Abstract
Now that genome-wide association studies (GWAS) are dominating the landscape of genetic research on neuropsychiatric syndromes, investigators are being faced with complexity on an unprecedented scale. It is now clear that phenomics, the systematic study of phenotypes on a genome-wide scale, comprises a rate-limiting step on the road to genomic discovery. To gain traction on the myriad paths leading from genomic variation to syndromal manifestations, informatics strategies must be deployed to navigate increasingly broad domains of knowledge and help researchers find the most important signals. The success of the Gene Ontology project suggests the potential benefits of developing schemata to represent higher levels of phenotypic expression. Challenges in cognitive ontology development include the lack of formal definitions of key concepts and relations among entities, the inconsistent use of terminology across investigators and time, and the fact that relations among cognitive concepts are not likely to be well represented by simple hierarchical “tree” structures. Because cognitive concept labels are labile, there is a need to represent empirical findings at the cognitive test indicator level. This level of description has greater consistency, and benefits from operational definitions of its concepts and relations to quantitative data. Considering cognitive test indicators as the foundation of cognitive ontologies carries several implications, including the likely utility of cognitive task taxonomies. The concept of cognitive “test speciation” is introduced to mark the evolution of paradigms sufficiently unique that their results cannot be “mated” productively with others in meta-analysis. Several projects have been initiated to develop cognitive ontologies at the Consortium for Neuropsychiatric Phenomics (www.phenomics.ucla.edu), in hope that these ultimately will enable more effective collaboration, and facilitate connections of information about cognitive phenotypes to other levels of biological knowledge. Several free web applications are available already to support examination and visualization of cognitive concepts in the literature (PubGraph, PubAtlas, PubBrain) and to aid collaborative development of cognitive ontologies (Phenowiki and the Cognitive Atlas). It is hoped that these tools will help formalize inference about cognitive concepts in behavioral and neuroimaging studies, and facilitate discovery of the genetic bases of both healthy cognition and cognitive disorders.
Keywords: Phenomics, Endophenotypes, Genetics, Cognition, Informatics
Introduction
We live in an exciting scientific era marked by daily discoveries about the biological bases of brain function, and are deluged with findings from functional imaging, large-scale genetic association studies, and other work of high relevance to cognitive neuropsychiatry from its many cognate disciplines. We suggest that the development of cognitive ontologies may be important to help manage and digest this knowledge, and that application of cognitive ontologies may yield unanticipated insights into brain-behavior relations. We first highlight some sources of this complexity that have arisen from efforts to find associations between genotypic variation and complex neuropsychiatric phenotypes, which have stimulated the development of bioinformatics and “phenomics” as new disciplines. It is then suggested that cognitive phenotypes may provide a useful level of analysis to help bridge the gap between genome and syndrome. Subsequent sections aim to familiarize the reader with ontologies as frameworks for formalizing knowledge about specific concepts and their inter-relations within a domain, and consider the unique challenges of designing ontologies to represent cognitive concepts, cognitive tests, and their relations to other kinds of knowledge. Finally, we describe works in progress that are designed to advance cognitive ontology development, the modeling of cognitive concepts, and the analysis of cognitive task effects, leveraging collaborative networks that have been enabled by the world wide web.
Managing complexity in the “Human Phenome Project”
In the “good old days” of neuropsychiatric genetics research -two years ago -the literature was bustling with reports suggesting association of “candidate genes” with a range of phenotypes. Some of the phenotypes were traditional diagnostic categories, while others were “endophenotypes”, including cognitive, electrophysiological, or neuroimaging measures. Now that we have reports from the first generation of genome-wide association studies (GWAS), the landscape is markedly different. Together with some disappointing failures to replicate prior candidate gene studies, the GWAS have prompted reconsideration and cast skepticism on findings that are not significant at a genome-wide level, and to consider findings “significant” today, suggested probability levels range from p<10-5 to p<10-7 (for more specific guidelines, see Freimer and Sabatti, 2004, Freimer and Sabatti, 2005). Some suggest that we currently possess no strong candidate genetic loci for neuropsychiatry research (Flint and Munafo, 2007). Meanwhile, recent GWAS outside neuropsychiatry are providing novel leads at a rate that strains human capacity for comprehension, and prompting critical re-evaluation of basic strategies for discovery in biomedicine (Frayling, 2007).
Initial GWAS results already make two critical points for neuropsychiatry research: (1) if we stick with conventional diagnostic categories as phenotypes, we are going to need very large samples to detect very small effects; (2) even if we are successful in defining “endophenotypes” or intermediate phenotypes, it remains unclear that these will possess “simpler genetic architecture.” Given the large number and scale of GWAS now targeting neuropsychiatric phenotypes, it seems inevitable that we will soon possess a large number of leads - both genetic and phenotypic - that will require follow-up. If there are hundreds, or perhaps thousands of genetic variants involved (not to mention their interactions with both other genetic variants and environmental effects), and myriad phenotypes to consider, how will we prioritize leads for mechanistic research that will inform theories of pathophysiology and the development of rational treatments?
Bioinformatics strategies are helping develop a “bottom-up” scaffold enabling researchers to make the initial steps upwards from the human genome to the human proteome, and already can help constrain hypotheses with knowledge of the biochemical signaling pathways that have been associated with key genomic variants. But the ultimate vision of connecting this basic biological knowledge to the “higher” and more complex phenotypes comprising neural systems functions, cognitive abilities, neuropsychiatric symptoms and syndromes, will require more systematic and formal descriptions of these entities and their relations to each other and their putative biological underpinnings. Much as the Gene Ontologies project has succeeded in advancing genomics research, we suggest that cognitive ontologies can help advance research in neuropsychiatric phenomics - the systematic study of neuropsychiatric phenotypes on a genome-wide scale.
New tools to manage complexity can help increase our odds of making bets with solid payoffs in biomedical discovery. We may soon be poised to overcome current obstacles in genotyping, including detection of copy number variations and rare variants; even epigenome-wide arrays may be available. But we are likely still to confront significant obstacles converting large lists of genetic variants into tractable biological research programs that will identify mechanisms by which these genes exert their effects on complex systems and syndromal levels. Within the Consortium for Neuropsychiatric Phenomics at UCLA (www.phenomics.ucla.edu), we have used a simple schematic scaffold for translational neuropsychiatric research from genome to syndrome, using seven levels (see Figure 1). Seven levels were selected not because we believe this reflects accurately the vast terrain connecting genome to syndrome, but rather because humans generally have difficulty maintaining in mind or discerning a larger number of categories.
Figure 1.
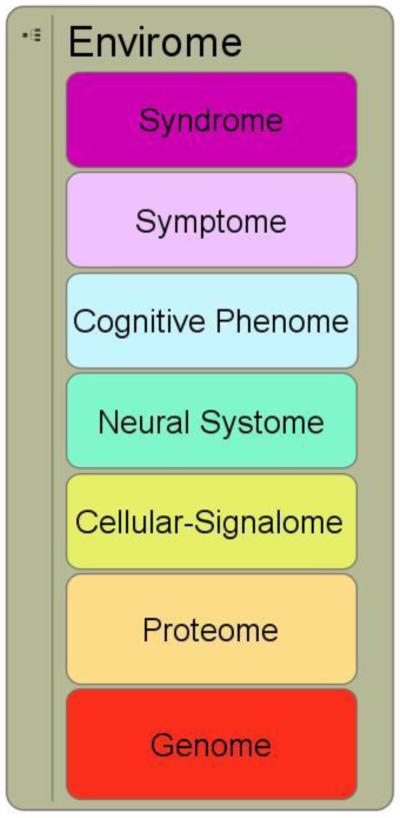
Simplified schematic of multilveled “-omics” domains for cognitive neuropsychiatry.
Even given this dramatic simplification, it is easy to see that relating genome to syndrome with rational mechanistic hypotheses is a vast task. Kendler graphically highlighted that genotype-to-phenotype relations are “many-to-many” (i.e., any given genotype influences multiple phenotypes (pleiotropy), and a given phenotype has multiple genotypic contributions)(Kendler, 2005). But these many-to-many relations exist between each level as we proceed from genome to syndrome (i.e., a single protein may affect multiple cellular systems, and many proteins are required in any one cellular system; a single kind of cellular system has ramifications in multiple neural systems, and each neural system depends on manifold cellular systems; and so on). The immediate conclusion is that there can be exponential expansion of effects as one follows paths up the hierarchy (pleiotropic expansion) and that any given phenotype may be affected by very large numbers of genes (polygenic expansion).
Simple back-of-the-napkin calculations help put in perspective the scope of the challenges. Starting with the simplest assumption that there is a single gene with syndromal manifestations, and conservative estimates of pleiotropy at each level (i.e., that a single variant may affect p number of proteins, that each protein affects c number of cellular systems or signaling pathways, and so on), the influence of any single gene explodes through the higher phenomic levels. For example, a single gene with 5-fold expansion across each of 7 levels of phenomic expression will yield 15,625 effects. If the expansion were 10-fold at each level, there would a million syndromal variants for each genetic variant.
