Abstract
The prognosis for patients diagnosed with mesothelioma is generally poor, and currently available treatments are usually ineffective. Therapies that specifically target tumor cells hold much promise for the treatment of cancers that are resistant to current approaches. We have previously selected phage antibody display libraries on mesothelioma cell lines to identify a panel of internalizing human single chain (scFv) antibodies that target mesothelioma-associated, clinically represented cell surface antigens, and further exploited the internalizing function of these scFvs to specifically deliver lethal doses of liposome-encapsulated small molecule drugs to both epithelioid and sarcomatous subtypes of mesothelioma cells. Here we report the identification of MCAM/MUC18/CD146 as the surface antigen bound by one of the mesothelioma targeting scFvs using a novel cloning strategy based on yeast surface human proteome display. Immunohistochemical analysis of mesothelioma tissue microarrays confirmed that MCAM is widely expressed by both epithelioid and sarcomatous types of mesothelioma tumor cells in situ but not by normal mesothelial cells. In addition, quantum dot-labeled anti-MCAM scFv targets primary meosthelioma cells in tumor fragment spheroids cultured ex vivo. As the first step in evaluating the therapeutic potential of MCAM-targeting antibodies, we performed single-photon emission computed tomography studies using the anti-MCAM scFv and found that it recognizes mesothelioma organotypic xenografts in vivo. The combination of phage antibody library selection on tumor cells and rapid target antigen identification by screening the yeast surface displayed human proteome could be a powerful method for mapping the targetable tumor cell surface epitope space.
Keywords: Mesothelioma, MCAM/MUC18/CD146, internalizing single chain antibody, yeast surface display of human proteome, SPECT/CT
INTRODUCTION
Mesothelioma is a deadly disease caused by malignant transformation of the mesothelium, the protective lining surrounding most of body’s internal organs. Mesothelioma is almost always associated with previous exposure to asbestos, and symptoms may not appear until 20-50 years post exposure (1). There is no generally accepted method for screening patients who have been exposed to asbestos, and diagnosis can be difficult since the symptoms of mesothelioma are similar to those caused by other conditions (2). There are three main types of mesothelioma: epithelioid, sarcomatoid, and mixed (3, 4). Epithelioid mesothelioma is the most common form, comprising between 50-70% of mesothelioma cases, and is the most likely to respond to treatment (4). Sarcomatoid mesothelioma accounts for 10-20% of mesothelioma cases and rarely responds to treatment (4, 5). Approximately 20-35% of mesothelioma cases are mixed type, which contains both epithelioid and sarcomatoid features and has an intermediate outlook (4, 6). Regardless of subtype, because diagnosis often occurs at a late stage of disease, the prognosis for malignant mesothelioma is generally poor, with median survival ranging from 8-14 months, and treatments are generally ineffective, especially in the case of sarcomatoid mesothelioma (7, 8). Thus, new diagnostic and therapeutic strategies are needed for mesothelioma, particularly the sarcomatoid type.
One promising area of antineoplastic drug development is to explore tumor susceptibility to targeted therapy (9-12). In principle, a variety of anti-tumor agents can be attached to tumor recognition molecules that target tumor-associated internalizing cell surface molecules to achieve intracellular delivery and targeted tumor killing (9, 11, 13). Currently, very few mesothelioma-associated cell surface markers that are expressed by all subtypes of mesothelioma are known (14). For example, mesothelin, a cell surface glycoprotein, has been shown to be a useful marker for epithelioid mesothelioma (15), but it is not expressed by the sarcomatous subtype of this disease (16). In addition, mesothelin is also expressed on normal mesothelial cells (16). Thus, the development of targeted therapies against mesothelioma will benefit from the identification of additional cell surface markers with more restricted expression on normal tissues and more specific associations with both epithelioid and sarcomatoid mesotheliomas.
