 2, and 3 represent 80% of the variance in the data. Genes were allocated to one of five distinct groups by way of k-means clustering. Large solid black circles denote the centroid of the corresponding group.
Table 3.
Gene clustering groupings. Resulted gene sorting according to extent and kinetics of expression. Specific gene allocation and centroid coordinates when all data are clustered.
| Group | Genes | Centroid Coordinates (PC1,PC2, PC3) |
|---|---|---|
| 1 | TGF-B, c-Fos, c-Jun, Timp-3, ADAMTS-5, MMP13, MMP3 | (-2.46, 2.37, -0.33) |
| 2 | Sox-9, HSP90, Timp-1, Link | (-3.10, 0.95, -0.07) |
| 3 | TNF-a, IL-1, IL-4, IL-6 | (-2.78, -1.50, -0.94) |
| 4 | IGF-2, MMP1, Fibronectin, Aggrecan | (-2.71, -0.24, 2.03) |
| 5 | IGF-1, Txnip, Timp-2, Collagen II | (0.32, 3.21, -0.19) |
Figure 4.
Five expression profiles represent the combination of 0%-compression 300 ng/ml of IGF-1, 50%-compression 0 ng/ml of IGF-1, and 50%-compression 300 ng/ml of IGF-1. Centroid profiles were calculated through the average projection coordinates of genes in each group, and transformed from principle component space through use of the calculated principle components. Mean ± SE (n varies based on group component number)
Table 4.
P-value of centroid profile separation. P-values were obtained through student T-test, comparing centroid to centroid Euclidean distance. Degrees of freedom were taken as the number of genes in each group.
| P-value of Centroid Separation | Centroid 1 | Centroid 2 | Centroid 3 | Centroid 4 |
|---|---|---|---|---|
| Centroid 2 | 0.032 | |||
| Centroid 3 | 0.017 | 0.034 | ||
| Centroid 4 | 0.007 | 0.073 | 0.016 | |
| Centroid 5 | 0.001 | 0.015 | 0.006 | 0.042 |
Group-1 genes (Table 3) were substantially up-regulated for all conditions and all time points (Fig. 4A). However, their response was particularly stimulated by 50%-compression. Group-2 genes showed early up-regulation for all conditions, followed by a return to control levels by 48 hours (Fig. 4B). In contrast, all the inflammatory cytokines grouped together (group 3) and showed substantial up-regulation by 24 hrs after IGF-1 alone (0%-compression) (Fig. 4C). This stimulatory effect of IGF-1 on cytokines was lessened by 50%-compression. Group-4 genes were initially up-regulated, then returned to control levels following 50%-compression with no IGF-1, opposite time evolution following IGF-1 at 0%-compression, and no change to combined IGF-1 plus 50%-compression (Fig. 4D). Group-5 genes showed decreasing expression with time at 50%-compression with or without IGF-1, though compression alone was initially stimulatory (Fig. 4E). IGF-1 alone caused minimal response.
To gain additional insight into the transcriptional responses to each treatment, all genes were additionally clustered by their response to IGF-1 alone, 50%-compression alone, and the combination of IGF-1 plus compression. The resulting gene groupings (Tables in Supplementary Appendix B) can be compared with clustering of all conditions and time points together (Table 3). Under IGF-1 alone (Appendix B.1), aggrecan, link and type II collagen grouped together as substantially upregulated, while Txnip, the only gene significantly down-regulated by IGF-1 treatment for multiple time points, was uniquely grouped. Clusters associated with compression alone (Appendix B.2) isolated IL-6 in its own group as highly non-responsive to compression. In addition, the highly up-regulated transcription factors grouped together, while transiently up-regulated proteinases grouped separately; both these groupings support previous reports on effects of compression alone[24]. Following combined compression plus IGF-1 (Appendix B.3), fibronectin partitioned to a unique group as non-responsive at all times, and the majority of proteinases also grouped together.
Taken together, we examined four different sets of clustered data: compression alone, IGF-1 alone, compression + IGF-1 (Appendix B), and all conditions and times clustered together (Table 3, Fig. 4). Importantly, three gene-pairings were consistently observed in all these multiple clustering approaches: MMP-1 and IGF-2; TIMP-3 and ADAMTS-5; and type II collagen and IGF-1.