This expansion logic also applies to top-down analyses of polygenic expansion. Imagine we seek genetic contributions to a single syndrome. Assuming the syndrome is defined by some number s of symptoms, and that each symptom has p cognitive underpinnings, and so forth, it is easy to appreciate that thousands of genes may contribute to complex phenotypes, and indeed that some brain-related phenotypes may be affected by substantial portions of the entire genome.
Similar conclusions are reached by computing the shared variance between levels in a 7-level hierarchy. For example, say we are interested in finding a gene that explains 25% of the variance in a complex syndromal phenotype. This demands that the average shared variance between levels is 80%, or in other words that each level must correlate approximately .9 with the next level. A more conservative but still optimistic estimate of 50% shared variance between levels (still demanding a correlation of .7, which is not far from the upward limits of reliability for psychiatric diagnosis, symptom rating scales, and cognitive phenotype measurements), yields shared variance between gene and syndrome of 1.6%. This might be considered the best case, possibly rational scenario. A more rational scenario is that in which approximately 20% of variance is shared across levels. This is more consistent so far with the typical correlations and effect sizes seen in psychiatry research, where correlations of cognitive measures to symptoms, or brain imaging parameters to cognitive dimensions, are not infrequently in the range of .4 to .5. In this scenario the shared variance of a genetic variant with a complex syndromal variant is .01%. A simple additive genetic model would thus require some 5000 genetic variants to explain a heritability of 50%, which is in the range of that observed for many cognitive, personality, or diagnostic phenotypes. The idea that this last scenario is more realistic than either of the former ones is supported by some reviews suggesting that genetic variation may explain only ∼20% of variation at the level of the transcript, and that less than 2.5% of variance was shared with “higher” phenotypes, regardless of their putative proximity to the genetic level (Flint and Munafo, 2007). This line of reasoning calls into question the notion that gene discovery will be advanced substantially by the study of any particular “endophenotype”, given that so far these have not often demonstrated a much more robust genetic signal, or “simpler genetic architecture.”
Two basic strategies have been proposed to overcome these daunting challenges. Plan A, which we might label the “massively univariate” approach is to increase sample sizes to detect the modest effects that appear most likely to characterize associations between genes and high-level phenotypes. It remains unclear how large these samples will need to be before we will find genetic variants associated with syndromal phenotypes like schizophrenia or bipolar disorder, but recent evidence suggests that such samples will likely exceed 10,000, and perhaps 50,000, before variants with robust genome-wide significance are observed. Two recent GWAS studies focusing on schizophrenia and bipolar disorder phenotypes, examining sample sizes exceeding 10,000 individuals, have reported a handful of strong targets (Ferreira et al., 2008, O’Donovan et al., 2008). Assuming that results from these studies stand up to replication, it is conspicuous that there is virtually no overlap with the long lists of candidate genes that have been suggested so far for these syndromes.
Plan B, which might be labeled the “multivariate” approach, or the “phenomics” approach, is to develop strategies to increase the magnitude of variance shared between genotype and phenotype through clever redefinition of genotype or phenotype, and the paths that relate genotype to phenotype. The cardinal premise of the “endophenotype” strategy is that there exist phenotypes “closer to the gene” that will share more variance with real gene effects. While the Flint and Munafo survey does not offer much cause for optimism, it should be recognized that this strategy has seldom been employed in GWAS studies so far, and that the potential increases in power of this strategy remain largely unexplored. Even when multiple phenotypes have been examined, these are often derived from a single level of analysis (e.g., multiple partially overlapping diagnostic schemata) that is far removed from putative biological substrates.
While not yet widely exploited for GWAS studies, some work with candidate genes has suggested that effect sizes may be considerably larger when examining neural system phenotypes (e.g., functional MRI measurements) relative to diagnostic or behavioral phenotypes (Egan et al., 2001, Hariri et al., 2003). It is also possible that more specific neurocognitive phenotypes may have strong relations with individual gene effects, by virtue of being more closely related to the physiological processes actually impacted by the genes. For example, traditional measures of “executive function” such as the Wisconsin Card Sorting Test perseverative error score shared less than 5% variance with the COMT val158met polymorphism, while more specific measures of cognitive set-shifting shared up to 40% variance with genotype (Bilder et al., 2002, Bilder et al., 2004, Nolan et al., 2004). Such high estimates might be inflated by chance given application of the candidate gene approach with small sample sizes, but highlight the possibility that refined phenotyping may yield greater promise not only by virtue of increasing statistical power, but further by enhancing insight into plausible mechanisms.
Even these theoretically more refined phenotypes are still derived from a single level of analysis, and it remains unclear what advantages might be obtained by defining new phenotypes that span different levels of investigation. For example, rather than identifying a new and improved neuroimaging or cognitive phenotype, we might find both more power and great mechanistic insight from combining a historical phenotype, an imaging phenotype, a cognitive phenotype, and a symptom phenotype. For example, perhaps a stronger genetic association might be found for individuals with poor premorbid social function, gray matter volume reduction, poor working memory, and negative symptoms, than could be found for any one of these alone. This may seem a counter-intuitive strategy from the experimental perspective, but certainly has parallels in other disease areas. For example, the diagnosis of cardiac valve dysfunction benefits greatly from combining history (e.g., of early rheumatic fever), with laboratory results (e.g., from EKG), and behavioral symptoms (e.g., shortness of breath on exertion).
The definition of novel multi-level, multivariate phenotypes may also benefit from advances in complexity theory, and confer substantially greater traction on what might seem to be insuperable obstacles. Particularly given the number of emergent properties putatively implicated in the traversal of biological levels from the genome to syndrome, our ability to identify convergences and self-organizing principles may help constrain the explosive expansion of possibilities to more manageable subsets. Thus, rather than attempting to explain the high heritability (perhaps 80%) of a syndrome such as schizophrenia by the additive effects of some 800 genetic variations each independently contributing 0.1% to phenotypic variance, there is hope that a smaller number of genetic variants might be identified that interact and converge on a more modest number of critical biological pathways. Stuart Kauffman has written eloquently about the “order for free” that characterizes large-scale networks, and applied these principles to problems as diverse as the origins of life and economics (Kauffman, 1995). Similar methods have been used to help understand self-organization in neural networks specifically, and biological networks more generally (Tononi et al., 1994, 1999, Sporns et al., 2005). Progress is already being made identifying the redundancy and degeneracy in gene networks and other biological systems using techniques that integrate information sciences and systems biology (Sridhar et al., 2007, Zheng et al., 2007, Centler et al., 2008). Application of similar methods to multi-level modeling of genome-to-syndrome hypotheses comprises a reasonable if not simple extension of these theoretical lines. Regardless of the specific methods that will be used, it is clear that novel strategies to effectively manage complexity of large scale networks spanning different levels of investigation will be critical to advance the emerging discipline of phenomics.
Why pick the cognitive phenome as a focal point?
If one accepts the basic principle that there exists some roughly hierarchical set of causal mechanistic links that lead from the genome to the syndrome, it is rational to ask: Where to start? The strategy of conducting “phenome-wide” scanning (measuring all possible phenotypic variations from mRNA through complex behavioral syndromes) is obviously impossible in practical terms, and arguably impossible even in theory. Given imperfections in our knowledge, we may never realize what truly comprises the human phenome; in contrast a complete sequence of an individual genome is straightforward, if currently a bit costly. Scientists must therefore choose a level of phenotypic complexity appropriate to their interest, to narrow the scope of possible investigation.
Some investigative teams are logically focusing on the human proteome, an attractive target given its “proximity” to the genome. But this does not offer clear traction for neuropsychiatry, given that so far few single protein anomalies have been identified as the culprits underlying complex psychiatric disease. To put this in perspective, consider the long and so-far frustrating search for an abnormal dopamine receptor protein in schizophrenia. Perhaps the recent GWAS findings implicating the L-type voltage gated calcium channel in bipolar disorder will prove to be more fruitful (Ferreira et al., 2008). Even if we do identify some critical proteins to help focus research, there will clearly remain substantial intervening biology to connect these protein variations via mechanistic hypotheses to neuropsychiatric symptoms and syndromes (Bilder, 2008).
In efforts to identify a rational starting point in the 7-layered schema, the CNP has suggested that the level of cognitive phenotype is attractive because: (a) the cognitive phenotypes may be rationally related to higher level symptoms and the syndromes that are defined by these; (b) cognitive phenotypes might offer links to neural systems level analysis, particularly given advances in functional neuroimaging that enable experimental manipulation of cognitive parameters and use of brain activation as the dependent variable; (c) at least some cognitive phenotypes appear to have homologs, or at least analogs, in other species, enabling links to a broader class of basic science studies.
