SUMMARY
The genome of the malaria parasite Plasmodium falciparum contains several multi-copy gene families, including var, rifin, stevor, and Pfmc-2TM. These gene families undergo expression switching and appear to play a role in antigenic variation. It has recently been shown that forcing parasites to express high copy numbers of transcriptionally active, episomal var promoters led to gradual down-regulation and eventual silencing of the entire var gene family, suggesting that a limiting titratable factor plays a role in var gene activation. Through similar experiments using rifin, stevor or Pfmc-2TM episomal promoters we show that promoter titration can be used as a general method to down-regulate multi-copy gene families in P. falciparum. Additionally, we show that promoter titration with var, rifin, stevor or Pfmc-2TM episomal promoters results in down-regulation of expression not only of the family to which the episomal promoter belongs, but also members of the other gene families, suggesting that the var-specific titratable factor previously described is shared by all four families. Further, transcriptionally active promoters from different families colocalize within the same subnuclear expression site, indicating that the role that nuclear architecture plays in var gene regulation also likely applies to the other multi-copy gene families of P. falciparum.
Keywords: transcription, gene expression, antigenic variation, promoter titration
INTRODUCTION
The genome of Plasmodium falciparum, the causative agent of the most severe form of human malaria, contains several multi-copy gene families including var, rifin, stevor, and Pfmc-2TM (Dzikowski, Templeton, and Deitsch, 2006). The members of these families are interspersed immediately adjacent to the telomeric repeats found at chromosome ends, as well as within large gene clusters located at several internal chromosomal loci (Kyes et al., 1999). While there are ~60 var genes within the haploid parasite genome, the rifin, stevor, and Pfmc-2TM gene families contain approximately 150, 35, and 13 members, respectively (Gardner et al., 2002). Of these four families, only var, which encodes the variable surface antigen called PfEMP1, has a known function.
Upon invasion of human red blood cells, the parasite places PfEMP1 on the surface of the infected cell (iRBC) (Kyes, Horrocks, and Newbold, 2001). This protein mediates cytoadherence of the iRBCs to various receptors on the vascular endothelium of the host’s circulatory system, removing them from the circulation and avoiding splenic clearance. The resulting vascular obstruction and localized immune response is thought to be responsible for many of the symptoms associated with severe malaria, and PfEMP1 is currently the sole known Plasmodium virulence factor (Miller et al., 2002). Due to its exposure on the iRBC surface, infected individuals readily produce antibodies to the form of PfEMP1 expressed by the parasites, however by limiting expression to a single var gene at a time, and by regularly switching between different var genes, parasites are able to avoid antibody recognition and maintain a lengthy, persistent infection. This process, called antigenic variation, requires strict mutually exclusive expression within the var gene family, a complex mechanism that remains poorly understood.
Like var genes, the rifin, stevor and Pfmc-2TM families have recently been shown to undergo expression switching (Lavazec, Sanyal, and Templeton, 2007;Niang, Yan, X, and Preiser, 2009;Kyes et al., 1999), leading to the suggestion that they may be involved, along with var genes, in antigenic variation. The proteins encoded by these gene families all belong to the two-transmembrane superfamily and have similar structures (Sam-Yellow e et al., 2004;Lavazec, Sanyal, and Templeton, 2006), and have been shown to localize to the RBC surface (Kyes et al., 1999;Niang, Yan, X, and Preiser, 2009;Lavazec, Sanyal, and Templeton, 2006), suggesting a role in interactions with the extracellular environment. However, despite this evidence, phenotypes associated with changes in expression within these multi-copy gene families have been difficult to ascertain and as of now, the RIFINs, STEVORS, and PfMC-2TMs have no known function.
Little is known about the mechanisms that regulate transcription in P. falciparum. Initial computational analysis of the Plasmodium genome identified few putative specific transcription factors (Gardner et al., 2002) leading to the suggestion that other mechanisms, such as promoter interactions, non-coding RNAs, chromatin modification and subnuclear architecture might play an unusually important role in regulating gene expression (Aravind et al., 2003). Studies of transcriptional regulation of the var gene family have identified many components that contribute to activation, silencing and mutually exclusive expression, including aspects of chromatin remodeling (Chookajorn et al., 2007;Lopez-Rubio et al., 2007;Freitas-Junior et al., 2005;Lopez-Rubio, Mancio-Silva, and Scherf, 2009), promoter-promoter interactions (Deitsch, Calderwood, and Wellems, 2001;Calderwood et al., 2003;Frank et al., 2006;Dzikowski et al., 2007) and noncoding RNAs (Su et al., 1995;Kyes et al., 2003;Epp et al., 2008). There is also evidence that subnuclear positioning might be important for var gene activation (Duraisingh et al., 2005;Voss et al., 2006;Ralph, Scheidig-Benatar, and Scherf, 2005), and transgenic parasites that have been genetically modified to activate more than one var promoter at a time have shown that active var promoters co-localize to a specific spot in the periphery of the parasite nucleus (Dzikowski et al., 2007), suggesting the presence of a var specific transcriptionally active site. This type of subnuclear localization has been shown to play a role in the mutually exclusive expression of variant surface glycoprotein (vsg) genes in African Trypanosomes (Navarro and Gull, 2001).
Despite the prominent involvement of chromatin modification and nuclear architecture in regulating variant gene expression in P. falciparum, it is likely that specific transcription factors also play a prominent role. Basal transcription factors including the TATA-box-binding protein have been identified, suggesting a transcription factor-based regulatory system (Callebaut et al., 2005), and DNA elements from the upstream regulatory regions of various genes display the proper timing of expression when placed on transfected episomal constructs (Koning-Ward et al., 1999;Dechering et al., 1999). Additionally, the complex nature of the Plasmodium lifecycle and its tightly regulated gene expression profiles suggest a need for specific transcriptional regulators (Bozdech et al., 2003;Le Roch et al., 2003). Recently, the ApiAP2 family of proteins, possessing weak homology to the plant AP2/ERF transcription factors, have been investigated as potential transcriptional regulators in P. falciparum (Balaji et al., 2005). One member of this family (PF14_0633) has been found by protein-binding microarray (PBM) and computational analysis to associate with var promoters (De Silva et al., 2008). Additionally, DNA binding assays using parasite nuclear extracts have identified protein complexes that bind to specific elements within the regulatory regions upstream of var promoters (Voss et al., 2003), and more recent studies have demonstrated the presence of a specific titratable nuclear factor required for var gene expression (Dzikowski and Deitsch, 2008). Titration of this factor by the presence of multiple transcriptionally active episomal var promoters is capable of repressing transcription of the entire endogenous multi-copy var gene family.
Since P. falciparum lacks the canonical RNAi pathway, and disrupting up to 150 genes in a multi-copy gene family is not feasible, knock-down of the rifin, stevor and Pfmc-2TM gene families and subsequent phenotypic analysis has been impossible to achieve. Here we show that down-regulation of gene expression by promoter titration as originally described by Iyer et al. (Iyer et al., 2007) and subsequently applied to the var gene family by Dzikowski et al. (Dzikowski and Deitsch, 2008) can be used as a general method to down-regulate multi-copy gene families, including rifin, stevor and Pfmc-2TMs, in P. falciparum. Additionally, we show via microarray and qRT-PCR that the var, rifin, stevor and Pfmc-2TM gene families share a common titratable factor necessary for expression. Episomal promoters of the four families are reciprocally capable of down-regulating the chromosomal members of the other families. The extent of the cross-down-regulation is dependent upon the temporal expression patterns of the gene families, with the most similarly expressed gene families showing the greatest reciprocal titration effect. Interestingly, although high copy numbers of episomal stevor promoters results in down-regulation of their own gene family, they up-regulate the Pfmc-2TM gene family, suggesting the possibility of functional redundancy and compensation between the proteins encoded by the two families. Finally, through DNA FISH, we show that a transcriptionally active episomal rifin promoter colocalizes with an active var promoter, indicating that these gene families utilize the same subnuclear expression site.