Discussion
In this study, we demonstrated that 48-hour sustained compression could markedly alter the effects of IGF-1 on expression of groups of genes involved in cartilage homeostasis, while IGF-1 was essentially unable to moderate certain transcriptional effects of compression alone. Temporal changes in expression following IGF-1 treatment were generally slower than those following compression. Our results enable comparison of the anabolic transcriptional response caused by IGF-1 under 0%-compression with the mixed anabolic and catabolic signals observed under IGF-1 coupled with 50%-compression. In addition, through PCA and clustering analyses, major co-expression trends were elucidated, grouping genes into highly responsive, non-responsive, and differentially active profiles (Fig. 4). Interestingly, aggrecan, link protein, and type II collagen transcription responded to IGF-1 in a compression-dependent manner, while fibronectin responded to IGF-1 in a compression-independent manner (Fig. 2F,G,H; Table 2). The gene pairs MMP-1 and IGF-2, TIMP-3 and ADAMTS-5, and type II collagen and IGF-1 were consistently co-expressed in most conditions, suggesting co-regulating and/or control relationships between members of each pair.
Effects of Compression on Transport and Intratissue concentration of IGF-1
When examining the extent and kinetics of the transcriptional responses to combined IGF-1 and compression, we note that both treatments were initiated simultaneously (Fig. 1). It is therefore important to consider whether compression could significantly alter transport of IGF-1 into the cartilage disks and, ultimately, the binding of IGF-1 to chondrocyte receptors. This question is also relevant to transport processes during joint loading in vivo. In a previous study using this explant and loading system, Bonassar[9] directly measured the transport of 125I-IGF-1 into same-sized disks of bovine calf cartilage under 0% and 50% radially unconfined compression. They reported a small but significant slowing of the characteristic diffusion time of 125I-IGF-1 into the tissue, from 10.0 hrs at 0% to 12.0 hrs at 50%-compression, and a small but significant decrease (12%) in the final intratissue concentration of IGF-1 caused by 50% relative to 0%-compression. However, when using 300 ng/ml of IGF-1 in the medium (as in the present study), such a small decrease in intratissue IGF-1 concentration caused little or no effect on proteoglycan biosynthesis (via 35S-sulfate incorporation)[9]. Here, we found that aggrecan and link protein gene expression were up-regulated by IGF-1 at 0%-compression, but dramatically down-regulated by IGF-1 at 50%-compression (Fig. 2G,F). However, the transcriptional effects of IGF-1 on TIMP-3 and fibronectin at 0% compression were not substantially altered by 50%-compression (Fig. 2C,H, Appendix A). Taken together, it is unlikely that the compression-dependence of aggrecan and link protein gene expression could be ascribed to effects of compression on transport of IGF-1.
Kinetics of aggrecan transcription vs biosynthesis
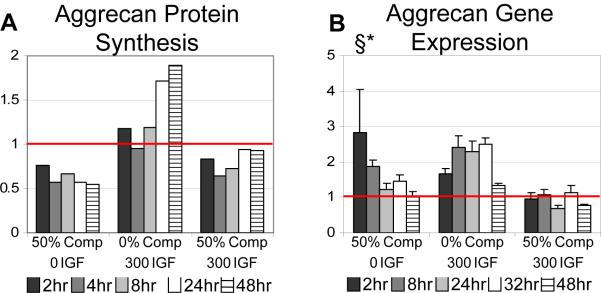
The transcription-time course data of Fig. 2 also enable quantitative comparison of the kinetics of gene expression versus protein synthesis in response to ±loading ±IGF-1; we focus on aggrecan as an important example (Fig. 5). At 0%-compression, 300 ng/ml IGF-1 increased aggrecan gene expression over the entire 2-48 hours of treatment compared to no-IGF-1 controls (Fig. 5B) and, correspondingly, increased aggrecan biosynthesis ~2-fold by 24-48 hours (Fig. 5A, adapted from[8]). Together, these data suggest that IGF-1 transcriptionally regulates aggrecan synthesis in uncompressed cartilage. In contrast, application of 50%-compression with no IGF-1 caused a significant decrease in aggrecan biosynthesis within the first 2 hours of compression (Fig. 5A), and this reduction was sustained during 48 hours of sustained compression. However, 50%-compression with no IGF-1 upregulated aggrecan gene transcription during at least the first 8 hours of compression (Fig. 5B), followed by a return to 0%-compression/no-IGF-1control levels by 24 hours. In a separate study[32], the time constant for such compression-induced inhibition of proteoglycan synthesis was calculated to be 40 min, again much shorter than the >8-hour period of increased aggrecan transcription. These results support the hypothesis[9] that decreased aggrecan synthesis caused by sustained compression with no IGF-1 is regulated by post-transcriptional machinery, e.g., possibly related to dramatic changes in chondrocyte rough endoplasmic reticulum and Golgi organelles caused by compression of cartilage[33].