Compelling arguments can be advanced for organizing research programs around alternate phenotypic nodes at other levels. For example, in the study of Alzheimer’s disease, the diagnostic entity in the DSM-IV (American Psychiatric, 1994) for which there is the strongest consensus about pathophysiology, it might be considered rational to focus on amyloid deposition or tau-opathies, as organizing phenotypes. Even in this example, however, it should be recognized that prior definitions of Alzheimer’s Disease may require revision, and that an extended range of related disease entities may emerge from this redefinition (Saura et al., 2005, Chiang et al., 2008). For disorders that are less understood, including psychotic disorders, mood disorders, anxiety disorders, and most disorders first appearing in childhood and adolescence, a focus on cognitive phenotypes may provide a helpful link between the high level syndromes identified as public health concerns, and brain biology. In order to connect cognitive phenotypes to the syndromal level, it is further explicit in this formulation that phenomics will benefit from dimensional representation of psychiatric problems at the symptom level, and this practice is supported by the advantages of dimensional over discrete categorical approaches to the characterization of psychiatric syndromes (Haslam and Kim, 2002, Haslam, 2003, Kraemer, 2007).
Narrowing the problem scope by selecting a phenotypic node as a starting point leaves us still with enormous complexity. To gain traction on the complex paths leading from genomic variation to syndromal manifestations, informatics strategies must be deployed to consolidate broader domains of knowledge and help researchers find the most important signals. Bioinformatics has grown enormously over the last decade, and generated repositories of biological knowledge at the genomic, transcript, proteomic, and even metabolomic levels. But higher phenomic levels are not yet well represented. Efforts so far to database higher level phenotypic knowledge (e.g., fMRI findings) have so far suffered, probably for multiple reasons, including concerns about the mechanisms and safeguards for data (and credit) sharing, and due to the complexity of the data themselves. But at least one substantive obstacle in these efforts has been the lack of a coherent conceptual system to organize these effects. Projects such as the Function Biomedical Informatics Network (FBIRN) have worked to resolve some of the problems with variability in data acquisition and data formatting parameters. It has proven more difficult to arrive at consensus on the optimal methods for describing the cognitive manipulations used as independent variables in these experiments. This poses a challenge for effective data sharing and synthesis, and further may make it difficult to generate appropriate inferences from neuroimaging data(Poldrack, 2006). A coherent consensus cognitive ontology could help such efforts realize their fullest potential.
What are ontologies and why might they help?
Can ontologies help to reduce the complexity of the enormous problem space we are confronting? While ontology development has seen enormous growth over the last decade, many investigators in cognitive neuropsychiatry are not very familiar with ontology development or how ontologies can be used. The term ontology may be most familiar given its long history of usage in philosophy to mean “...the study of the nature of being, existence, or reality in general and of its basic categories and their relations” (Wikipedia, accessed 10/4/2008). The term ontology has a different usage in computer and information sciences, as “... a formal representation of a set of concepts within a domain and the relations between those concepts. It is used to reason about the properties of that domain, and may be used to define the domain” (Wikipedia, accessed 10/4/2008). A more concise definition of the term was offered by Gruber (2003): “an ontology is a specification of a conceptualization” (page 199). Gruber further emphasizes the purpose of such explicit specification is to enable “knowledge sharing and reuse.” It is precisely this capacity for developing shared representations of knowledge regarding cognitive phenotypes that we believe is of paramount importance for advancing the discipline so that emerging knowledge can be more readily shared within the cognitive neuroscience community, and so that links can be forged to other levels of both clinical and biological knowledge.
There are various degrees of formalization possible in ontology development; at least four levels of progressively increased elaboration in ontology definition have been identified (Grethe et al., 2003):
Controlled Vocabularies: The first stage in most ontology development is the development of a lexicon, or set of terms representing the concepts in a specific domain. The vocabulary is “controlled” by establishing operational definitions of the terms, identification of synonyms, and identification of other properties of the terms (for example, part of speech, pluralization, and so on). Ideally this will lead to a set of “standard names” for key concepts in the domain, which can then be used to foster greater agreement in concept representation by the users.
Simple Taxonomies: Once a set of terms reflecting the conceptual entities of the domain is identified, the next level of refinement is often the classification of these entities into a taxonomy, sometimes referred to as a “class hierarchy.” These class hierarchies are often represented as directed acyclic graphs, which enforce a unidirectional “flow” from superordinate categories to subordinate category members, and use a specific type of relation sometimes described as “is a” relationships. This kind of class hierarchy works well in certain concept areas (i.e., phylogenies of plants and animals; for example, a cat is a mammal, a mammal is an animal, and so forth).
Graph-like Ontologies: Many conceptual domains are not well modeled by simple class hierarchies, and more complex types of relation are required to represent the structure of the domain. It is likely that the domain of cognition is a case in point. Thus, while there might be some consensus that “episodic memory” is a sub-class of “memory”, other concepts such as “working memory” might be considered a component of “memory” or a component of “executive function”, emphasizing different aspects of this concept. Such ontologies may possess many unique relation types (”has a”, “contained in”, “causes”, “activates”, etc.). In cognitive ontologies, there are several particularly useful relation types. For example, cognitive task indicators (the specific variables that summarize performance on aspects of the task) are considered to measure certain cognitive concepts. Conversely, cognitive concepts may be “measured by” task indicators. Such ontologies may be rich with different kinds of relations but lack more formal semantics.
Logic-based Ontologies: More formal logic-based ontologies permit inference of relations that are logical consequences of formal concept definitions that in turn arise from formalized syntax and semantics. The goal of such ontologies is to afford automated inference or reasoning within the domain of the ontology. There are efforts to establish conventions for logic-based languages that standardize syntax and semantics for information interchange, including: CLIF - Common Logic Interchange Format; CGIF - Conceptual Graph Interchange Format; and XCL - eXtended Common Logic Markup Language, based on XML (for more information, see www.obitko.com).
For the domain of “cognition” there does not exist yet any agreed upon lexicon, and a lexicon is necessary for developing any more elaborate ontology, so this represents a rational starting point. It is also clear that the concepts in the domain of cognition are unlikely to be well expressed by simple taxonomic structure, although such representations may be useful intermediate steps to help clarify relations among key concepts (a few examples are provided below). Ultimately, we believe the most functional ontologies may be graph-like, offering a high degree of flexibility in ontological specification, and acknowledging that our understanding of this domain may long remain only quasi-structured and the conceptual entities themselves subject to revision as we learn more about relations between brain and behavior.
Cognitive Ontology Nuts and Bolts: How Can We Anchor Abstract Cognitive Concepts?
To address this question it may help to flesh out more concretely what elements comprise a simple ontology. At the base of ontologies are specific examples or Instances of the things that exist in the domain. These can be concrete things like individual people, animals, cars, or bottles of wine; or the things may be abstract like words or numbers (i.e., “thing” in this context does not signify a concrete object, but instead signifies existence - a “thing” can be distinguished from nothing).1 Ontologies usually also contain Classes or categories of things, and these are often arranged hierarchically, as noted above for simple taxonomies. Relations are used in ontologies to formalize the ways in which instances or classes can relate to each other (the classic “is a” relation is widely used to denote class membership, e.g., a cat is a mammal). Attributes, sometimes referred to as properties or features, are qualifiers that may apply to the things in the ontology. These may include attributes important to identifying class membership (e.g., “has sweat glands” and “has hair” are attributes of animals that help identify them as mammals), or attributes that help distinguish instances within a class (e.g., “is red” may distinguish one set of cars from others of the same make and model, but other makes and models may share this attribute).
For cognitive ontologies the instances are relatively abstract given that mental entities are not observable. For example, consider what comprises an “instance” of the concept “memory”: is it a subjectively experienced “memorandum”? Is it a process? Is it a neural system-level representation reflecting connection weights among a set of neurons linked in a network? We believe there is a need for ontologies to help organize and sharpen definitions for the abstract concepts that are used in cognition research (e.g., is “working memory” a kind of “memory”, and what are the exact distinctions among the terms “short term memory” and “working memory” or between “episodic memory” and “declarative memory”?). Given the imprecision, inconsistency, and temporal instability in usage of cognitive concept terms, we suggest it is important to anchor these concepts at the measurement level. If this suggestion is accepted, instances are the results of specific measurements made on individual animals (including humans). Thus, while capturing useful aspects of our shared conceptualizations of cognitive processes on the one hand, an ontology of cognitive concepts can be usefully rooted in more concrete and stable measurement methods that provide the experimental and observational basis for the abstractions used in our discipline.