RESULTS
Promoter titration can be used to down-regulate multi-copy gene families in Plasmodium falciparum
Recently Iyer et al. showed that transfection of the rodent parasite P. yoellii with an episomal plasmid carrying the upstream regulatory region and promoter of the 235 kDa rhoptery protein led to an unexpected down-regulation of the endogenous, chromosomal copy of the 235 kDa gene (Iyer et al., 2007). The authors speculated that this down-regulation could be the result of promoter competition for a limiting transcription factor, as has been described in other systems (Schodin et al., 1995). Dzikowski et al. subsequently extended this technique to the study of var gene expression and showed that by forcing parasites to carry large numbers of transcriptionally active episomal var promoters, it is possible to down-regulate and ultimately silence all endogenous, chromosomal var gene expression (Dzikowski and Deitsch, 2008). This was achieved by transfecting parasites with a plasmid carrying a blasticidin-S-deaminase (bsd) cassette driven by a var promoter that was made constitutively active by separating it from a regulatory element found in var introns. By using the bsd selectable marker, it is possible to regulate the copy number of episomally replicating plasmids, with exposure to increasing blasticidin concentrations resulting in larger concatamers (Epp, Raskolnikov, and Deitsch, 2008). This in turn leads to greater levels of repression of the chromosomal genes and suggests the presence of a titratable nuclear factor necessary for endogenous var gene expression (Dzikowski and Deitsch, 2008). The ability to knock-down expression of entire multi-copy gene families in P. falciparum would be a useful tool since current technology for down-regulating genes is limited to knockout strategies, which are not applicable to large gene families. We therefore decided to investigate whether promoter titration could be used as a general method for down-regulation of multi-copy gene families in P. falciparum.
The rifin, stevor and Pfmc-2TM multi-copy gene families were chosen for further investigation. The var promoter in the plasmid pVBH (Dzikowski et al., 2007) was replaced with a representative promoter from each of the three gene families and the resulting constructs were transfected into two recently sub-cloned lines, C3 and B3, which were derived from the parasite isolate NF54. The C3 parasite line predominantly expresses a single var gene (PFD1005c) (Frank et al., 2007) while the B3 parasite line primarily expresses one stevor (MAL7P1.227) and one Pfmc-2TM (PFA0680c) (Lavazec, Sanyal, and Templeton, 2007). Stably transformed parasites were initially selected using 2μg/ml blasticidin, a concentration anticipated to select for low plasmid copy numbers. The cultures were then split and one was subjected to treatment with 10μg/ml blasticidin for two weeks or until normal growth rates were restored. RNA was then harvested from synchronized cultures at eighteen, twenty-four or twenty-eight hours post invasion for investigation into var, stevor, or Pfmc-2TM transcript levels, respectively and genomic DNA was collected to determine episomal plasmid copy numbers (Figure 1A). qRT-PCR assays were performed on each pair of cDNA samples using gene specific primer sets to the var, stevor, or Pfmc-2TM families. The large size of the rifin gene family and our lack of gene specific primers to all 135 members precluded us from assessing expression levels of this family by qRT-PCR. Therefore analysis of rifin gene expression was determined by hybridization to a microarray of P. falciparum 3D7coding regions.
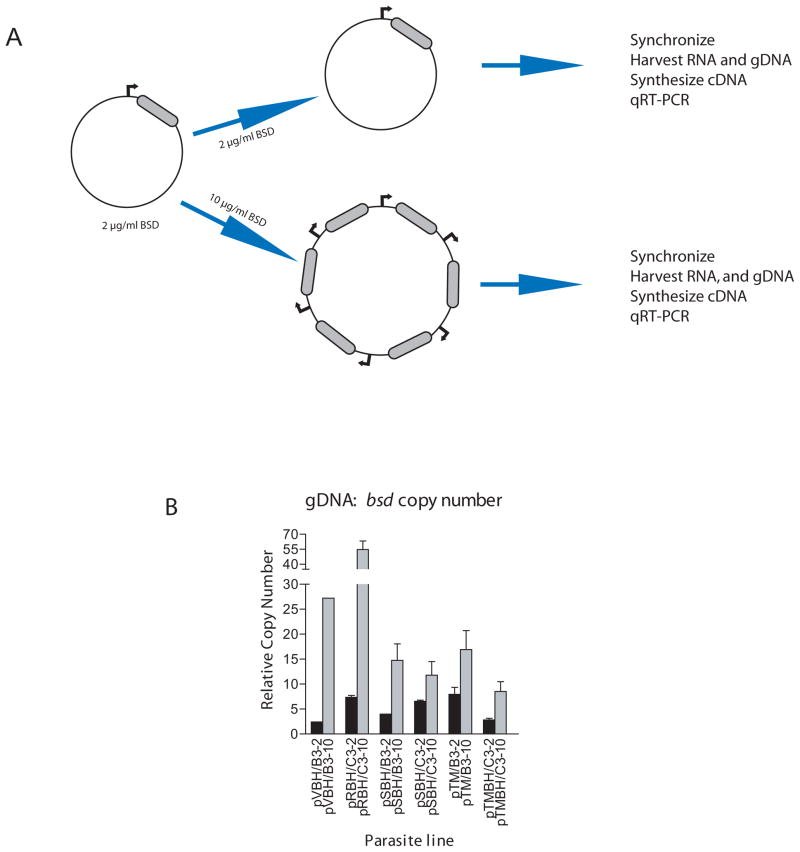
Figure 1.
Method of Blasiticidin Titration. (a) Plasmids carrying either a var (Dzikowski et al., 2007), rifin (PF10_0398), stevor (PFF1550w), or Pfmc-2TM (PFF0060w) promoter driving a bsd cassette (grey region) were transfected into parasites and selected with 2 μg/ml BSD. Cultures were then split and one subculture was subjected to higher levels of drug, forcing the parasite to carry higher copy numbers of the episomal promoter. Cultures were then synchronized, RNA extracted, cDNA synthesized, and qRT-PCR performed. (b) Genomic bsd copy number in each transformed parasite line under low (black bars) and high (grey bars) drug doses. In each line, the number of bsd copies, and therefore promoter copies, increases under higher drug pressure. Copy numbers were determined using q-PCR and comparison to the single-copy genes P60-seryl-tRNA synthetase (PF07_0073), P61-fructose bisphosphate aldolase (PF14_0425) and P100-actin (PFL2215).
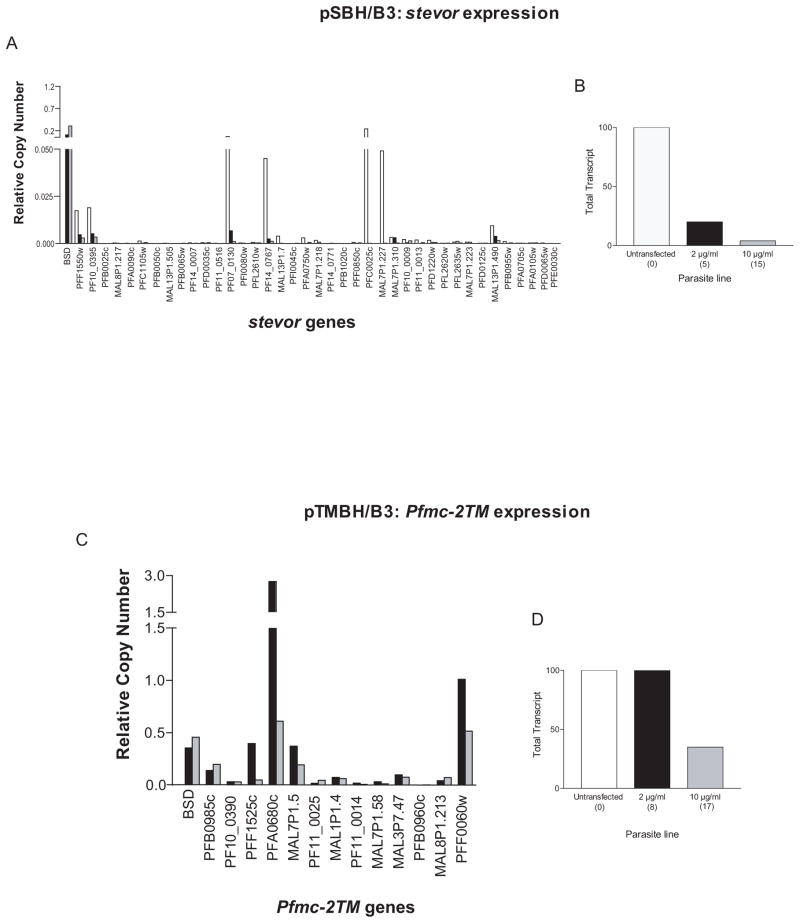
As expected, growth of the transformed parasites lines under high concentrations of blasticidin resulted in a corresponding increase in the copy number of episomal plasmids containing var, rifin, stevor and Pfmc-2TM promoters (Figure 1B). The differences in plasmid copy numbers between the different transgenic lines are most likely due to differences in promoter strengths and thus the number of bsd cassettes required to provide sufficient deaminase activity to survive the selection pressure. In order to determine if increasing the number of active episomal stevor and Pfmc-2TM promoters resulted in down-regulation of the endogenous members of these gene families, the B3 transfectants (pSBH/B3 and pTMBH/B3, respectively) grown under low and high doses of blasticidin were assayed by qRT-PCR and the levels of endogenous gene expression compared. In the SBH/B3 line, endogenous stevor transcript levels were strongly repressed in the transfected lines, with parasites grown under high doses of blasticidin displaying greater repression than those grown under low doses (Figure 2A). When the total endogenous stevor gene expression levels of the parasites under high doses of blasticidin were compared to that of the untransfected B3 line, a reduction of approximately 95% was observed (Figure 2B). The relatively strong repression in parasites grown under low levels of blasticidin suggests that the presence of even the small concatamers may be repressing endogenous gene expression to a significant degree. Similarly in the TMBH/B3 line, high copy numbers of Pfmc-2TM episomal promoters down-regulated total endogenous Pfmc-2TM transcript levels by 65% (Figure 2D). However, unlike stevor genes, transfected parasites grown under low doses of blasticidin showed no down-regulation as the gene expression levels were comparable with those from untransfected B3 parasites (Figure 2D). In all cases, levels of repression were more tightly correlated with increases in episomal promoter copy numbers rather than levels of bsd transcripts, consistent with the idea that it is the total number of episomal promoters that is important for titration rather than the number of transcripts being produced. Despite the reduced gene expression in the lines grown under high doses of blasticidin, parasite transfected with either plasmid displayed no obvious phenotype outside of the initial delayed growth previously reported (Dzikowski and Deitsch, 2008).