Figure 5.
Aggrecan protein synthesis compared to aggrecan gene expression. (A) Aggrecan protein synthesis as measured by 35S radiolabel incorporation normalized to 0%-compression 0 IGF-1 adapted from Bonassar et al. [9]. Mean plotted, significance taken from Bonassar. (B) Aggrecan gene expression normalized to 18s and plotted relative to 0%-compression 0 IGF-1. § denotes a significant effect of IGF-1 on gene expression; * denotes a significant effect of compression on gene expression; see Table 2. (p-values < 0.05) Mean ± SE (n=4 animals)
Comparison to Previous Studies
The observation that collagen II and aggrecan were initially upregulated but subsequently down-regulated to nearly control levels by 24-48 hrs of 50%-compression alone (Fig. 2G,H) is consistent with the literature[18,20,23,24]. While IGF-1 caused upregulation of collagen II expression by isolated chondrocytes from young goat (8 months)[14] and normal adult human articular cartilage[7,15], we found only modest upregulation of collagen II by IGF-1 treatment of explants maintained at the control 0%-compression (Fig. 2H, Table 2). These differences may be due to cartilage age, species, and/or the response of isolated cells versus intact tissue. Similarly, MMP-1, MMP-3, and MMP-13 expression by isolated adult human chondrocytes was unaffected by addition of IGF-1 after 48 hours[7,15], while IGF-1 treatment of immature bovine explants, here, caused a substantial increase in MMP-1 and MMP-13 over 4-5 time points (Fig. 2I,B). IGF-1 mRNA levels were maximum at 32 hours (Appendix A); similarly, previous studies using monolayer cultures of immature equine chondrocytes cultured in 100 ng/ml IGF-1 reported peak IGF-1 mRNA levels at 24 hours within a 72-hour culture period[5]. Sox-9 expression was significantly upregulated by IGF-1 (Appendix A), while previous studies[15] using monolayers of adult human chondrocytes showed little change in Sox-9 at 72 hours of culture with 100 ng/mL IGF-1.
Co-Expressed Gene Pairs - TIMP-3 and ADAMTS-5
TIMPs act in a stochiometric fashion to reversibly inhibit MMPs[34]. Of the four TIMPs(1-4), TIMP-3 is a strong inhibitor of aggrecanase-1 (ADAMTS-4) and aggrecanase-2 (ADAMTS-5), with Ki values in the subnanomolar range[35,36]. When added exogenously to bovine nasal and porcine cartilage, TIMP-3 inhibited aggrecanase activity induced by catabolic factors[37]. In the present study, TIMP-3 was significantly up-regulated by exogenous IGF-1 in a compression-independent manner (Fig. 2C). Although aggrecan can be degraded by members of the MMP family, ADAMTS-5 was recently reported to be the primary aggrecanase responsible for aggrecan degradation in a murine model of osteoarthritis[38]. The finding that TIMP-3 and ADAMTS-5 expression profiles were grouped together in multiple clustering approaches suggests a biological control of aggrecan turnover, and possibly regulation of anabolic and catabolic factors that govern this turnover. Consistent with this hypothesis, ADAMTS-5 and TIMP-3 were also reported to cluster together in a previous study of the transcriptional effects of 50%-compression alone (no IGF-1) in the presence and absence of an intracellular calcium chelator (BAPTA-AM) and an inhibitor of cyclic AMP-activated protein kinase A (Rp-cAMP) used to identify mechanotransduction mechanisms[24].