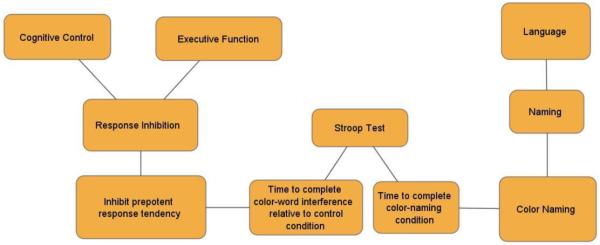
Figure 2 illustrates a simple class hierarchy for a cognitive concept and its relation to a cognitive measurement. The figure suggests that verbal declarative memory is a kind of declarative memory, and that declarative memory is a kind of memory. The figure also suggests that verbal list learning procedures can offer measures of verbal declarative memory (and thus also of declarative memory, and memory by inheritance), but that this relation is mediated specifically via an indicator (number of words recalled) on a specific verbal list learning test. While it may be true that all verbal list learning tests possess indicators of verbal declarative memory, other measures derived from these tests may not measure verbal declarative memory (for example, indicators for semantic clustering or proactive interference, although derived from the same test, may measure different constructs). The importance of this distinction is still more apparent with an example where indicators map to more distinctive concepts. Figure 3 shows how two different indicators derived from the Stroop Test measure two divergent cognitive concepts (one is the tendency to inhibit prepotent responses via the “classic Stroop effect” which is itself a contrast effect between an “interference” condition and a control condition; the other is simple color naming skill). Thus it would be incorrect to say simply that the “Stroop Test” measures “response inhibition” without reference to the specific observable indicator that measures this function.
Figure 2.
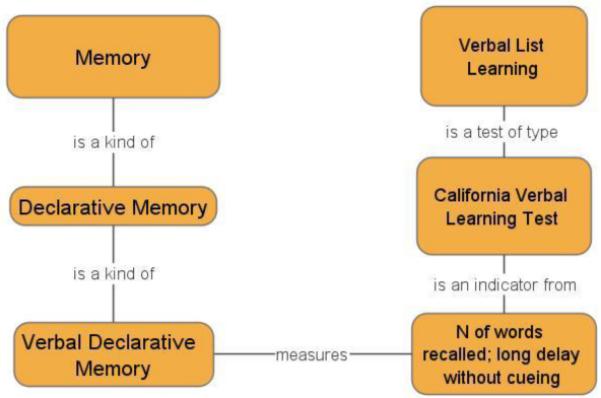
Alignment of cognitive concept and task hierarchies via measurement models.
Figure 3.
Alignment of cognitive concept and task hierarchies with multiple divergent indicators, and possible controversy over class membership within the concept hierarchy.
Figure 3 further highlights that some concepts (e.g., response inhibition) may be considered subclasses of multiple higher level concepts (e.g., is response inhibition better considered a kind of “cognitive control” or an “executive function”?). In prior work we have shown that there can be dramatic increases in usage of some concept labels (such as the term “cognitive control”), while there is considerably greater stability at the measurement level (for example, in the literature on heritability of cognitive control, there was not a single instance where the cognitive control construct was measured using an indicator that had not previously been considered an indicator of “working memory”, “response inhibition”, “task switching/set shifting”, or “response selection”)(Sabb et al., 2008). Anchoring cognitive concepts to observable indicators promotes greater consensus and interpretation at appropriate levels of abstraction, with greater immunity from variability in concept labeling.
Anchoring cognitive concepts to the measurement level further enables cognitive ontologies to serve as the organizational foundation for repositories of quantitative, empirical data that can in turn be used for automated meta-analysis and construction of multi-level causal models. For example, assume there are multiple indicators of a specific concept, and that we possess information about the covariance of those test indicators. Then the validity of the concept can be examined using the methods of covariance structure analysis (i.e., structural equation modeling, where the concept is defined as a latent construct, and the observed variables are the indicators of that construct). While structural equation modeling typically uses case-level data where each observation represents an individual subject and their scores on relevant indicators, there is an emerging literature on Meta-Analytic Structural Equation Modeling, which enables analyses to be pooled at the study result level (Furlow and Beretvas, 2005, Riley et al., 2007, Cheung, 2008). There are multiple challenges to assuring the validity of results derived from such analyses, centering on the degree to which results from different studies can reasonably be pooled. Given appropriate caution, however, there is promise that these tools will help foster our capacity to synthesize results from widely divergent studies and develop estimates of models spanning multiple empirical investigations.
It is logical to worry that anchoring cognitive concepts to the measurement level might reify concept definitions and constrain the specification of of novel concepts. This is not a limitation, however, if the system for ontology specification is flexible and can be modified by individual investigators. For example, an investigator may have a completely novel theory and unique set of concept labels. This investigator could construct an individual ontology containing these new concepts and their inter-relations, and also link these to existing empirical data, or provide the rationale as to why the existing empirical data are not relevant to these concepts, thus highlighting the need for new data to assess the validity of the novel theory. Strategies for coping with conflicting theoretical views are discussed further in a subsequent section. It should also be recognized that concept ontologies can be advanced without being anchored to any empirical data, and therefore it is entirely possible to pursue cognitive ontology development in a “top-down” fashion (i.e., from concept to task). On the other hand, there is a risk, in developing a “top-down” ontological model of a concept domain, that if it is not linked to empirical data, it will remain essentially unfalsifiable.
Do we really need Cognitive Ontologies?
It is reasonable to ask: Why bother with ontology development? There are multiple reasons for supporting development of cognitive ontologies to advance research on the genetics of, and treatments for psychiatric disorders.
Connections are important. Links to other repositories of biological knowledge will be possible only if the knowledge regarding complex cognitive and neuropsychiatric phenotypes are rationally constructed and appropriately indexed. Current indices are too crude to enable meaningful inferences to be drawn. For example, the Medical Subject Headings (MeSH) that are the most widely used indexing system for medical literature contain only 6 terms under the concept “Cognition.” These terms do not reflect very well current research in cognitive psychology and neuroscience (the complete list is: Awareness; Cognitive Dissonance; Comprehension; Consciousness; Imagination, which includes the sub-concepts: Dreams and Fantasy; and Intuition). Similarly, the components of ontologies in other repositories of biological knowledge are underdeveloped from the perspective of those interested in cognition (for example, searching for the term “cognition” in the National Library of Medicine’s (NLM’s) Entrez GENE database currently yields a total of only 83 hits and the same search in NLM’s OMIM (Online Mendelian Inheritance in Man) yields only 54 hits [as of 10/8/2008]). These searches are clearly failing to show high sensitivity to a large literature already in existence. Good cognitive ontologies would enable literature reports and data to be linked far more effectively with relevant biological constructs and support both validation of existing hypotheses and exploration for development of novel hypotheses. As molecular libraries and proteomics knowledgebases continue to expand, cognitive ontologies may become critical links also in drug discovery by helping to narrow the range of potential targets and their possible mechanisms of action.
Formalization is important. The “structure” of cognitive and neuropsychiatric phenotypes remains too frequently the target of subjective speculation, and is strongly influenced by conceptual and semantic factors that are often independent of the underlying data. Perhaps in part due to the “fuzziness” of these conceptual distinctions, their imperfect relations to measurement models, and their loose validation with respect to underlying biological processes, it is easy for conclusions to be drawn that are inappropriately generalized, lead to inertia in certain research areas, along with replication failures and unproductive controversy. It is also important to the disciplines of psychology and psychiatry for the concepts we use to be clearly operationalized, to foster more effective public communication. A challenge to our disciplines is that many terms we use in special ways may overlap with lay language, and this can too often result in misinterpretation. Adoption of stricter definitions can help avoid misappropriation of our language.
Sharing is important. Currently ideas are shared in publications, conferences, lab meetings, and on the internet; these are the primary means for learning, communicating viewpoints, generating ideas, and designing hypotheses and experiments about cognition. But despite some widely read textbooks, there remains no collective repository of formalized knowledge about the structure of cognition, the methods used to measure its facets, or the relations of cognitive functions to behavioral disorders, brain function, or other biological characteristics. Given the demonstrated efficiency of collaborative networking applications like Wikipedia, we believe that is both technically feasible and desirable for a shared knowledge-base reflecting current thinking and argumentation about cognition and its relations to be developed.
If cognitive psychology and neuroscience were simple disciplines, independent of other areas of brain and biological sciences, there would be less need for ontology development. But cognitive psychology and neuroscience are intrinsically interdisciplinary sciences, and there is hope in coming years that substantial breakthroughs may emerge specifically at the interdisciplinary edges between cognitive sciences and other disciplines, particularly given the growth trajectories of genetics and neuroimaging. Cognitive ontologies will be important to maximize the potential for realizing such breakthroughs.
Challenges for Cognitive Ontology Development: Conflict Resolution and Lability of Concept Labels
Several of the challenges facing cognitive ontology development were mentioned above, including conspicuously the lack of formal definitions of key concepts and relations among entities, and the fact that cognitive concepts are not likely to be well represented by simple hierarchical “tree” structures. There are additional challenges because the usages of the concepts differ among investigators, and even the tasks used to measure cognitive constructs evolve over time. Some examples may clarify these points.