Figure 2.
Downregulation of endogenous stevor and Pfmc-2TM expression by increased copy numbers of the plasmids pSBH and pTMBH, respectively. (a) Change in stevor RNA levels following increase in copy number of pSBH. Expression levels are shown of endogenous stevor genes in pSBH/B3 under low (black bars) and high (grey bars) blasticin doses. Untransfected B3 parasites (carrying no episome) are represented by white bars. All RNA samples were taken at 24 hours post invasion. (b) Representation of data depicted in (a). Total stevor transcript levels of untransfected B3 parasites (white bar) and pSBH transfected B3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. (c) Change in Pfmc-2TM RNA levels following increase in copy number of pTMBH. Expression levels are shown of endogenous Pfmc-2TM genes in pTMBH/B3 parasites under low (black bars) and high (grey bars) blasticidin doses. All RNA samples were taken at 28 hours post invasion. (d) Representation of data depicted in (c). Total Pfmc-2TM transcript levels of untransfected B3 parasites (white bar) and pTMBH transfected B3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. In both parasite lines, the episomal promoters repress expression of their own endogenous gene family. Relative copy numbers are measured with respect to the control housekeeping gene P61-fructose bisphosphate aldolase (PF14_0425) using the formula 2−ΔΔCT with NF54 gDNA as the calibrator.
To determined if the reduced endogenous gene expression was simply a side-effect of the increased blasticidin dose, parallel experiments were performed using the plasmid pHBIRH in which bsd is driven by a var intron promoter (Epp, Raskolnikov, and Deitsch, 2008). No reduction in var expression was observed (data not shown), demonstrating that the effect was not a non-specific result of large plasmid copy numbers or exposure to high blasticidin concentrations. Thus, the promoter titration method can be used as a general tool for the specific down-regulation of multi-copy gene families in P. falciparum.
var, rifinin, stevor and Pfmc-2TM gene families share a common titratable factor
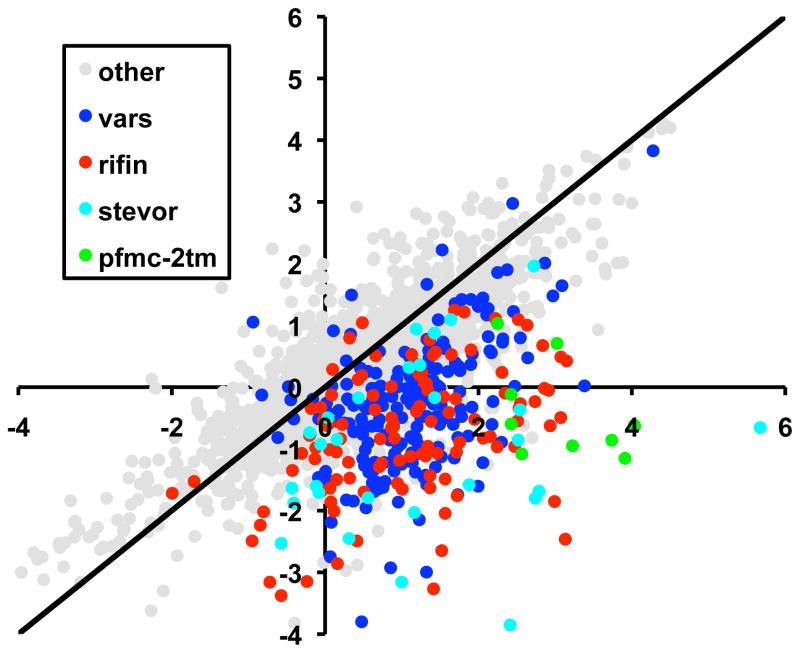
Previous work using episomal var promoters to down-regulate expression of the var gene family indicated that other non-var, housekeeping genes were not affected (Dzikowski and Deitsch, 2008). However, it is possible that certain subsets of genes share specific transcription factors, and thus the transcriptional down-regulation observed with high copy numbers of episomes carrying var promoters might not be exclusive to the var gene family. In order to assess the effect of var promoter titration on expression of all other genes in the P. falciparum genome, microarray analysis was performed on untransfected parasites and those carrying pVBH in the presence of 10μg/ml blasticidin. RNA was taken from a parasite population 14–20 hours after RBC invasion at a time when both var and rifin genes are highly expressed. The overall abundance of the majority of mRNA transcripts was relatively unchanged when comparing the two parasite populations (Pearson correlation value 0.85–0.96), confirming that the transfection and blasticidin selection did not result in large scale alterations in the parasite transcriptome, and that the parasite populations were at roughly the same point in the asexual cycle when the RNA was harvested. As expected, members of the var gene family were substantially down-regulated (Pearson correlation 0.54), as previously observed by qRT-PCR. Notably, expression of rifin, stevor and Pfmc-2TM family members was also significantly reduced between 10 μg/ml and no blasticidin control, suggesting they may share a titratable factor with the var gene family (Figure 3). These gene families share several attributes, including location within specific sub-telomeric and internal chromosomal clusters as well as transcriptional activation and silencing in a clonally variant fashion (Dzikowski, Templeton, and Deitsch, 2006). We therefore investigated whether the var, rifin, stevor and Pfmc-2TM multi-copy gene families share a limited, titratable nuclear factor and thus can be can be co-repressed through promoter titration.
Figure 3.
Downregulation of expanded subtelomeric gene families when selecting for increased copy number of the pVBH plasmid using high blasticidin concentrations. The ratios of transcript signals obtained by DNA microarray under no versus 10 μg/ml blasticidin are plotted. In gray are the intensity values for all genes excluding members of the var, rifin, stevor and Pfmc-2tm gene families. Overall, transcript levels intensities remain unchanged under drug selection and are evenly distributed around a line with a slope = 1 (black). However, the majority of the var, rifin, stevor and pfmc-2tm show a clear decrease in intensity upon selection with 10 μg/ml blasticidin. Both axes represent log2(Cy3/Cy5) intensity values.