MMP-1 and IGF-2
MMP-1 (collagenase-1) cleaves key ECM molecules including collagen II, fibronectin, and link protein[39]. MMP-1 also plays a role in the regulation of paracrine signals through the degradation of cytokines such as IL-1β[40]. MMP-1 can also degrade IGF binding proteins (IGFBP-3,5) which can indirectly increase the presence of free (unbound) IGFs. IGF-1 and IGF-2 bind IGFBP-3, the most abundant IGF binding protein in human serum[41]. IGF-2 stimulates DNA and proteoglycan synthesis by chondrocytes[42], and can act in an autocrine fashion[43], e.g., by stimulating type-1 IGF receptor (a key receptor for IGF-1 and, with lesser affinity, IGF-2)[44,45]. The observed co-expression of MMP-1 and IGF-2 under all clustering approaches suggests a balance through their known anabolic and catabolic activities and, additionally, the capability of a combined anabolic response due to the stimulatory effects of MMP-1 degradation of IGFBPs[46]. The co-expression of MMP-1 and IGF-2 has recently been observed in tumorigenesis (via microarray analysis)[47]: MMP-1 associated with invasion and IGF-2 with cell proliferation. To our knowledge, this is the first observation of such clusters of co-expression in chondrocytes subjected to exogenous IGF-1 and static compression.
Collagen II and IGF-1
Type II collagen adds structure and strength to cartilage, and IGF-1 can elevate levels of type II collagen synthesis and gene expression under different conditions[7,14,48]. The co-expression of type II collagen and IGF-1 under all compression and/or IGF-1 treatments suggests a positive feedback loop between them. Under IGF-1 treatment both are up-regulated (Fig. 2), while compression down-regulates both, consistent with data suggesting that IGF-1 acts in an autocrine fashion[5,44]. Further studies using promoter analysis must be performed to confirm if these pairings are co-expressed.
The combined effects of sustained compression and IGF-1 treatment on transcription of genes involved in ECM maintenance have been demonstrated in this study. It is possible to speculate that growth factor treatment for cartilage repair in vivo must account for the effects of cartilage loading on growth factor-mediated chondrocyte transcription. Ongoing studies focus on the effects of growth factor treatment of cartilage following injurious compression that might result from joint injury in vivo.
Supplementary Material
Acknowledgements
Funded by NIH Grant R01-AR33236 (AJG), R01-HG003352 (DMR) and P42-ES04699 (DMR); CAW was supported by the NIH Training Grant T32-EB006348.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunziker EB. Articular cartilage repair: basic science and clinical progress. A review of the current status and prospects. Osteoarthritis Cartilage. 2002;10:432–63. doi: 10.1053/joca.2002.0801. [DOI] [PubMed] [Google Scholar]
- 2.Trippel SB, Ghivizzani SC, Nixon AJ. Gene-based approaches for the repair of articular cartilage. Gene Ther. 2004;11:351–9. doi: 10.1038/sj.gt.3302201. [DOI] [PubMed] [Google Scholar]
- 3.Gelse K, von der Mark K, Aigner T, Park J, Schneider H. Articular cartilage repair by gene therapy using growth factor-producing mesenchymal cells. Arthritis Rheum. 2003;48:430–41. doi: 10.1002/art.10759. [DOI] [PubMed] [Google Scholar]
- 4.Grande DA, Mason J, Light E, Dines D. Stem cells as platforms for delivery of genes to enhance cartilage repair. J Bone Joint Surg Am. 2003;85-A(Suppl 2):111–6. doi: 10.2106/00004623-200300002-00015. [DOI] [PubMed] [Google Scholar]
- 5.Nixon AJ, Saxer RA, Brower-Toland BD. Exogenous insulin-like growth factor-I stimulates an autoinductive IGF-I autocrine/paracrine response in chondrocytes. J Orthop Res. 2001;19:26–32. doi: 10.1016/S0736-0266(00)00013-9. [DOI] [PubMed] [Google Scholar]