Monoclonal antibodies (mAbs) are able to recognize antigenic determinants of diverse chemical composition with high affinity and specificity, and are therefore promising candidates for the development of targeted cancer therapies. Antibodies targeting tumor-associated epitopes could be utilized in applications such as induction of antibody-dependent cell cytotoxicity or inhibition of signaling pathways involved in tumor cell migration, growth, and survival. In addition, antibodies targeting internalizing tumor epitopes could be exploited to achieve specific intracellular delivery of therapeutic agents (9, 17, 18).
We have previously selected a naïve phage antibody display library on mesothelioma cell lines derived from both epithelioid and sarcomatous subtypes, and identified a panel of internalizing mAbs that target cell surface antigens associated with both subtypes of mesothelioma (19). Most importantly, immunohistochemistry (IHC) studies showed that these scFvs bind to mesothelioma cells in situ, thereby recognizing clinically represented tumor antigens. We have further exploited the internalizing function of these scFvs to deliver immunoliposomes encapsulating the small molecule drug topotecan specifically to mesothelioma cells, and demonstrated targeted killing of both epithelioid and sarcomatous mesothelioma cells in vitro (19). To facilitate further therapeutic development, we have begun to identify antigens recognized by this panel of phage antibodies. We have previously reported the construction of a large yeast surface-displayed human cDNA library, which was used to identify cellular proteins binding to post-translational modifications (20) and small signaling molecules (21). In this report we describe the identification of one of the target antigens, MCAM/CD146/MUC18, by screening the yeast surface human cDNA display library with a mesothelioma-targeting phage antibody. Mesothelioma tissue microarray studies showed that MCAM is overexpressed on > 80% of both epithelioid and sarcomatous mesothelioma tissues but not normal mesothelium. Finally, using single-photon emission computed tomography/computed tomography (SPECT/CT), we showed that the technetium (99mTc)-labeled anti-MCAM scFv was able to detect tumor cells in mesothelioma organ xenografts in vivo, suggesting that this scFv may be useful for the development of targeted immunotherapies against mesothelioma.
MATERIALS AND METHODS
Materials
Reagents for mammalian cell transfection: Lipofectamine™ 2000 and Opti-MEM (Invitrogen, Carlsbad, CA). Reagents for scFv purification and characterization: nitrilotriacetic acid-nickel (Ni-NTA) agarose beads (Qiagen, Hilden, Germany), EZ-Link Sulfo-NHS-LC-Biotin (Pierce, Rockfold, IL), and streptavidin Qdot 705 conjugate (Invitrogen). Reagents for FACS and immunohistochemistry: streptavidin-phycoerythrin (SA-PE) (Invitrogen/BioSource, Camarillo, CA), streptavidin-alexa 488 and 647 (SA-488 and SA-647) (Invitrogen/Molecular Probes, Eugene, OR), affinity purified anti- MCAM/CD146 antibody (Invitrogen), anti-human cytokeratin monoclonal antibody AE1/AE3 (Dako, Carpinteria, CA), anti-CD34 monoclonal antibody (Chemicon/Millipore, Billerica, MA), biotin-labeled rabbit anti-fd bacteriophage (Sigma-Aldrich, St. Louis, MO), streptavidin horseradish peroxidase (SA-HRP) (Sigma-Aldrich), HRP-conjugated goat anti-mouse and HRP-conjugated goat anti-human (heavy and light chain) antibodies (Jackson ImmunoResearch, West Grove, PA), 2,2’-azinobis(3-ethylbenzothiazoline-6-sulfonate) (ABTS), diaminobenzedine tetrahydrochloride (DAB) (Sigma-Aldrich), Antigen Unmasking Solution and hematoxylin (Vector Laboratories, Burlingame, CA), Optimal Cutting Temperature (OCT) compound (Sakura Finetec USA, Torrance, CA).
Human tissues
The protocol for tissue acquisitions was approved by the Institutional Review Board and in accordance with an assurance filed with and approved by the Department of Health and Human Services. Surgically removed mesothelioma tissues were either embedded in paraffin to create tissue microarrays (22, 23), or maintained as organ cultures (tumor fragment spheroids) as previously described (24).