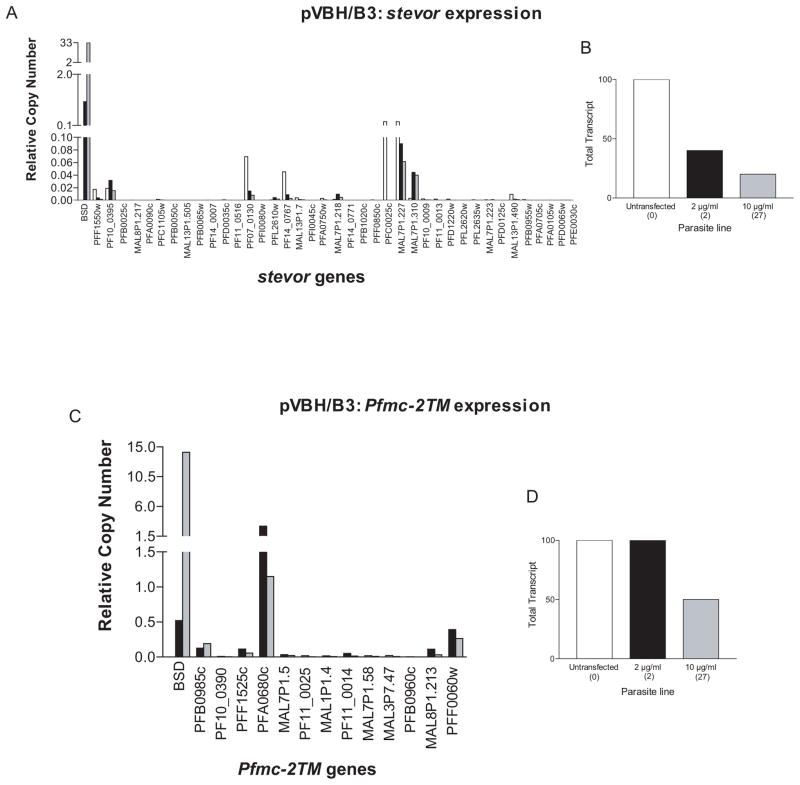
B3 parasites transfected with pVBH (pVBH/B3) were assayed by qRT-PCR for changes in endogenous stevor and Pfmc-2TM expression. In these experiments, untransfected parasites were also included for comparison. RNA was harvested from synchronized cultures at twenty-four or twenty-eight hours post invasion for investigation into stevor or Pfmc-2TM transcript levels, respectively. Both families showed significant repression, with greater repression when parasites were grown under high doses of blasticidin (Figure 4A and 4C). When compared to expression levels in untransfected B3 parasites, stevor transcript levels showed an 80% reduction (Figure 4B). Similar to the TMBH/B3 line (Figure 2D), Pfmc-2TM transcript levels showed no repression under low levels of blasticidin, but displayed an approximately 50 percent reduction in expression levels when the high dose parasites were compared to the untransfected parasites (Figure 4D). Taken together with the microarray data indicating a var promoter-induced down-regulation of the rifin family, it appears that these four families share a titratable nuclear factor necessary for their expression.
Figure 4.
Episomal var promoters downregulate endogenous stevor and Pfmc-2TM expression. (a) Change in stevor RNA levels following increase in copy number of pVBH. Expression levels are shown of endogenous stevor genes in pVBH/B3 parasites under low (black bars) and high (grey bars) blasticin doses. Untransfected B3 parasites (carrying no episome) are represented by white bars. All RNA samples were taken at 24 hours post invasion. (b) Representation of data depicted in (a). Total stevor transcript levels of untransfected B3 parasites (white bar) and pVBH transfected B3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. (c) Change in Pfmc-2TM RNA levels following increase in copy number of pVBH. Expression levels are shown of endogenous Pfmc-2TM genes in pVBH/B3 parasites under low (black bars) and high (grey bars) blasticin doses. All RNA samples were taken at 28 hours post invasion. (d) Representation of data depicted in (c). Total Pfmc-2tm transcript levels of untransfected B3 parasites (white bar) and pVBH transfected B3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. Relative copy numbers are measured with respect to the control housekeeping gene P61-fructose bisphosphate aldolase (PF14_0425) using the formula 2−ΔΔCT with NF54 gDNA as the calibrator.
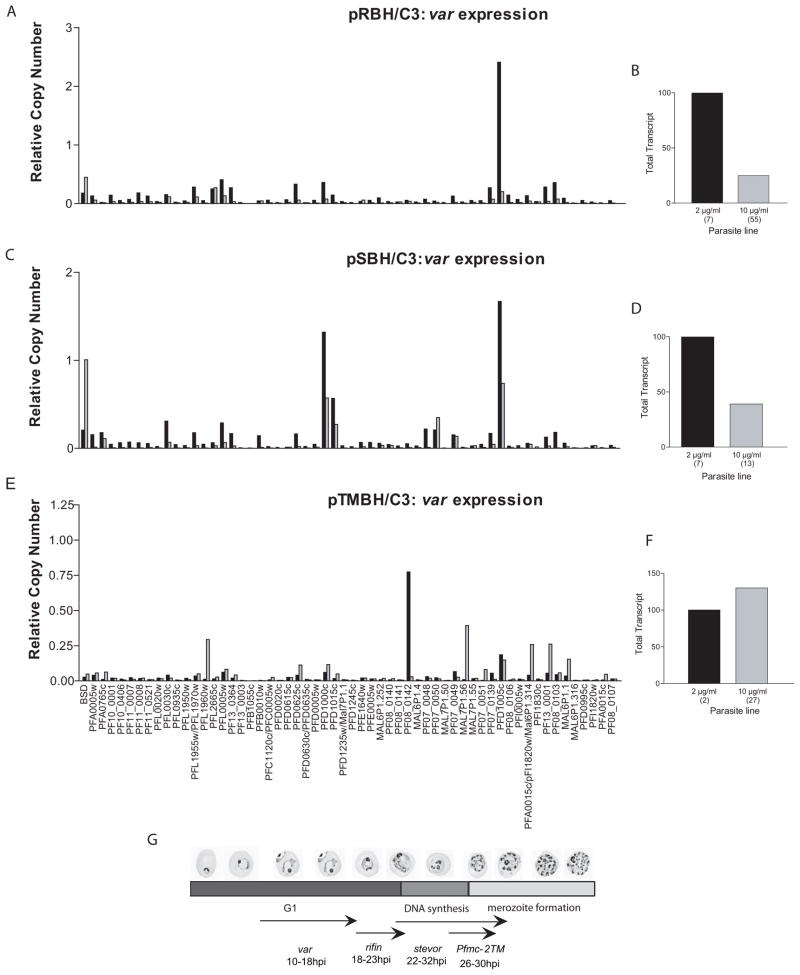
In the reciprocal experiment, C3 parasites carrying the rifin, stevor, or Pfmc-2TM promoter constructs (pRBH/C3, pSBH/C3, and pTMBH/C3, respectively) were assayed at 18 hours after invasion by qRT-PCR for changes in endogenous var gene expression. Increased copy numbers of pRBH and pSBH caused a 75% and 61% reduction in var gene expression, respectively (Figure 5B and 5D). Interestingly, titration with pTMBH had no effect on total var gene expression (Figure 5E), although there is some indication that var gene switching may have occurred. The idea that switching could have been induced by titration with pTMBH seems unlikely, but cannot be ruled out in this experiment. The extent of repression of the var family by these promoters correlates with their timing of peak expression within the 48 hour replicative cycle of the parasite (Figure 5G). Temporally, var genes are expressed first within the cycle (16–18 hours post invasion (hpi)), followed by rifin (18–23 hpi), stevor (22–32 hpi), and finally Pfmc-2TM genes (26–30 hpi) (Kyes et al., 2003;Kaviratne et al., 2002;Lavazec, Sanyal, and Templeton, 2007). Of these families, rifin expression overlaps the greatest with var family expression and we observed the greatest effect on var expression in response to high copy numbers of rifin promoters. stevor family expression overlaps that of var less, and we observed a correspondingly smaller degree of down-regulation of var expression by the episomal stevor promoters. Finally, the Pfmc-2TM family promoters, with peak expression 10–12 hours following var, show no effect. In order to confirm that the co-regulatory effects were specific to these gene families and not simply temporal in nature, qRT-PCR primer sets were made to sbp1 (PFE0065w) and the gene encoding antigen 332 (PF11_0507). Both of these genes are expressed during late rings, coincident with var genes. If the co-regulation was exclusively temporal in nature, one would expect a var promoter to down-regulate these similarly timed genes. When pVBH/C3 under low and high doses of blasticidin were assayed, no change in expression for either of these genes was observed (data not shown). Taken together, as the temporal expression patterns of the rifin, stevor and Pfmc-2TM gene families moved away from that of the var family, the episomal promoter-induced down-regulation of var genes became less pronounced. Thus, there appears to be a temporal aspect to this transcriptional co-regulation of these specific gene families.