- 6.Sah RL, Chen AC, Grodzinsky AJ, Trippel SB. Differential effects of bFGF and IGF-I on matrix metabolism in calf and adult bovine cartilage explants. Arch Biochem Biophys. 1994;308:137–47. doi: 10.1006/abbi.1994.1020. [DOI] [PubMed] [Google Scholar]
- 7.Bau B, McKenna LA, Soeder S, Fan Z, Pecht A, Aigner T. Hepatocyte growth factor/scatter factor is not a potent regulator of anabolic and catabolic gene expression in adult human articular chondrocytes. Biochem Biophys Res Commun. 2004;316:984–90. doi: 10.1016/j.bbrc.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Starkman BG, Cravero JD, Delcarlo M, Loeser RF. IGF-I stimulation of proteoglycan synthesis by chondrocytes requires activation of the PI 3-kinase pathway but not ERK MAPK. Biochem J. 2005;389:723–9. doi: 10.1042/BJ20041636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonassar LJ, Grodzinsky AJ, Srinivasan A, Davila SG, Trippel SB. Mechanical and physicochemical regulation of the action of insulin-like growth factor-I on articular cartilage. Arch. Biochem. Biophys. 2000;379:57–63. doi: 10.1006/abbi.2000.1820. [DOI] [PubMed] [Google Scholar]
- 10.Smith P, Shuler FD, Georgescu HI, Ghivizzani SC, Johnstone B, Niyibizi C, et al. Genetic enhancement of matrix synthesis by articular chondrocytes: comparison of different growth factor genes in the presence and absence of interleukin-1. Arthritis Rheum. 2000;43:1156–64. doi: 10.1002/1529-0131(200005)43:5<1156::AID-ANR26>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Studer RK, Bergman R, Stubbs T, Decker K. Chondrocyte response to growth factors is modulated by p38 mitogen-activated protein kinase inhibition. Arthritis Res Ther. 2004;6:R56–R64. doi: 10.1186/ar1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veilleux N, Spector M. Effects of FGF-2 and IGF-1 on adult canine articular chondrocytes in type II collagen-glycosaminoglycan scaffolds in vitro. Osteoarthritis Cartilage. 2005;13:278–86. doi: 10.1016/j.joca.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Im HJ, Pacione C, Chubinskaya S, Van Wijnen AJ, Sun Y, Loeser RF. Inhibitory effects of insulin-like growth factor-1 and osteogenic protein-1 on fibronectin fragment-and interleukin-1beta-stimulated matrix metalloproteinase-13 expression in human chondrocytes. J Biol Chem. 2003;278:25386–94. doi: 10.1074/jbc.M302048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darling EM, Athanasiou KA. Growth factor impact on articular cartilage subpopulations. Cell Tissue Res. 2005;322:463–73. doi: 10.1007/s00441-005-0020-4. [DOI] [PubMed] [Google Scholar]
- 15.Aigner T, Gebhard PM, Schmid E, Bau B, Harley V, Poschl E. SOX9 expression does not correlate with type II collagen expression in adult articular chondrocytes. Matrix Biol. 2003;22:363–72. doi: 10.1016/s0945-053x(03)00049-0. [DOI] [PubMed] [Google Scholar]
- 16.Messai H, Duchossoy Y, Khatib AM, Panasyuk A, Mitrovic DR. Articular chondrocytes from aging rats respond poorly to insulin-like growth factor-1: an altered signaling pathway. Mech Ageing Dev. 2000;115:21–37. doi: 10.1016/s0047-6374(00)00107-x. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Wan L, Kozanek M. Determination of real-time in-vivo cartilage contact deformation in the ankle joint. J Biomech. 2008;41:128–36. doi: 10.1016/j.jbiomech.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Ragan PM, Badger AM, Cook M, Chin VI, Gowen M, Grodzinsky AJ, et al. Down-regulation of chondrocyte aggrecan and type-II collagen gene expression correlates with increases in static compression magnitude and duration. J. Orthop. Res. 1999;17:836–42. doi: 10.1002/jor.1100170608. [DOI] [PubMed] [Google Scholar]