Production of scFvs
To produce soluble scFvs, genes encoding scFvs were cloned into an expression vector imparting a c-myc and a hexahistidine tag at the C-terminus (25, 26). Following IPTG induction, bacterial cells were harvested by centrifugation, resuspended in 200 mg/ml sucrose, 1 mM EDTA, 30 mM Tris-HCl, pH 8.0, on ice for 30 min, and centrifuged again to collect the supernatant. The pellet was resuspended in 5 mM MgSO4 on ice for 30 min, and centrifuged to collect the supernatant. Both supernatants were pooled and loaded on a Ni+-NTA column pre-equilibrated with 15 mM imidazole/PBS, and washed with 20 mM imidazole/PBS (26, 27). Bound scFvs were eluted with 250 mM imidazole/PBS, dialyzed against PBS, and analyzed by spectrophotometry (BioMini, Shimadzu, Japan).
Tissue microarray study
Mesothelioma tissue microarrays were treated with xylene to remove paraffin, re-hydrated in 100%, 95% and 70% ethanol, and boiled in a pressure cooker in Antigen Unmasking Solution for 5 min. The slides were then stained with an anti-MCAM rabbit antibody at RT for 1h, washed three times with PBS, further incubated sequentially with biotin-labeled goat anti-rabbit antibody and SA-HRP, and bound antibodies were detected using DAB substrate (26). To detect blood vessels, some slides were incubated separately with mouse anti-CD34 monoclonal antibody followed by goat anti-mouse HRP, and bound antibodies were detected using DAB. The stained tissues were counter-stained with hematoxylin, dried in 70%, 95% and 100% ethanol, mounted and analyzed. A scFv (N3M2) with no detectable binding to mesothelioma cells by FACS analysis was used as the control for all experiments.
Antigen identification by screening a yeast surface displayed human cDNA library
The yeast surface human cDNA display library (20, 21) was grown in SR-CAA (2% raffinose, 0.67% yeast nitrogen base and 0.5% casamino acids) at 30 °C to an OD600 of approximately five. To induce expression of cDNA products on the yeast surface, the yeast were re-inoculated at an OD600 of 0.5 in SRG-CAA (SR-CAA+2% galactose) and grown at 30 °C for 16-36 h. Induction was monitored by an anti-Xpress™ monoclonal antibody (Invitrogen). For the first round of sorting, about 108 induced yeast cells in 500 μl PBS were incubated with biotinylated phage antibodies for 4 h at 4 °C. Unlabeled helper phage was added to compete away non-specific binding to phage particles. Cells were washed twice with PBS, incubated with 500 μl of 1/500 diluted SA-PE for 20 min at 4 °C, sorted by FACS (FACSAria, BD Biosciences, San Jose, CA) and recovered on SD-CAA plates. Approximately 5 × 107 cells were analyzed in the first round selection. In subsequent rounds, SA-647 was alternated with SA-PE to minimize the selection of clones that bind the detection reagent. After three rounds of sorting, individual clones were picked, induced, and tested for phage binding by FACS. Plasmids were recovered from yeast clones exhibiting phage antibody binding using a modified QIAprep Spin Miniprep protocol that incorporates a glass bead cell lysis step (Qiagen, Hilden, Germany). Isolated plasmids were transformed into DH5α cells, purified, and the cDNA inserts were sequenced. Public gene and protein databases were searched for matches to each cDNA insert.
Ectopic expression of MCAM in mammalian cells
Plasmids containing full-length human MCAM cDNA (pCMV-MCAM, OriGene, Rockville, MD) or a control human cDNA, GLG1, (pCMV-GLG1) under control of the CMV promoter was mixed with lipofectamine™ 2000 and Opti-MEM according to manufacturer’s instructions, and incubated with BPH-1 cells growing at 80% confluency in 24 well plates. All experiments were done in triplicate. Expression of MCAM was checked at day 3 using an anti-MCAM antibody (Invitrogen). Following confirmation, M1 phage antibody and control helper phage were incubated with transfected cells, and binding was detected by biotin-labeled anti-fd bacteriophage followed by SA-PE.