Figure 5.
var, rifin, stevor, and Pfmc-2TMs are share a common factor required for efficient expression. (a) Change in var RNA levels following increase in copy number of pRBH. Expression levels are shown of endogenous var genes in pRBH/C3 parasites under low (black bars) and high (grey bars) blasticin doses. (b) Representation of data depicted in (a). Total var transcript levels of pRBH transfected C3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. (c) Change in var RNA levels following increase in copy number of pSBH. Expression levels are shown of endogenous var genes in pSBH/C3 parasites under low (black bars) and high (grey bars) blasticidin doses. (d) Representation of data depicted in (c). Total var transcript levels of pSBH transfected C3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. (e) Change in var RNA levels following increase in copy number of pTMBH. Expression levels are shown of endogenous var genes in pTMBH/C3 parasites under low (black bars) and high (grey bars) blasticin doses. (f) Representation of data depicted in (e). Total var transcript levels of pTMBH transfected C3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. All RNA was harvested at 18 hours post invasion (hpi). Relative copy numbers are measured with respect to the control housekeeping genes P60-seryl-tRNA synthetase (PF07_0073) using the formula 2−ΔΔCT with NF54 gDNA as the calibrator. (g) Temporal expression pattern of var, rifin, stevor, and Pfmc-2TMs during the 48 hour P. falciparum life cycle. Peak expression of var genes is at 16–18 hpi, followed by rifin at18–23 hpi, stevors at 22–32 hpi, and finally Pfmc-2TMs at 26–30 hpi. Episomal rifin, stevor, and Pfmc-2TM promoters downregulate endogenous var expression. The level of co-regulation, however, correlates with the temporal order of gene expression of these families during the 48 hour replicative cycle.
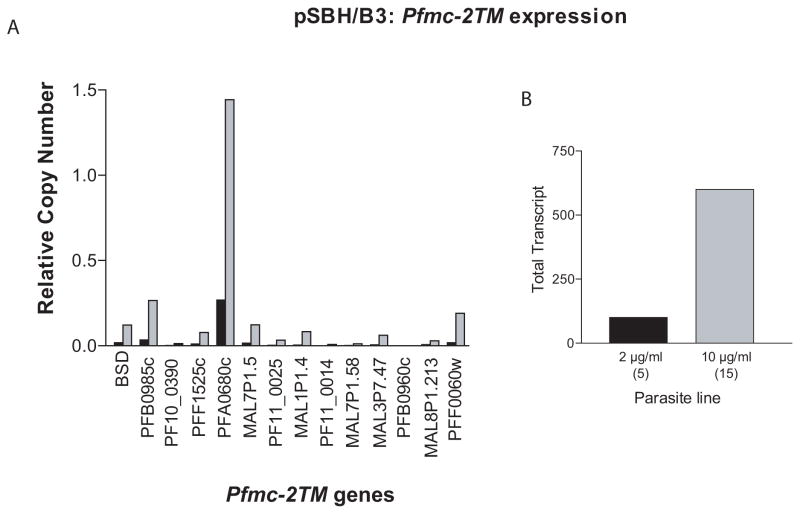
The function of the proteins encoded by the rifin, stevor, and Pfmc-2TM multi-copy gene families are not known. Proteins from all three families have similar domain structures comprised of signal peptide, PEXEL/HT erythrocyte trafficking motifs, and two predicted transmembrane domains. Moreover, the genes are found in similar genomic locations, suggesting that they might share common mechanisms of transcriptional regulation. We therefore investigated what effect down-regulating members of the stevor gene family would have on Pfmc-2TM expression, and vice versa. The parasite lines pTMBH/B3 and pSBH/B3 were selected on 10μg blasticidin and assayed by qRT-PCR for down-regulation of endogenous stevor and Pfmc-2TM genes, respectively. Though pTMBH/B3 showed no change in stevor expression (data not shown), pSBH/B3 exhibited a 3–6 fold up-regulation of Pfmc-2TM transcripts (Figure 6). The up-regulation of Pfmc-2TM expression in response to stevor promoter titration was consistent and reproducible in several independent experiments and suggests two possible interpretations. First, the episomal stevor promoter could be titrating a factor that represses Pfmc-2TM expression. Alternatively, the proteins encoded by the two families may be functionally redundant and therefore the Pfmc-2TM family could be up-regulated in pSBH/B3 in order to compensate for the down-regulation of stevors (shown above, Figure 2A). The possibility that expression of members of these families might be required for parasite viability is supported by our inability to drive down endogenous Pfmc-2TM expression to less than 65% (Figure 2C), suggesting an important role for the corresponding protein.
Figure 6.
pSBH upregulates Pfmc-2TM expression. (a) Expression levels of endogenous Pfmc-2TM genes in pSBH/B3 parasites under low (black bars) and high (grey bars) blasticin doses. (b) Representation of data depicted in (a). Total Pfmc-2tm transcript levels of pSBH transfected B3 parasites under low (black bar) and high blasticidin doses (grey bar). The approximate copy number of the transfected episome for each drug concentration is shown in parentheses. RNA was harvested at 28 hpi and relative copy numbers were measured with respect to the control housekeeping gene P100-actin (PFL2215) using the formula 2−ΔΔCT with NF54 gDNA as the calibrator.
Active rifin and var promoters colocalize at a subnuclear expression site
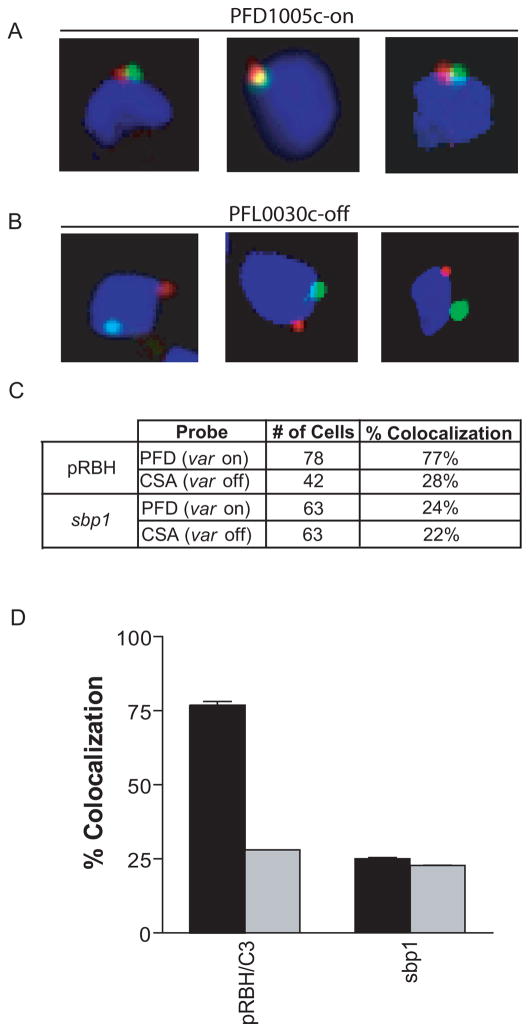
It has been suggested that localization to a subnuclear expression site might play a role in control of var gene expression (Duraisingh et al., 2005;Ralph, Scheidig-Benatar, and Scherf, 2005;Voss et al., 2006;Lopez-Rubio, Mancio-Silva, and Scherf, 2009). Recently, it has been demonstrated that active var promoters co-localize greater than 90% of the time at a specific spot in the parasite nucleus (Dzikowski et al., 2007). Since var, rifin, stevor, and Pfmc-2TM genes appear to share a titratable nuclear factor, we were interested to determine if active promoters from these four families co-localize to the same subnuclear position. For var and rifin genes, the time periods during the asexual cycle in which they are expressed overlap, therefore we performed DNA-FISH (fluorescent in situ hybridization) on the parasite line pRBH/C3, which like its parent line C3, predominantly expresses the var gene PFD1005c (Frank et al., 2007). Localization of biotin-labeled probes specific to the active var gene PFD1005c or inactive var gene PFL0030c was compared with a fluorescein-labeled blasticidin probe which specifically hybridized with the episomal plasmid pRBH. The active episomal rifin promoter co-localized with the active endogenous var gene, PFD1005c, 77% of the time compared to 28% with the inactive var promoter PFL0030c (Figure 7) indicating that these two active promoters generally occupy the same subnuclear position. Similar to the data reported by Dzikowski et al., the two spots were usually found immediately adjacent to one another, rather than completely overlapping, suggesting a fairly large transcriptionally active region (Dzikowski et al., 2007). Attempts to do similar experiments with pSBH/C3 and pTMBH/C3 were complicated by the fact that the periods of active transcription of stevors and Pfmc-2TMs do not substantially overlap with that of var genes and because in both of these cultures significant var gene switching had occurred (Figure 5), making the var gene expression pattern heterogenous. Therefore no conclusions can be drawn regarding these promoters.