- 19.Ragan PM, Chin VI, Hung HH, Masuda K, Thonar EJ, Arner EC, et al. Chondrocyte extracellular matrix synthesis and turnover are influenced by static compression in a new alginate disk culture system. Arch. Biochem. Biophys. 2000;383:256–64. doi: 10.1006/abbi.2000.2060. [DOI] [PubMed] [Google Scholar]
- 20.Hunter CJ, Imler SM, Malaviya P, Nerem RM, Levenston ME. Mechanical compression alters gene expression and extracellular matrix synthesis by chondrocytes cultured in collagen I gels. Biomaterials. 2002;23:1249–59. doi: 10.1016/s0142-9612(01)00245-9. [DOI] [PubMed] [Google Scholar]
- 21.Guilak F, Meyer BC, Ratcliffe A, Mow VC. The effects of matrix compression on proteoglycan metabolism in articular cartilage explants. Osteoarthr. Cartilage. 1994;2:91–101. doi: 10.1016/s1063-4584(05)80059-7. [DOI] [PubMed] [Google Scholar]
- 22.Kim YJ, Grodzinsky AJ, Plaas AH. Compression of cartilage results in differential effects on biosynthetic pathways for aggrecan, link protein, and hyaluronan. Arch. Biochem. Biophys. 1996;328:331–40. doi: 10.1006/abbi.1996.0181. [DOI] [PubMed] [Google Scholar]
- 23.Valhmu WB, Stazzone EJ, Bachrach NM, Saed-Nejad F, Fischer SG, Mow VC, et al. Load-Controlled Compression of Articular Cartilage induces a transient stimulation of Aggrecan gene expression. Arch. Biochem. Biophys. 1998;353:29–36. doi: 10.1006/abbi.1998.0633. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald JB, Jin M, Dean D, Wood DJ, Zheng MH, Grodzinsky AJ. Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. Journal of Biological Chemistry. 2004;279:19502–19511. doi: 10.1074/jbc.M400437200. [DOI] [PubMed] [Google Scholar]
- 25.Plumb MS, Treon K, Aspden RM. Competing regulation of matrix biosynthesis by mechanical and IGF-1 signalling in elderly human articular cartilage in vitro. Biochim Biophys Acta. 2006;1760:762–7. doi: 10.1016/j.bbagen.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Fitzgerald JB, Dimicco MA, Grodzinsky AJ. Mechanical injury of cartilage explants causes specific time-dependent changes in chondrocyte gene expression. Arthritis Rheum. 2005;52:2386–95. doi: 10.1002/art.21215. [DOI] [PubMed] [Google Scholar]
- 27.Baayen R, Davidson D, Bates D. Mixed-effects modeling with crossed random effects for subjects and items. Journal of Memory and Language. 2008;59:390–412. [Google Scholar]
- 28.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 2000;97:10101–6. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holter NS, Mitra M, Maritan A, Cieplak M, Banavar JR, Fedoroff NV. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci U S A. 2000;97:8409–14. doi: 10.1073/pnas.150242097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eleswarapu SV, Leipzig ND, Athanasiou KA. Gene expression of single articular chondrocytes. Cell Tissue Res. 2007;327:43–54. doi: 10.1007/s00441-006-0258-5. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman L, aPJR . Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons Ltd.; Chinchester, New York, Weinheim: 1990. p. 342. [Google Scholar]
- 32.Sah RLY, Kim YJ, Doong JYH, Grodzinsky AJ, Plaas AHK, Sandy JD. Biosynthesis Response to Cartilage Explants to Dynamic Compression. J. Orthop. Res. 1989;7:619–636. doi: 10.1002/jor.1100070502. [DOI] [PubMed] [Google Scholar]