In Figure 3 a simple example was presented, in which the concept “response inhibition” was considered a kind of “cognitive control”, but also a kind of “executive function.” While resolving such a conflict might seem a simple matter of taste and of little consequence, such choices ultimately can impact research directions, selection of methods, and approaches to synthesizing results both within and across studies. It would be possible to define a class hierarchy and enforce unique parent-child relations, based on selected expert opinions. We believe that there is sufficient imprecision and ambiguity in the definition of cognitive concepts and their inter-relations that such mixed parentage should be allowed, and open to argumentation. Thus we are developing infrastructure to represent precisely such conflicts and enable active argumentation about the “correct” structure as perceived by the field, which we hope will be resolved by empirical evidence supporting one or the other point of view. Absent empirical substantiation, it is also possible to incorporate “voting” summaries, which can indicate which are majority and minority views, with annotation describing the rationale supporting each perspective. We hope that some approaches to classification will be pruned early. For example, the utility of a cognitive concept representing “frontal lobe functions” might be considered redundant, and could be obviated by creating relations of certain cognitive processes specifically to external validating measures of “frontal lobe function” as measured by fMRI activation or single unit activity in anatomically defined regions. Such tasks are simplified in part by reasonably well developed taxonomies of neuroanatomic structure (e.g., Foundational model of anatomy, Neuronames, and the modification of this utilized in the PubBrain application: www.PubBrain.org).
An interesting feature of cognitive concept labels is that these are not stable in use over time, and sometimes appear to reflect changes in fashion more than changes in either the underlying concept or the underlying measurements. For example, Jacobsen is usually credited with the initial observations of deficits in animals’ ability to perform delayed response tasks following lesions to the dorsolateral prefrontal cortex (Jacobsen, 1936). Subsequent studies revealed that this ability was associated with sustained activation in frontostriatal systems, and with unique firing patterns of pyramidal cells in the frontal lobe, during intervals in which animals were performing the delayed response tasks. Considerable work using this delayed response paradigm was conducted by Pribram and colleagues (for review see Pribram and McGuinness, 1975), Fuster and colleagues (e.g., Fuster and Alexander, 1971), Goldman-Rakic and colleagues (reviewed in Goldman-Rakic, 1987), and many others subsequently. Jacobsen referred to the delayed response behavior as “immediate memory”; Pribram referred to it as “activation”; Fuster called it “short term memory”; and Goldman-Rakic called it “working memory”. Experiments and theory focusing on these processes also have used the terms “cognitive control” (Posner and Snyder, 1975, Miller and Cohen, 2001). Interestingly, “cognitive control” is considered in Wikipedia a synonym for “executive function”, and within cognitive control some theorists have identified a key element as “context processing”, which may include “maintenance” and “updating” components (Braver and Barch, 2002). While each of these terms possess unique connotations, this example was used to emphasize that results of a single experiment (e.g., on a simple delayed response task) might be subject to a plethora of semantic labels, and is problematic precisely because these labels have distinct connotations.
The idea that popularity of concept labels may also affect usage even when connotations are not different is supported by a recent analysis that aimed to determine heritability for measures of “cognitive control” (Sabb et al., 2008). Since there was little genetic literature specifically mentioning “cognitive control”, we enumerated a series of five terms most frequently co-occurring with “cognitive control” in the PubMed literature over the last decade (these were “working memory,” “response inhibition,” “response selection,” and “task switching/set shifting”). We then sampled 30 papers that used each of these terms to determine which specific cognitive task measures were employed to assess each construct. The surprising result was, despite a clear ascendancy in use of the term “cognitive control” to describe results, there was not a single cognitive test that was uniquely associated with “cognitive control”; all of the specific measures had previously been used as an indicator of one of the other five concept labels. These findings further underscore the importance of anchoring cognitive concept labels at the measurement level.
Task Evolution and Task Speciation
Even if we are able to achieve higher consistency in labeling of results that are in fact consistent, there remains a challenge of identifying appropriate labels for tasks that have changed over time. Most tasks used in cognitive psychology research are not identical across different laboratories or even within the same laboratory over time. A major advantage of anchoring cognitive ontologies to the measurement level is that the strategy for determining changes in task properties is easier than tracking changes in concept definitions and usage. The process is easier because task parameters are usually (if not always) operationalized objectively, offering a clear basis to judge divergence in methods. The process is also easier because most tasks are based on prior tasks, and thus can more readily be considered descendants in a phylogenetic sense. The capacity to trace task modifications via their historical roots confers further advantages, by providing a phylogenetic perspective that may complement other methods for task classification. Thus for example, rather than classifying tasks by the putative cognitive processes that these measure in a “top-down” manner, it may be possible in many cases to trace origins to some smaller set of “founder” tasks. By tracing the lineage of a specific task implementation reported in the literature, it is feasible to determine more readily exactly which parameters changed between versions, so that any differences in associated effects may be more rationally attributed to specific manipulations.
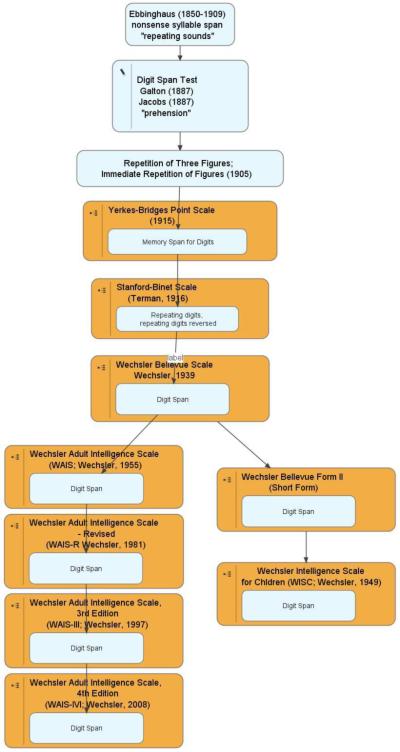
While there are not yet many detailed historical surveys on the origins of cognitive tasks, when available these offer unique insights into the current “state of the art”, and can offer perspective also on construct redefinitions that have emerged over time. For example, there is a very interesting analysis of the origins of the Wechsler IQ scales (Boake, 2002). We have constructed a diagram that summarizes that history (available online at www.phenomics.ucla.edu); Figure 4 extracts only 12 of the 95 elements from that diagram that pertain specifically to the familiar Digit Span test (please see Boake [2002] for complete citation information). Figure 4 illustrates how the digit span procedure has evolved over the last century, and further how concepts may have shifted regarding what functions are tapped by this test. According to Boake, Ebbinghaus considered this a measure of repeating sounds, while Galton and Jacobs felt this process was best labeled “prehension.” Through earlier versions of the Wechsler Scales, factor analytic studies initially led digit span performance to be considered an index of “attention”, while in studies of children a similar factor was labeled “freedom from distractibility.” More recent factor analytic studies suggest digit span scores load most on a “working memory” factor (although this followed inclusion of another “working memory” test, the Letter-Number Sequencing test, into these analyses). The digits backward condition also may be the most widely cited example of the “manipulation” component of working memory, or “working with memory.” The ephemeral quality of these labels belies the observation that key aspects of the actual test procedure have remained constant for 120 years.
Figure 4.
History of the “Digit Span” task (adapted from Boake, 2002), starting with the work of Ebbinghaus in the 19th century, through the most recent release of the WAIS-IV in 2008. Note: citation details are provided by Boake, 2002).
If tasks can be classified based on their phylogenetic origins, it is then reasonable to ask when a task has evolved significantly from its predecessors, to the point that it might be considered a new task “species”? We believe this process can be operationalized by introducing the concept of cognitive task “speciation.” Task speciation can be used to mark those events in test development that generate paradigms sufficiently unique that their results cannot be “mated” productively with others in meta-analysis. This determination remains a matter of some subjectivity, and rests with the individual investigator as to whether two sets of test results are sufficiently similar to be considered reasonable targets for meta-analytic combination. But by providing an objective framework, through a systematic analysis of specific task properties that have changed between versions, it may be superior to the methods used currently, which are usually not fully specified.
Taking the digit span procedures as an example (Figure 4), should we actually consider the WAIS-IV version to be the same task species as the original Ebbinghaus procedure? Multiple features of the task have changed, including the number of sequences, the rate of administration, the inclusion of the digits backwards condition, and of course the exact number that are to be repeated. Thus for many purposes one might argue that these are clearly different species, that should not be combined in meta-analyses. But what about results from the WAIS-III versus results from the WAIS-IV tasks with the same name? The WAIS-IV did modify the task to eliminate rhyming numbers; but did this really change the construct measurement significantly? For some investigators this difference might be trivial, but for someone interested in phonological processing and its effects, this difference might be of primary interest.
Despite such differences of opinion, we believe it is likely that clear taxonomies reflecting the origins of tasks and specifying the changes that have been introduced in different versions can be illuminating and of value in determining how best to synthesize data for specific purposes. Particularly important is the fact that this taxonomic development is ideally suited for collaborative, incremental development. Such work might be facilitated by the forthcoming release of the American Psychological Association’s PSYCTest database, which may offer a substantial target collection of psychological test names and associated annotation.