Figure 7.
Active var and rifin promoters colocalize at a transcriptionally active subnuclear spot. For DNA-FISH, the nuclei are stained with DAPI (blue), and the pRBH episomes are detected with a bsd probe (green). Endogenous var genes (PFD1005c-on, or PFL0030c (CSA)-off) were detected with gene-specific probes (red). Ring stage parasites transfected with pRBH were probed with the bsd probe plus either the PFD1005c-on, or PFL0030c-off probe (a) Examples of cells scored as colocalizing. (b) Examples of cells scored as non-colocalizing. (c) Quantification of colocalization of the active episome (green) or the control gene sbp1 with either the active var gene (PFD) or inactive var gene (CSA) (red) in each parasite line. Percentages and cell numbers are an average of three separate, blinded counts. (d) Graphical representation of data in (c). Black bars represent colocalization with active var gene (PFD). Grey bars represent colocalization with inactive var gene (CSA).
From this data, it appears that the active episomal rifin promoter does indeed localize to the same position in the nucleus as an active var promoter. The reliance of the two families on the same titratable factor, along with their similar temporal expression patterns supports this finding. Previously, we observed that an unrelated gene (PFF1125c) also colocalized with active var promoters ~75% of the time, suggesting that there might be a limited number of active transcription sites within the nucleus, and thus transcriptionally active genes might tend to colocalize frequently regardless of promoter type. To investigate this phenomenon further, we performed FISH with probes to sbp1 (PFE0065W), a ring-stage specific gene, and the active or inactive var genes PFD1005c and PFL0030c. However, unlike PFF1125c, colocalization between the active sbp1 gene and the active var gene was only ~25%, suggesting that the subnuclear spot is not occupied equally by all active promoters. What dictates the subnuclear position of different promoters at different times therefore remains difficult to predict and deserves additional investigation.
DISCUSSION
The ability to investigate gene function through knockouts has proven to be a powerful tool for understanding the biology of malaria parasites. However, in instances where knockouts are lethal or where numerous genes have redundant functions, simple gene knockouts are not informative. In the case of multi-copy gene families, with the exception var genes, it has not been possible to simultaneously knockout the expression of all members of a family. For example, targeting up to 150 genes for disruption, as would be necessary in the case of the rifin family, is not possible with current technology. The lack of RNAi in Plasmodium makes the use of this type of RNA-based approaches also infeasible. Here we have shown that the promoter titration method can be used for the specific down-regulation of large multi-copy gene families. In addition, promoter titration allows stepwise down-regulation of gene expression, thereby potentially enabling the detection of intermediate phenotypes that might be obscured by lethality in a complete knockout. Specific down-regulation of large gene families may give insight into gene phenotypes which we have as of yet been unable to observe. This technique, though very useful, does have caveats that need to be considered. For example, although promoter titration with a var promoter caused no detectable change in expression of ~90% of P. falciparum genes, down-regulation of other multi-copy families was observed. Thus, it is necessary to ensure the specificity of the episomal promoter chosen, keeping in mind that gene families which share promoter sequences could also share limited regulatory factors.
In the case of the vars, rifins, stevors and Pfmc-2TMs, the possibility that they share a limiting nuclear factor necessary for expression might be due to the similarities in how the genes are regulated and in the roles the encoded proteins play in RBC modification. All are multi-copy gene families primarily located in sub-telomeric regions which undergo expression switching (Dzikowski, Templeton, and Deitsch, 2006). Additionally, like PfEMP1 encoded by var genes, both RIFINs and STEVORS have been localized to the infected red blood cell surface and this, along with their hypervariability and abundance in the genome, has implicated them in antigenic variation (Kyes et al., 1999;Niang, Yan, X, and Preiser, 2009). Furthermore, the RIFINs, STEVORs, and PfMC-2TMs share a common protein structure, potentially suggesting a common function (Lavazec, Sanyal, and Templeton, 2006). It might not be surprising, therefore, that the four families share a common titratable nuclear factor necessary for their transcription. Although it has been shown that the pattern of rifin, stevor and Pfmc-2TM expression is not linked to that of var genes (Sharp et al., 2006;Lavazec, Sanyal, and Templeton, 2007), it is possible that similar mechanisms play a hand in regulating their expression. Chromatin modification, subnuclear localization, and interactions with DNA regulatory elements have all been shown to impact var gene expression and silencing, and some or all of these mechanisms may require the titratable factor detected here to regulate transcription of these other multi-copy gene families.
It has been hypothesized that the members of the two transmembrane superfamily might play a role in antigenic variation and cytoadherence, rosetting, and/or solute pore structure (Kyes et al., 1999;Helmby et al., 1993). The specific down-regulation of these families by promoter titration had the potential to elucidate a phenotype; unfortunately, the down-regulation of the rifin, stevor and Pfmc-2TM families showed no obvious phenotype in cultured parasites other than an initial decrease in growth rate that is likely due to the blasticidin pressure. PfEMP1, encoded by var genes, is widely recognized as the major P. falciparum virulence factor and its expression is thought to be necessary for parasite persistence in vivo. Nonetheless, both Voss et al. (2006) and Dzikowski et al. (2006) showed that parasites that do not express PfEMP1 also display no detectable phenotype in vitro (Dzikowski, Frank, and Deitsch, 2006;Voss et al., 2006). The similar lack of an in vitro phenotype may suggest a role for RIFINs, STEVORs, and PfMC-2TMs in direct interactions with the host. It is intriguing that parasites in which stevor expression was down-regulated repeatedly exhibited a notable up-regulation of Pfmc-2TM transcripts, and that we were unable to completely abolish Pfmc-2TM expression by promoter titration. It is possible that the proteins encoded by the two gene families are functionally redundant and necessary for parasite viability, and thus Pfmc-2TM family up-regulation compensates for the down-regulation of the stevors in the same line. Inter-family compensation could also explain the lack of phenotype in the titrated parasites. In this light it would be interesting to observe the phenotype of parasites with complete knockdowns of all three families (rifins, stevors, and Pfmc-2TMs), if indeed the triple-knockdown proves to be viable.
The fact that the expression of all four of the gene families studied here can be altered through promoter titration provides added emphasis to their promoters/upstream regulatory regions and the titratable factor that controls them. These elements might not only be involved in transcriptional activation, but also in the mechanism that controls mutually exclusive expression. However, the nature or identity of the titratable factor described here and by Dzikowski & Deitsch remains to be determined. Few identifiable putative specific transcription factors have been annotated in the P. falciparum genome (Aravind et al., 2003). One notable candidate family is that of the apicomplexan AP2 (Apetala2), or ApiAP2, proteins. The P. falciparum genome contains twenty-six members of this family which show weak homology to a family of plant AP2/ERF DNA-binding proteins (Balaji et al., 2005). The proteins vary both in structure and lifecycle stage of expression (De Silva et al., 2008). Several members have been shown to bind var promoter sequences by protein binding microarrays (PBMs). One in particular, PF14_0633, is of considerable interest as its predicted consensus binding sequence is present in the var, stevor, and Pfmc-2TM promoters used for promoter titration in this paper (De Silva et al., 2008;Voss et al., 2003). Additionally, this protein is expressed during the late ring stage, coincident with var family expression. It remains to be determined if this ApiAP2 family is involved in var transcriptional regulation. Further, it should be noted that several titratable factors might be involved in the regulation of these gene families, and thus the titration phenomenon could be the result of a limiting amount of any protein or protein complex.