- 33.Szafranski JD, Grodzinsky AJ, Burger E, Gaschen V, Hung HH, Hunziker EB. Chondrocyte mechanotransduction: effects of compression on deformation of intracellular organelles and relevance to cellular biosynthesis. Osteoarthritis Cartilage. 2004;12:937–46. doi: 10.1016/j.joca.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Edwards D, Clendeninn NJ, Appelt K. Cancer drug discovery and development. Humana Press; Totowa, N.J.: 2001. The Tissue Inhibitors of Metalloproteinases (TIMPs): Biology and Regulation. Matrix metalloproteinase inhibitors in cancer therapy; pp. 67–84. [Google Scholar]
- 35.Kashiwagi M, Tortorella M, Nagase H, Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5) J Biol Chem. 2001;276:12501–4. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
- 36.D'Lima DD, Hashimoto S, Chen PC, Colwell CW, Jr., Lotz MK. Human chondrocyte apoptosis in response to mechanical injury. Osteoarthritis Cartilage. 2001;9:712–9. doi: 10.1053/joca.2001.0468. [DOI] [PubMed] [Google Scholar]
- 37.Gendron C, Kashiwagi M, Hughes C, Caterson B, Nagase H. TIMP-3 inhibits aggrecanase-mediated glycosaminoglycan release from cartilage explants stimulated by catabolic factors. FEBS Lett. 2003;555:431–6. doi: 10.1016/s0014-5793(03)01295-x. [DOI] [PubMed] [Google Scholar]
- 38.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–8. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 39.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ushiki T, Hitomi J, Ogura S, Umemoto T, Shigeno M. Atomic force microscopy in histology and cytology. Arch. Histol. Cytol. 1996;59:421–31. doi: 10.1679/aohc.59.421. [DOI] [PubMed] [Google Scholar]
- 41.Ali O, Cohen P, Lee KW. Epidemiology and biology of insulin-like growth factor binding protein-3 (IGFBP-3) as an anti-cancer molecule. Horm Metab Res. 2003;35:726–33. doi: 10.1055/s-2004-814146. [DOI] [PubMed] [Google Scholar]
- 42.Davenport-Goodall CL, Boston RC, Richardson DW. Effects of insulin-like growth factor-II on the mitogenic and metabolic activities of equine articular cartilage with and without interleukin 1-beta. Am J Vet Res. 2004;65:238–44. doi: 10.2460/ajvr.2004.65.238. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Kubo T, Kobayashi K, Imanishi J, Takigawa M, Arai Y, et al. Hydrostatic pressure influences mRNA expression of transforming growth factor-beta 1 and heat shock protein 70 in chondrocyte-like cell line. J Orthop Res. 1997;15:150–8. doi: 10.1002/jor.1100150122. [DOI] [PubMed] [Google Scholar]
- 44.Loeser RF, Shanker G. Autocrine stimulation by insulin-like growth factor 1 and insulin-like growth factor 2 mediates chondrocyte survival in vitro. Arthritis Rheum. 2000;43:1552–9. doi: 10.1002/1529-0131(200007)43:7<1552::AID-ANR20>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 45.Armstrong DG, Gutierrez CG, Baxter G, Glazyrin AL, Mann GE, Woad KJ, et al. Expression of mRNA encoding IGF-I, IGF-II and type 1 IGF receptor in bovine ovarian follicles. J Endocrinol. 2000;165:101–13. doi: 10.1677/joe.0.1650101. [DOI] [PubMed] [Google Scholar]
- 46.Fowlkes JL, Serra DM, Nagase H, Thrailkill KM. MMPs are IGFBP-degrading proteinases: implications for cell proliferation and tissue growth. Ann N Y Acad Sci. 1999;878:696–9. doi: 10.1111/j.1749-6632.1999.tb07765.x. [DOI] [PubMed] [Google Scholar]
- 47.Iyengar P, Combs TP, Shah SJ, Gouon-Evans V, Pollard JW, Albanese C, et al. Adipocyte-secreted factors synergistically promote mammary tumorigenesis through induction of anti-apoptotic transcriptional programs and proto-oncogene stabilization. Oncogene. 2003;22:6408–23. doi: 10.1038/sj.onc.1206737. [DOI] [PubMed] [Google Scholar]
- 48.Pufe T, Petersen W, Fandrich F, Varoga D, Wruck CJ, Mentlein R, et al. Programmable cells of monocytic origin (PCMO): a source of peripheral blood stem cells that generate collagen type II-producing chondrocytes. J Orthop Res. 2008;26:304–13. doi: 10.1002/jor.20516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.