Labeling of scFv with near infrared emitting quantum dots
A near infrared fluorescent nanometer crystal with a polymer shell directly coupled to streptavidin (Qdot streptavidin 705 conjugate, Invitrogen) was conjugated to the anti-MCAM scFv or control scFv in two steps. First, the scFv was biotin-labeled with Sulfo-NHS-LC-Biotin for 30 min at RT according to manufacture’s instructions and purified by elution with PBS (pH 7.2) through a gel-filtration PD-10 column containing Superdex-G25 (GE Healthcare, Piscataway, NJ). Next, the purified biotin-labeled scFvs were incubated with the streptavidin-Qdot 705 for 30 min at RT to form the final conjugates, which were purified by eluting with PBS through a PD-10 column containing Superdex-200. By measuring the molar extinction coefficient at 280 and 705 nm, the final concentration of scFvs and Qdot 705 was estimated at 0.8 and 0.5 μM, respectively.
Incubation of scFv with human tumor fragment spheroids ex vivo
Tumor fragment spheroids (24) were incubated with Qdot 705-conjugated anti-MCAM or control scFvs at 50 nM for 4 h at 37 °C. Tumor fragments from three tumors were used (2 epithelial, 1 mixed). Ten spheroids were incubated with each antibody. After 4 h, the spheroids were washed with media, allowed to sediment, embedded in OCT, and frozen in liquid nitrogen for later sectioning. Cryosectioned specimens (10 μm thickness) were viewed by confocal microscopy in the near infrared spectrum using a Zeiss LSM510 microscope (Carl Zeiss Microimaging, Thornwood, NY). In separate staining, the tumor fragments were stained with anti-human cytokeratin AE1/AE3 antibodies to confirm the presence of mesothelioma cells.
Preparation of [99mTc(CO)3(OH2)3]+
The IsoLink® kit (Tyco/Mallinckrodt, Saint Louis, MO) was used to prepare the [99mTc(CO)3 ] moiety. A 10 mL penicillin vial containing potassium boranocarbonate (8.5 mg, 63 μmol), sodium tetraborate·10H2O (2.9 mg, 8.0 μmol), Na-tartrate (15.0mg, 53 μmol), and Na2CO3 (4.0 mg, 38 μmol) was fitted with a rubber septum and the vial flushed with N2(g) for 15 min. 99mTcO4- eluted from the 99Mo/99mTc generator (GE healthcare, CA) (370 MBq, 10-20 mCi) in 1000 μL of saline was added by a syringe, and the solution was heated to 100 °C for 30 min. After cooling on ice, the alkaline solution was neutralized to final pH 6.0-6.5 by the addition of 180-200 μL of 1 M HCl. Quality control was performed by reverse-phase HPLC.
Radiolabeling of scFv
An aliquot (20-30 μL) of scFv solution at 5 mg/mL was mixed with 100-500 μL [99mTc(CO)3(OH2)3]+ solution and the mixture was heated at 37 °C for 60 min. The reaction mixture was cooled down to RT and the product isolated using a PD-10 column containing Superdex-G25 with PBS (pH 7.2) as eluant (28). Both the anti-MCAM scFv, M1, and the control scFv, N3M2, which was randomly picked from the unselected naïve phage antibody library and tested for lack of binding to tumor cell lines by FACS, were labeled and purified in the same way. The specific activities of these labeled scFvs were similar (within standard deviations).