Recently it has been shown that active var genes localize to a specific subnuclear expression site (Duraisingh et al., 2005;Voss et al., 2006;Ralph, Scheidig-Benatar, and Scherf, 2005;Dzikowski et al., 2007). This type of location-dependent allelic exclusion mechanism has been established for the vsg genes in African Typanosomes (Navarro and Gull, 2001). It is possible that the titratable factor necessary for the expression of the four gene families discussed here is not a molecule or transcription factor per se, but rather access to this subnuclear compartment. The large promoter-carrying episomes may simply displace the endogenous genes in this expression site, leading to their transcriptional repression. Here, an active episomal rifin promoter was shown to co-localize with an active endogenous var promoter, and episomal rifin promoters also displayed the greatest repressive effect on var gene expression, followed by lesser effects by stevors and Pfmc-2TM promoters. It is possible that all four promoter types may localize to the same subnuclear position, perhaps containing a specific transcriptional factory, at different times during the replication cycle. If this is the case, the active genes would move into and out of the transcriptional region throughout the 48 hour asexual cycle at times corresponding with the expression pattern of the each gene family. Since rifin family expression closely follows and overlaps that of the var family, one might expect a high level of colocalization between these two families and a greater repressive effect on var gene expression than stevor and Pfmc-2TM genes, which is indeed the result observed here.
EXPERIMENTAL PROCEDURES
Parasite Culture
All parasites used were derivatives of the NF54 parasite line and were cultivated at 5% hematocrit in RPMI 1640 medium, 0.5% Albumax II (Invitrogen), 0.25% sodium bicarbonate, and 0.1 mg/ml gentamicin. Parasites were incubated at 37°C in an atmosphere of 5% oxygen, 5% carbon dioxide and 90% nitrogen. The C3, clonally expressing the var gene PFD1005c, and B3, predominantly expressing one stevor (MAL7P1.227) and one Pfmc-2TM (PFA0680c), lines have been described previously (Frank et al., 2007;Lavazec, Sanyal, and Templeton, 2007).
Parasite cultures were synchronized using percoll/sorbitol gradient centrifugation as previously described (Aley, Sherwood, and Howard, 1984;Calderwood et al., 2003). Briefly, infected RBCs were layered on a step gradient of 40%/70% percoll containing 6% sorbitol. The gradients were then centrifuged at 12,000 g for 20 minutes at room temperature. Highly synchronized, late stage parasites were recovered from the 40%/70% interphase, washed twice with complete culture media and placed back in culture. To verify that this process resulted in populations of parasites that were at similar points in the cell cycle, microarray gene expression data was compared between two separate cultures that were equivalently synchronized. This analysis showed that the overall abundance of the majority of mRNA transcripts were virtually identical when comparing the two parasite populations (Pearson correlation value 0.85–0.96), indicating that they were at equivalent points in the 48 hour replicative cycle.
Plasmid Construction and Parasite Transfection
The plasmids pRBH, pSBH and pTMBH were constructed by replacing the var promoter in pVBH, as described (Dzikowski et al., 2007), with a 1.6 kb portion of the promoter region from a rifinin (PF10_0398), stevor (PFF1550w), or Pfmc-2TM (PFF0060w). The promoter regions were amplified by PCR and the cloning sites Hpa1 (3′) and Kpn1 (5′) introduced using the following primer sets: PF10_0398, 5′-ATATAGGTACCCACTCATTAAGATTACCTTG-3′, 5′-ATATAGTTAACGTGGAACCACATTTGCCC-3′; PFF1550w, 5′-ATATAGGTACCGAACATCTCCTAACATATATG-3′, 5′-ATATAGTTAACGATAAGCGGAAATTTTGTTG-3′; PFF0060w, 5′-ATATAGGTACCCTCAAATATCATAGATTCTAC-3′, 5′-ATATAGTTAACGTGATATATGAAAAAGTGCTGG-3′. Parasites were transfected as described (Deitsch, Driskill, and Wellems, 2001). Briefly, 0.2cm electroporation cuvettes were loaded with 0.175 ml of erythrocytes and 100 μg of plasmid DNA in incomplete cytomix solution. Stable transfectants were initially selected on 2μg/ml blasticidin (Invitrogen, USA). In order to obtain parasites carrying large plasmid copy numbers, these cultures were then subjected to 10 μg/ml blasticidin for two weeks or until normal growth rates were observed.
Genomic DNA extraction, RNA extraction and cDNA synthesis
Genomic DNA was extracted as described (Dzikowski and Deitsch, 2008). Briefly, iRBCs were pelleted and lysed with Saponin. Parasites were pelleted, washed with PBS, and taken up in 200 μl TSE buffer (100 mM NaCl, 50 mM EDTA, 20 mM Tris, pH 8), 40 μl of 10% SDS and 20 μl 6M NaClO4. This suspension was rocked for twelve hours and genomic DNA was extracted with phenol/chloroform. The resulting DNA was taken up in 100 μl dH2O.
RNA extraction and cDNA synthesis was performed as described (Dzikowski and Deitsch, 2006). Briefly, RNA was extracted from synchronized parasite cultures at 18, 24, or 28 hours post invasion, corresponding with the peak RNA expression levels for determination of var, stevor, or Pfmc-2TM transcript levels, respectively. RNA was extracted with the TRIZOL LS Reagent® as described (Kyes, Pinches, and Newbold, 2000) and purified on PureLink column (Invitrogen) according to manufacturer’s protocol. Isolated RNA was then treated with Deoxyribonuclease I® (Invitrogen) to degrade contaminating gDNA. cDNA synthesis was performed from 800 ng total RNA with Superscript II Rnase H reverse transcriptase ® (Invitrogen) with random primers ® (Invitrogen) as described by the manufacturer.
DNA Microarray Hybridizations
For DNA microarray hybridizations, cDNA with amino-allyl incorporated dUTPs was generated from RNA as described (Bozdech et al., 2003). As an arbitrary reference, we used pooled RNA from hourly timepoints of a synchronized 48-hour culture of 3D7 strain parasites used. Coupling to Cy3 (reference pool) and Cy5 (experimental sample) (GE Healthcare) and array hybridizations were performed as described using a recently designed new-generation P. falciparum-specific long oligonucleotide DNA microarray (Hu et al., 2007). The arrays were scanned using an Axon 4200A scanner and images analyzed using Axon GenePix software (Axon Instruments, Union City, CA, USA). Data were normalized by the global signal intensity across both channels. The resulting data were further analyzed using the Princeton University Microarray database (PUMAdb, http://puma.princeton.edu) for cluster analysis.
Real-time qRT-PCR
For qRT-PCR reactions to detect transcription from all var, stevor, and Pfmc-2TM genes present in the 3D7 genome, we used the primer sets published by Salanti et. al. (2003), Lavazec et al. (2007), and Sharp et al. (2006) respectively (Salanti et al., 2003;Lavazec, Sanyal, and Templeton, 2007;Sharp et al., 2006). PCR primers used to amplify sbp1 (PFE0065W) were: 5′-ACGAACCAACACAATTACAG-3′, 5′-AGCTGCTTCTCCACTAAAC-3′, while those used to amplify antigen 332 (Pf11_0507) were: 5′-CGAAGAAAATGTCTCATTCA-3′, 5′ ′-TCCCGAATTTTTATCTTCAT-3′. Transcript copy numbers were determined using the formula 2−ΔΔCT as described in the Applied Biosystems User Bulletin 2 using NF54 gDNA as the calibrator. Specifically, relative copy number was calculated as 2 exponential negative ((Ct target gene in cDNA – Ct reference gene in cDNA)-(Ct target gene in gDNA-Ct target gene in gDNA)). Copy numbers of episomal promoters were calculated using qRT-PCR of gDNA and comparing the ΔCTof bsd to that of the single copy housekeeping gene P60-seryl-tRNA synthetase (PF07_0073), P61-fructose bisphosphate aldolase (PF14_0425) or P100-actin (PFL2215) as described (Frank et al., 2006). The control gene used for the graphs shown in the figures was chosen arbitrarily and is specified in the figure legends. All qRT-PCR assays were performed at least in duplicate for each template with no apparent differences, and each experiment was completed at least twice in its entirety, again with no significant differences.