Lessons from Gene Ontologies
The Gene Ontology (GO; www.geneontology.org) project stands as a major success story in ontology development, and provides a useful example for development of schemata to represent higher levels of phenotypic expression. GO started as an effort to foster better compatibility among databases containing information about three model organisms (worms, flies, and mice). It has grown to include more than 25,000 terms in three ontologies spanning: biological processes, cellular components and molecular functions. GO has emerged as a research standard completely through interest of participating scientists (rather than through any efforts to impose or enforce this standardization). Bada and colleagues have emphasized a number of the key elements that fostered the successful development, deployment, and adoption of GO by its community (Bada et al., 2004).
Elements of this success included
community engagement; clear goals; limited scope; simple intuitive structure; continuous evolution; active curation; and early adoption. We enumerate below how each of these features of successful ontology development may apply to the development of cognitive ontologies.
Community engagement: Gene ontologies benefited from an eager and active group of investigators interested in developing methods to cope with the burgeoning data that were emerging from high-throughput genotyping efforts. It was not created by external engineers, but rather emerged within the biology research community from the individuals who were facing challenges finding and managing large amounts of information. We have at hand a comparably complex if not more daunting challenge, and will need to gather input from multiple disciplines, including but not limited to: cognitive neuroscience; neuropsychology; behavioral genetics; and neuroimaging research communities. Different levels of expertise will also be needed. Certain features of cognitive ontology development will likely benefit from the input of highly experienced experts in the field. Particularly the initial postulates, assertions, and arguments about these may appropriately be generated by “opinion leaders” who possess extensive prior knowledge. But the ultimate success in fleshing out relevant literature, detailing specific effects, and links to data will demand wide-ranging effort from individuals at all levels of training. Doctoral and postdoctoral trainees may be ideally suited to participate as critical stake holders given their engagement in digesting novel corpora of literature, and their efforts to develop coherent frameworks for digesting the evidence.
Clear goals and limited scope: The GO project benefited from having goals limited to the specification of gene products so that there would be consistent annotation of biological databases. There is a risk that development of a cognitive ontology could founder on efforts to make it too comprehensive, and thus it will be important to limit the scope of concepts that will be target of cognitive ontology development. As noted above, specification of any ontology requires minimally a lexicon (the collection of terms that describe the entities contained in the ontology) and specification of relation types (the actual semantic descriptions of how entities relate to each other). The scope of the cognitive concept list need not be exhaustive for proof of concept work, but should be reasonably comprehensive in order to support early applications such as unbiased literature retrieval. The Consortium for Neuropsychiatric Phenomics and Cognitive Atlas projects at UCLA have generated lists of some 5000 concepts representing “cognition”, and through automated and manual editing narrowed this lexicon to some 2000 terms that we believe offer a reasonable starting point in the representation of cognitive concepts and cognitive tasks. Specifying relations is complex, but there is likely a need to identify: (a) relations of cognitive concepts to specific task indicators (variables) used to measure these concepts (internal validity indicators); (b) relation of task indicators to the tasks themselves (it is possible to identify both general and task-specific context effects on these indicators; for example “response time” may have broad relations, across multiple tasks to some other external variables like aging, but also may mean markedly different things in one task relative to another); (c) relation of task indicators to other variables (external validity indicators). While there may be many types of external validity indicators, we believe it is useful to consider as starting points: (i) effect sizes for group difference (e.g., patients versus controls) and (ii) correlations (e.g., test variable correlates with age or cortical gray matter volume; notably other measures of association with variables that are categorical may also be transformed conveniently into correlation coefficients). The CNP work on cognitive ontologies is targeting selectively cognitive concepts and the tasks used to measure these. Elaboration of these concepts and relations to other levels is currently kept minimalistic, but open for individual user annotations to input more elaborate qualifications, and these may suggest future directions for extension.
Simple intuitive structure: GO benefited from the use of some simple, intuitively appealing class hierarchies and annotation links to decrease the barriers to use. The CNP and CA work on cognitive ontologies is also based on simple class hierarchies of cognitive concepts, and association graphs that help to illustrate how task indicators relate to the cognitive concepts, and how the cognitive concepts relate to each other. This fosters relatively straightforward navigation, and the capacity to browse readily at varying depths.
Early use, active curation, and continuous evolution: Key elements in the success of GO included development from initial goals with stepwise, meaningful additions, active engagement of its initial authors and a growing user base. It is too soon to tell what early use and adoption patterns will be for ontologies representing cognitive concepts and their inter-relations. Usage patterns will clearly depend on many of the factors listed above, particularly the perceived value to users for their work, and removing barriers to entry and use. Active curation will depend on adequate support, which will in turn likely depend on usage and perceived utility to the user community. Continuous evolution will depend on both use, diversity of inputs, and flexibility of the initial design, so that it readily accommodates change.
Cognitive Ontology Development: Frameworks for Literature Mining and Collaborative Knowledge-Building
We have initiated several projects to help develop cognitive ontologies, which we hope ultimately will both foster increased understanding and sharing of conceptual frameworks important to cognitive neuroscience, and further enable connections of information about cognitive phenotypes to other levels of biological knowledge. Some of this development took place under the aegis of the Center for Cognitive Phenomics, which was supported by a planning grant from the NIH Roadmap Initiative program in interdisciplinary research (P20RR020750). We are continuing this work within the Consortium for Neuropsychiatric Phenomics, particularly in the project “Hypothesis Web Development for Neuropsychiatric Phenomics” (RL1LM009833; D. Stott Parker, PI), and also in a new project “The Cognitive Atlas” (R01MH082795, Russ Poldrack, PI). Links to these projects and associated resources are maintained at www.phenomics.ucla.edu.
Among these developments are multiple freely available web applications. Several of these web-based applications support examination and visualization of cognitive concepts in the scientific literature as this is represented in PubMed (PubGraph, PubAtlas, and PubBrain). The scientific literature is essentially the phenomics database we possess today. From a conceptual standpoint, this can be considered a very large database that is minimally structured, and thus requires new literature mining technology to extract useful information. The phenomics information it contains is complex - far more complex than genomics data. Genomics data are linked explicitly with sequences, and hence can use sequences as an indexing framework for retrieving information. Phenomics is more challenging because phenotypes involve diverse measurements that lack a natural organization or indexing framework. Phenotype measurements in the literature are communicated using phrases, obliging us to retrieve information using simple language queries and other tools for literature mining. PubGraph, PubBrain, and PubAtlas are all attempts to provide “phenotype maps” based on the literature in PubMed/MEDLINE.
PubGraph allows a general specification of phenotypes, and maps phenotypes corresponding to individual literature queries into a topology (graph) reflecting strength of association. PubGraph permits users to either select from a number of pre-established lexica, or to input their own lexica, as the queries to PubMed. Nodes in these graphs reflect results of individual query expressions, and edges in these graphs represent measures of association between the nodes (e.g., the Jaccard coefficient, which is the intersection divided by the union of the queries).
PubAtlas emphasizes scale, using a grid to map associations of larger sets of phenotypes, and it also supports development of lexica to formalize hierarchies of phenotypes in the same way that the BLAST sequence alignment tool uses databases to formalize related sets of genetic sequences. These lexica are themselves elementary ontologies. PubAtlas offers similar capabilities to PubGraph, but with additional features, including thresholding by association strength, along with “velocity plots” showing the co-occurrences history by year. For example, Figure 5 illustrates co-occurrences of the term “digit span” with a lexicon of 900 cognitive concept terms, thresholded to reveal only the strongest associations (natural log of Jaccard coefficient > 10-4). The association table shows that the strongest associations (other than associations with “digit span” itself, are with “attention span” and “memory span”). Also shown in Figure 5 is the history plot for association of “digit span” with the concept “working memory”, which reveals a steady increase in the late 1980’s and an acceleration of use in the 21st century. By clicking on any of the table entries, PubAtlas invokes PubMed and generates the query of interest so that the user can inspect the relevant literature directly.
Figure 5.
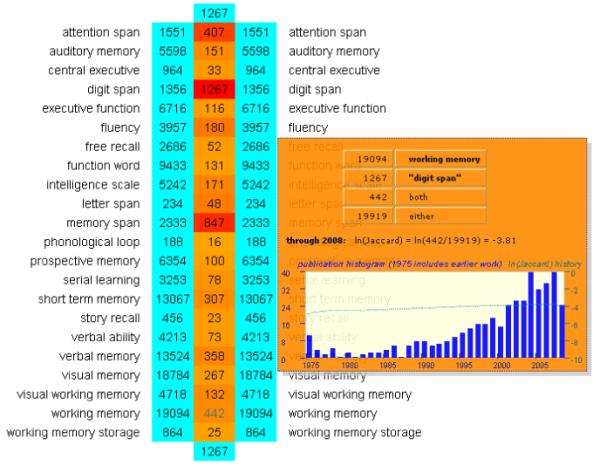
PubAtlas output for the intersection of “digit span” with a lexicon of 900 cognitive concepts, thresholded to show only associations with ln Jaccard coefficient >-4.0. The “heat map” reveals strong associations with “memory span”, “attention span” and “working memory”. The insert shows the history in use of the term “working memory” together with “digit span”.