Fluorescent in situ hybridization
DNA probes were labeled with biotin and fluorescein using Roche High-Prime Kits. Biotin was detected using streptavidin AlexaFluor 594 (Molecular Probes). Synchronous ring-stage parasites were fixed in 4% paraformaldehyde solution as described in Methods in Malaria Research (Mancio-Silva, Freitas-Junior, Scherf). Slides were washed twice with 2X SSPE and DNA FISH was carried out on ring stage parasites as described by Freitas-Junior et al. (Freitas-Junior et al., 2000) with modifications. Briefly, probes were hybridized overnight (94°C, 2 min., 37°C, 16 hours). Slides were washed in 2X SSPE, 37°C, 1 hour; 0.2X SSPE 50°C, 1 hour; 0.2X SSPE 37°C, 30 min.; maleic acid buffer, room temp.,10 min.; 1% blocking reagent in maleic acid buffer (Roche), room temp., 1 hour; 1% blocking reagent in maleic acid buffer (Roche) plus AlexaFluor 594 (Molecular Probes), room temp., 30 min; 3 times maleic acid buffer, room temp, 20 min. After washing, the slides were mounted in antifade medium and visualized using an Olympus M081 fluorescent microscope. Composite images were produced using Photoshop 6.0 and the images blindly counted by three individuals.
The PFD1005c probe was generated using the following primers: 5′-CGGATGATGGTGATACTG-3′, 5′-CTCCACATGATGGTAGAAGAC-3′. The resulting 680 bp PCR fragment from the 3′ region of exon 1 was gel-extracted and biotin labeled using a Roche High-Prime kit. The PFL0030c (var2CSA) probe was generated using nested primers to the uORF region of this gene. Primary reaction primers: 5′-GGTACCTGAACGCTTAAAGAAACAAGG-3′, 5′-CTGCAGCATTTTGTCCAACCATTTACA-3′. Secondary reaction primers: GCAATGTATAACAAAAATATAAG-3′, GACACCAAATATTCCTATGTGC-3′. The resulting 1 kb PCR product was gel-extracted and biotin labeled using a Roche High-Prime kit. The bsd probe was made by labeling whole pCR®2.1-TOPO (Invitrogen) plasmid carrying a bsd cassette with fluoroscein using a Roche High-Prime kit. The sbp1 (PFE0065w) probe was generated using the following primers: 5′-ATGTGTAGCGCAGCTCGAGC-3′, 5′-TTAGGTTTCTCTAGCAACTG-3′. The resulting 1 kb PCR product was gel-extracted and fluoroscein labeled using a Roche High-Prime kit.
Acknowledgments
The authors would like to thank Dr. William Howitt and Lori Bond-Widner for assistance in scoring FISH images, and Sohini Sanyal for suggestions regarding experimental design. This work was supported by grant AI 52390 from the National Institutes of Health to KWD. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of the William Randolph Hearst Foundation. RD is a Golda Meir Scholar and supported by the Marie Curie International Reintegration Grant (IRG) [203675] and the German Israeli Foundation [2163-1725.11/2006]. KWD and RD are supported by a grant from the United States-Israel Binational Science Foundation [2007350].
REFERENCE LIST
- Aley SB, Sherwood JA, Howard RJ. Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J Exp Med. 1984;160:1585–1590. doi: 10.1084/jem.160.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: Genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, Derisi JL. The transcriptome of the intraerythrocytic developmental cycle of. Plasmodium falciparum PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. Journal of Biological Chemistry. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Prat K, Meurice E, Mornon JP, Tomavo S. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics. 2005;6:100. doi: 10.1186/1471-2164-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, Llinas M. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci U S A. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechering KJ, Kaan AM, Mbacham W, Wirth DF, Eling W, Konings RN, Stunnenberg HG. Isolation and functional characterization of two distinct sexual-stage-specific promoters of the human malaria parasite. Plasmodium falciparum Mol Cell Biol. 1999;19:967–978. doi: 10.1128/mcb.19.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Deitsch KW, Driskill CL, Wellems TE. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Research. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Deitsch K. Antigenic variation by protozoan parasites: insights from Babesia bovis. Mol Microbiol. 2006;59:364–366. doi: 10.1111/j.1365-2958.2005.05007.x. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Deitsch KW. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J Mol Biol. 2008;382:288–297. doi: 10.1016/j.jmb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Templeton TJ, Deitsch K. Variant antigen gene expression in malaria. Cell Microbiol. 2006;8:1371–1381. doi: 10.1111/j.1462-5822.2006.00760.x. [DOI] [PubMed] [Google Scholar]
- Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite. Plasmodium falciparum RNA. 2008;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp C, Raskolnikov D, Deitsch KW. A regulatable transgene expression system for cultured Plasmodium falciparum parasites. Malar J. 2008;7:86. doi: 10.1186/1475-2875-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Amulic B, Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol. 2007;64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Constantini D, Amulic B, Burdougo E, Deitsch K. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J Biol Chem. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite. Plasmodium falciparum Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmby H, Cavelier L, Pettersson U, Wahlgren M. Rosetting Plasmodium falciparum-infected erythrocytes express unique strain-specific antigens on their surface. Infection and Immunity. 1993;61:284–288. doi: 10.1128/iai.61.1.284-288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Llinas M, Li J, Preiser PR, Bozdech Z. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics. 2007;8:350. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer JK, Amaladoss A, Genesan S, Preiser PR. Variable expression of the 235 kDa rhoptry protein of Plasmodium yoelii mediate host cell adaptation and immune evasion. Mol Microbiol. 2007;65:333–346. doi: 10.1111/j.1365-2958.2007.05786.x. [DOI] [PubMed] [Google Scholar]
- Kaviratne M, Khan SM, Jarra W, Preiser PR. Small variant STEVOR antigen is uniquely located within Maurer’s clefts in Plasmodium falciparum-infected red blood cells. Eukaryot Cell. 2002;1:926–935. doi: 10.1128/EC.1.6.926-935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning-Ward TF, Speranca MA, Waters AP, Janse CJ. Analysis of stage specificity of promoters in Plasmodium berghei using luciferase as a reporter. Mol Biochem Parasitol. 1999;100:141–146. doi: 10.1016/s0166-6851(99)00042-0. [DOI] [PubMed] [Google Scholar]
- Kyes S, Horrocks P, Newbold C. Antigenic variation at the infected red cell surface in malaria. Annu Rev Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Christodoulou Z, Raza A, Horrocks P, Pinches R, Rowe JA, Newbold CI. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Molecular Microbiology. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes SA, Rowe JA, Kriek N, Newbold CI. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc Natl Acad Sci USA. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazec C, Sanyal S, Templeton TJ. Hypervariability within the Rifin, Stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Nucleic Acids Res. 2006;34:6696–6707. doi: 10.1093/nar/gkl942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazec C, Sanyal S, Templeton TJ. Expression switching in the stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Mol Microbiol. 2007;64:1621–1634. doi: 10.1111/j.1365-2958.2007.05767.x. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, Zhou YY, Blair PL, Grainger M, Moch JK, Haynes JD, De la Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez RR, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome Wide localization of heterochromatin couples clonally variant gene regulation to perinuclear repressive centers in malaria parasites. Cell Host and Microbe. 2009 doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- Niang M, Yan YX, Preiser PR. The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathog. 2009;5:e1000307. doi: 10.1371/journal.ppat.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci USA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, Hviid L, Theander TG. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Molecular Microbiology. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Florens L, Johnson JR, Wang T, Drazba JA, Le Roch KG, Zhou Y, Batalov S, Carucci DJ, Winzeler EA, Yates JR., III A Plasmodium gene family encoding Maurer’s cleft membrane proteins: structural properties and expression profiling. Genome Res. 2004;14:1052–1059. doi: 10.1101/gr.2126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodin DJ, Zhuang Y, Shapiro DJ, Katzenellenbogen BS. Analysis of mechanisms that determine dominant negative estrogen receptor effectiveness. J Biol Chem. 1995;270:31163–31171. doi: 10.1074/jbc.270.52.31163. [DOI] [PubMed] [Google Scholar]
- Sharp S, Lavstsen T, Fivelman QL, Saeed M, McRobert L, Templeton TJ, Jensen ATR, Baker DA, Theander TG, Sutherland CJ. Programmed transcription of the var gene family, but not of stevor, in Plasmodium falciparum gametocytes. Eukaryotic Cell. 2006;5:1206–1214. doi: 10.1128/EC.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JV, Peterson DS, Ravetch JV, Wellems TE. A large and diverse gene family (var) encodes 200–350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Voss TS, Kaestli M, Vogel D, Bopp S, Beck HP. Identification of nuclear proteins that interact differentially with Plasmodium falciparum var gene promoters. Molecular Microbiology. 2003;48:1593–1607. doi: 10.1046/j.1365-2958.2003.03528.x. [DOI] [PubMed] [Google Scholar]