PubBrain maps the literature into phenotype space -- by displaying PubMed hits in the 3D phenotype geometry of brain anatomical regions. PubBrain performs this using a “blast” query of any arbitrary PubMed search with respect to an ontology representing neuroanatomic terms (adapted from the Foundational Model of Anatomy and its NeuroNames lexicon). After identifying co-occurrences of a specific term with the anatomic terms, PubBrain then projects the co-occurrence statistics onto a three-dimensional probabilistic atlas of the human brain, which was developed in collaboration with the Laboratory of NeuroImaging (LONI) at UCLA (Shattuck et al., 2008). Figure 6 provides the PubBrain output for the query “digit span”, illustrating the retrieval results for an arbitrary location (crosshairs positioned in the cingulate gyrus), and generating links to 32 publications that were used to generate this map.
Figure 6.
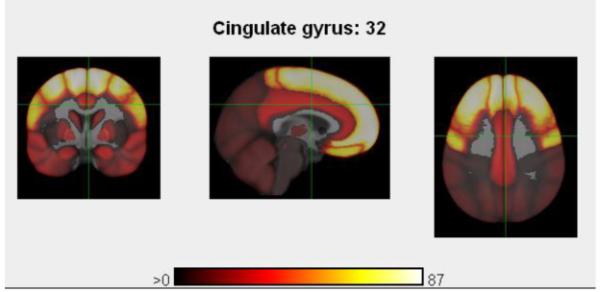
PubBrain output showing the projection of the query “digit span” on a three-dimensional probabilistic atlas of the brain; this view was centered on the cingulate gyrus (32 hits). Other frontal cortical regions had up to 87 co-occurrences.
These systems (PubGraph, PubBrain, and PubAtlas) illustrate how phenomics requires new tools centered on the phenotype concept. These tools now rest on PubMed, but they will be able to evolve as phenomics becomes better formalized, and curated phenomic databases become important. Thus, they not only show the importance of visual interfaces for understanding complex phenotype data, but also give us a view of “phenotype mapping” tools of the future.
Several other projects are underway to foster collaborative development of ontologies and collect annotation about cognitive concepts. The explosion of social collaborative networking and collaborative knowledge development during the “Web 2.0” era resulted in some dramatic achievements. The Wikipedia is one of the most widely appreciated of these deliverables, now containing more than 2.5 million articles. Wikipedia already is almost 40 times larger than the Encyclopædia Britannica, and at least some investigations have suggested the quality of scientific articles is not markedly different between these two sources of information (Giles, 2005). Concern about the validity of Wikipedia’s content is not preventing its widespread use and continued growth. While only a decade ago “the wisdom of crowds” might have seemed an oxymoron, today there is concrete evidence that Web 2.0 has fulfilled at least part of its promise. The development of the web itself spawns futuristic thinking, and Web 3.0 already has been a topic of discussion since at least 2006 (this coinage is attributed to John Markoff of the New York Times, according to Wikipedia). According to some, the way forward will be paved with key features of the semantic web, which would add semantic structure and conventions to web contents to facilitate information storage, retrieval, and sharing. Ontology development is among the key elements of the Web 3.0 vision. In both the Consortium for Neuropsychiatric Phenomics and Cognitive Atlas projects, collaborative knowledge-building is a central goal.
The Phenowiki (www.phenowiki.org) was developed to serve as a collaborative database for phenotype annotation. It contains wiki-like descriptions for a series of cognitive tasks and concepts, and these task and concept entries are linked to an underlying database that contains quantitative annotation about specific study results. For example, it already includes annotations about 8 publications that used the Digit Span test, including quantitative results about heritability coefficients, effect sizes for group differences between various patient and comparison groups, and reliability coefficients for the specific measures that are derived for this test. Some additional applications of the Phenowiki were described by Sabb and colleagues (Sabb et al., 2008). The Phenowiki already can serve as a collaborative repository for storing quantitative effect size information about group differences, treatment effects, and other psychometric properties of individual tasks, and outputs of the Phenowiki can be used directly to generate meta-analytic summary statistics. Future developments of this tool are aimed to enable automated meta-analysis of selected topic areas and for specific task results.
The Hypothesis Web project aims further to develop collaborative tools for the specification, visualization, and sharing of scientific hypotheses with quantitative annotation. Currently, most hypotheses are laid out in scholarly discussions as a series of verbal postulates without much quantification of each assertion. Instead, each assertion is assigned a dichotomous value (i.e., “true”) based on scientific findings that are almost always only true to some degree of statistical certainty. We might take for example the “COMT hypothesis of schizophrenia” (this example chosen because at least one of us [RMB] is guilty of such unquantified theoretical speculation). The original hypothesis linking the val158met polymorphism in the COMT gene with schizophrenia included some tantalizing elements: the COMT gene affects the COMT enzyme; the COMT enzyme affects dopamine (DA) metabolism; antipsychotic drugs affect DA metabolism; antipsychotic drugs ameliorate symptoms of schizophrenia; DA metabolism affects frontal function and cognitive function; velocardiofacial syndrome involves a “knockout” of the COMT gene and is associated with schizophrenia; and more (for a review, see Williams et al., 2007). This hypothesis has now been the subject of more than 250 papers, which “virtually exclude” the likelihood of a simple link between the most studied Val158Met polymorphism and schizophrenia (Williams et al., 2007). Imagine that from the beginning of this quest to find links of a genetic variant to a complex syndrome such as schizophrenia, we had conditioned each assertion with clearer explication of quantitative effect sizes (and appropriate confidence intervals) linking each level of this hypothesis to the next. By stating more explicitly that the COMT gene has only partial effect on the enzyme, the enzyme only partial effect on DA concentrations in the PFC, DA concentrations in PFC only partial effect on neural network function, and so on, our failure to find associations with the complex syndrome might seem less surprising, and help keep us focused on the critical dimensions of the hypothesis that may possess validity and be important for cognitive neuroscience. The Hypothesis Web project aims to bring to users convenient methods for specifying and manipulating assertions, with quantitative annotation, in a collaborative framework. While still early in its development, the PubAtlas application already enables users to generate literature-based “models” that specify the nodes of a particular hypothesis as articulated via PubMed queries, and to assess the associations between nodes in terms of literature co-occurrence statistics. It is hoped that one day applications like the Hypothesis Web will help revolutionize the way we present hypotheses (for example, in the Background and Significance sections of grant applications, and in the Introduction sections of scientific publications). Rather than presenting arguments as a series of partially qualified statements, hypotheses of the future may include more formal specification and quantification of their evidentiary bases.
The Cognitive Atlas project specifically aims to provide the infrastructure for collaborative development of cognitive ontologies. The central organizing framework for this development will be a web application in which cognitive concepts are represented. It will allow networks of researchers to collaborate on assertions about mental concepts, including how these are measured, how the concepts relate to each other, and how the concepts relate to other entities (such as diagnostic groups, drug effects, or neural system activations). A key feature of this infrastructure will be its capacity to represent uncertainty about assertions, and to represent explicitly disagreement and controversy. This will involve verbal arguments for and against key assertions, but will further enable representation of quantitative evidence supporting the validity (or lack thereof) regarding assertions. For example, a specific “node” in the cognitive atlas representing the validity of a concept (”the phonological buffer”) might be supported by some literature (Baddeley and Hitch, 1974) but not supported by other literature (Jones et al., 2004). Users will be able to “vote” as to which of these two contradictory opinions they find most persuasive, providing a quantitative index of community-perceived validity, and add comments supporting their opinions. Additional quantitative indices regarding cognitive constructs will be provided by associating empirical data with the edges in the conceptual network. For example, the assertion that “working memory is impaired in schizophrenia” might be supported by evidence showing that patients with the diagnosis of schizophrenia showed deficits relative to a healthy comparison group on the “N-back task”, and that this relative deficit is associated with a quantitative effect size (such as Cohen’s d), of a specific magnitude, and with quantified confidence intervals around that effect size. With the accrual of a sufficient database of empirical findings, it should become possible to test competing ontologies by comparing their implied covariance structure to the actual covariance structure of these data, using methods like the meta-analytic structural equation modeling approach described above. The knowledge base developed in the Cognitive Atlas will provide a formal conceptual infrastructure for the annotation of behavioral and neuroscientific data, allowing the kind of large-scale data mining that has become central in bioscience but has not yet penetrated cognitive neuroscience.
Summary & Conclusion
We now face unprecedented challenges and opportunities to pursue the new transdiscipline of phenomics and relate the human genome to complex neuropsychiatric syndromes. Cognitive phenotypes may offer a rational focus of inquiry and help bridge knowledge from neural systems to symptomatic levels. To enable these connections cognitive neuropsychiatry needs cognitive ontologies -- structured representations of cognitive concept domains and their measurements. This paper summarizes some of the specific challenges faced in developing cognitive ontologies, and offers strategies to help overcome barriers posed by the abstract nature and temporal instability of cognitive concepts. Examples of works-in-progress, and opportunities for collaborative development are presented. It is hoped that these efforts will help advance cognitive neuroscience and ultimately our understanding and treatment of neuropsychiatric syndromes.
Acknowledgements
This work was supported by USPHS grants including the NIH Roadmap Initiative Consortium for Neuropsychiatric Phenomics, including linked awards UL1DE019580 (Bilder, Sabb), RL1MH083268 (Freimer), RL1LM009833 (Parker, Chu, Fox), and The Cognitive Atlas (R01MH082795; Poldrack, Kalar). The development of this work was also supported by the NIH Roadmap exploratory center for interdisciplinary research (P20RR020750), and the Tennenbaum Center for the Biology of Creativity.
Footnotes
Since ontologies are created to enable the classification of instances, they may not actually contain the instances themselves; nevertheless it is important to consider the actual “things” that the ontology aims to classify.
References
- American Psychiatric A . Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- Bada M, Stevens R, Goble C, Gil Y, Ashburner M, Blake JA, Cherry JM, Harris M, Lewis S. (A short study on the success of the Gene Ontology. J Web Semantics. 2004;1:235–240. [Google Scholar]
- Baddeley AD, Hitch G. Working memory. In: Bower GH, editor. The psychology of learning and motivation: Advances in research and theory. Vol. 8. Academic Press; New York: 1974. pp. 47–89. [Google Scholar]
- Bilder RM. Phenomics: building scaffolds for biological hypotheses in the post-genomic era. Biological psychiatry. 2008;63:439–440. doi: 10.1016/j.biopsych.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Czobor P, Malhotra AK, Kennedy JL, Ni X, Goldman RS, Hoptman MJ, Sheitman B, Lindenmayer JP, Citrome L, McEvoy JP, Kunz M, Chakos M, Cooper TB, Lieberman JA. Neurocognitive correlates of the COMT Val(158)Met polymorphism in chronic schizophrenia. Biological psychiatry. 2002;52:701–707. doi: 10.1016/s0006-3223(02)01416-6. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004;29:1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- Boake C. From the Binet-Simon to the Wechsler-Bellevue: tracing the history of intelligence testing. J Clin Exp Neuropsychol. 2002;24:383–405. doi: 10.1076/jcen.24.3.383.981. [DOI] [PubMed] [Google Scholar]
- Braver TS, Barch DM. A theory of cognitive control, aging cognition, and neuromodulation. NeurosciBiobehavRev. 2002;26:809–817. doi: 10.1016/s0149-7634(02)00067-2. [DOI] [PubMed] [Google Scholar]
- Centler F, Kaleta C, di Fenizio PS, Dittrich P. Computing chemical organizations in biological networks. Bioinformatics. 2008;24:1611–1618. doi: 10.1093/bioinformatics/btn228. [DOI] [PubMed] [Google Scholar]
- Cheung MW. A model for integrating fixed-, random-, and mixed-effects meta-analyses into structural equation modeling. Psychol Methods. 2008;13:182–202. doi: 10.1037/a0013163. [DOI] [PubMed] [Google Scholar]
- Chiang PK, Lam MA, Luo Y. The many faces of amyloid beta in Alzheimer’s disease. Curr Mol Med. 2008;8:580–584. doi: 10.2174/156652408785747951. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, MacIntyre DJ, MacLean AW, St CD, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Ferrier IN, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37:163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- Furlow CF, Beretvas SN. Meta-analytic methods of pooling correlation matrices for structural equation modeling under different patterns of missing data. Psychol Methods. 2005;10:227–254. doi: 10.1037/1082-989X.10.2.227. [DOI] [PubMed] [Google Scholar]
- Fuster JM, Alexander GE. Neuron activity related to short-term memory. Science. 1971;173:652–654. doi: 10.1126/science.173.3997.652. [DOI] [PubMed] [Google Scholar]
- Giles J. Internet encyclopaedias go head to head. Nature. 2005;438:900–901. doi: 10.1038/438900a. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Handbook of physiology Vol5 The nervous system. American Physiological Society; Bethesda, MD: 1987. Circuitry of primate prefrontal cortex and regulation of behavior by representational memory; pp. 373–417. [Google Scholar]
- Grethe J, Gupta A, Ludascher B, Martone ME. BIRN ontologies session. Vol. 2008. 2003. p Powerpoint slide show. [Google Scholar]
- Gruber TR. Knowledge Acquisition. Vol. 5. 2003. A translation approach to portable ontologies; pp. 199–220. [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. Journal of Neuroscience. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam N. Categorical versus dimensional models of mental disorder: the taxometric evidence. Australian and New Zealand Journal of Psychiatry. 2003;37:696–704. doi: 10.1080/j.1440-1614.2003.01258.x. [DOI] [PubMed] [Google Scholar]
- Haslam N, Kim HC. Categories and continua: A review of taxometric research. Genetic Social and General Psychology Monographs. 2002;128:271–320. [PubMed] [Google Scholar]
- Jacobsen CF. Studies of cerebral function in primates, I. The function of the frontal association areas in monkeys. JCompPsychol. 1936;13:3–60. [Google Scholar]
- Jones DM, Macken WJ, Nicholls AP. The phonological store of working memory: is it phonological and is it a store? J Exp Psychol Learn Mem Cogn. 2004;30:656–674. doi: 10.1037/0278-7393.30.3.656. [DOI] [PubMed] [Google Scholar]
- Kauffman S. At Home in the Universe: The Search for Laws of Self-Organization and Complexity. Oxford; New York: 1995. [Google Scholar]
- Kendler KS. Psychiatric genetics: a methodologic critique. American Journal of Psychiatry. 2005;162:3–11. doi: 10.1176/appi.ajp.162.1.3. [DOI] [PubMed] [Google Scholar]
- Kraemer HC. DSM categories and dimensions in clinical and research contexts. International Journal of Methods in Psychiatric Research. 2007;16:S8–S15. doi: 10.1002/mpr.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Nolan KA, Bilder RM, Lachman HM, Volavka J. Catechol O-methyltransferase Val158Met polymorphism in schizophrenia: differential effects of Val and Met alleles on cognitive stability and flexibility. AmJ Psychiatry. 2004;161:359–361. doi: 10.1176/appi.ajp.161.2.359. [DOI] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Poldrack RA. Can cognitive processes be inferred from neuroimaging data? Trends Cogn Sci. 2006;10:59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Posner MI, Snyder CRR. Attention and cognitive control. In: Solso RL, editor. Information Processing and Cognition. Erlbaum; Hillsdale, NJ: 1975. pp. 55–85. [Google Scholar]
- Pribram KH, McGuinness D. Arousal, activation, and effort in the control of attention. Psychological review. 1975;82:116–149. doi: 10.1037/h0076780. [DOI] [PubMed] [Google Scholar]
- Riley RD, Abrams KR, Sutton AJ, Lambert PC, Thompson JR. Bivariate random-effects meta-analysis and the estimation of between-study correlation. BMC Med Res Methodol. 2007;7:3. doi: 10.1186/1471-2288-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabb FW, Bearden CE, Glahn DC, Parker DS, Freimer N, Bilder RM. A collaborative knowledge base for cognitive phenomics. Molecular psychiatry. 2008;13:350–360. doi: 10.1038/sj.mp.4002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Chen G, Malkani S, Choi SY, Takahashi RH, Zhang D, Gouras GK, Kirkwood A, Morris RG, Shen J. Conditional inactivation of presenilin 1 prevents amyloid accumulation and temporarily rescues contextual and spatial working memory impairments in amyloid precursor protein transgenic mice. J Neurosci. 2005;25:6755–6764. doi: 10.1523/JNEUROSCI.1247-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S, Dhamdhere K, Blelloch G, Halperin E, Ravi R, Schwartz R. Algorithms for efficient near-perfect phylogenetic tree reconstruction in theory and practice. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:561–571. doi: 10.1109/TCBB.2007.1070. [DOI] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5033–5037. doi: 10.1073/pnas.91.11.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM. Measures of degeneracy and redundancy in biological networks. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3257–3262. doi: 10.1073/pnas.96.6.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Owen MJ, O’Donovan MC. Is COMT a susceptibility gene for schizophrenia? Schizophr Bull. 2007;33:635–641. doi: 10.1093/schbul/sbm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Yeh CW, Yang CD, Jang SS, Chu IM. On the local optimal solutions of metabolic regulatory networks using information guided genetic algorithm approach and clustering analysis. J Biotechnol. 2007;131:159–167. doi: 10.1016/j.jbiotec.2007.06.019. [DOI] [PubMed] [Google Scholar]