In vivo SPECT/CT and biodistribution studies
Animal studies were approved by the Institutional Review Board and adhered to the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals. Tumor fragment spheroids (1 × 2 × 2 mm3 size) generated from human mesothelioma tissues were injected into the peritoneal space of the nude mice (NCr nu/nu, Taconic, Hudson, NY), about four weeks before the imaging experiment (24). Ten nude tumor-bearing mice were each injected via the tail vein with 18.5 MBq of the 99mTc anti-MCAM scFv (50 μg) in 100 μL PBS. As a control, ten nude tumor-bearing mice were each injected with 18.5 MBq of the 99mTc-labeled control scFv. The mice were imaged with a combined modality SPECT/CT (X-SPECT, Gamma Medica, Northridge, CA) at 2h, 4h, 6h, and 8h and then sacrificed and dissected for IHC and biodistribution studies. For IHC, a fraction of the excised tumor was embedded in paraffin and analyzed by IHC using anti-AE1/AE3 mAb and an anti-MCAM antibody (Invitrogen) to confirm the presence of tumors, and HRP-conjugated goat anti-human antibodies to confirm the presence of human scFvs. For biodistribution studies, tumors, blood, and major organs were collected and weighed wet. The radioactivity in these samples was measured using a gamma counter, calibrated against a known quantity of the injected dose, and presented as percentage injected dose per gram (%ID/g).
Statistics
The two-tailed Student t test was used to analyze a pair of variables, and a p value less than 0.05 was considered statistically significant. Where appropriate, the data are presented as the mean ± SD.
RESULTS
The mesothelioma-targeting M1 phage antibody binds MCAM
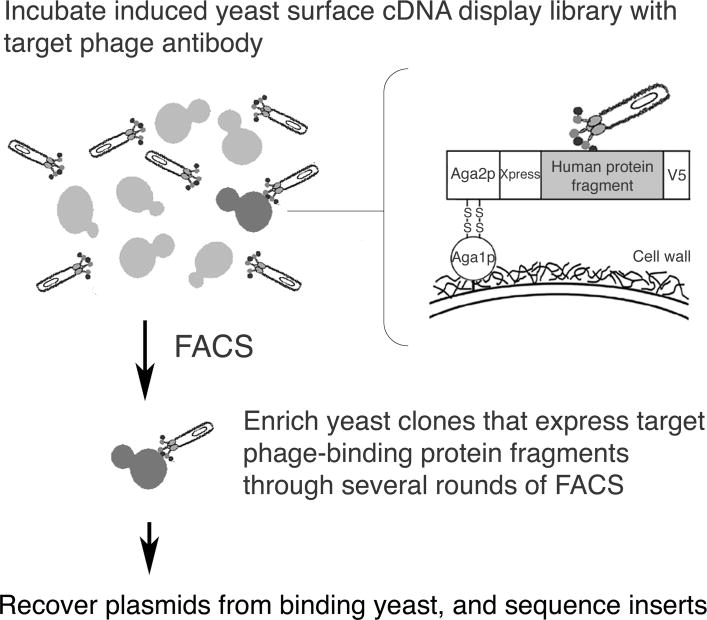
Using our recently developed expression cloning strategy based on yeast surface human proteome display (20, 21), we have begun to systematically identify mesothelioma cell surface antigens bound by our panel of internalizing phage antibodies. We initially focused our identification efforts on a scFv, M1, which binds to a broad panel of tumor cell lines and may thus recognize a commonly expressed tumor cell surface antigen. We have previously constructed an inducible library of human protein fragments displayed on the yeast surface as C-terminal fusions to the yeast a-agglutinin subunit, Aga2p, and demonstrated utility of this library in mapping protein-ligand interactions (20, 21). We used a similar strategy (Figure 1) to identify the M1-targeted mesothelioma antigen using the M1 phage antibody as the “bait” to select binding clones from the yeast surface cDNA display library by FACS (20, 21).
Figure 1.
Outline of the antigen identification strategy based on yeast surface cDNA display. A yeast library displaying human protein fragments on the cell surface was incubated with the target M1 phage antibody. Yeast that bind specifically to the M1 phage antibody were identified by FACS-based screening, and the plasmids carried by these yeast were harvested and sequenced to identify the human cDNA fragments.
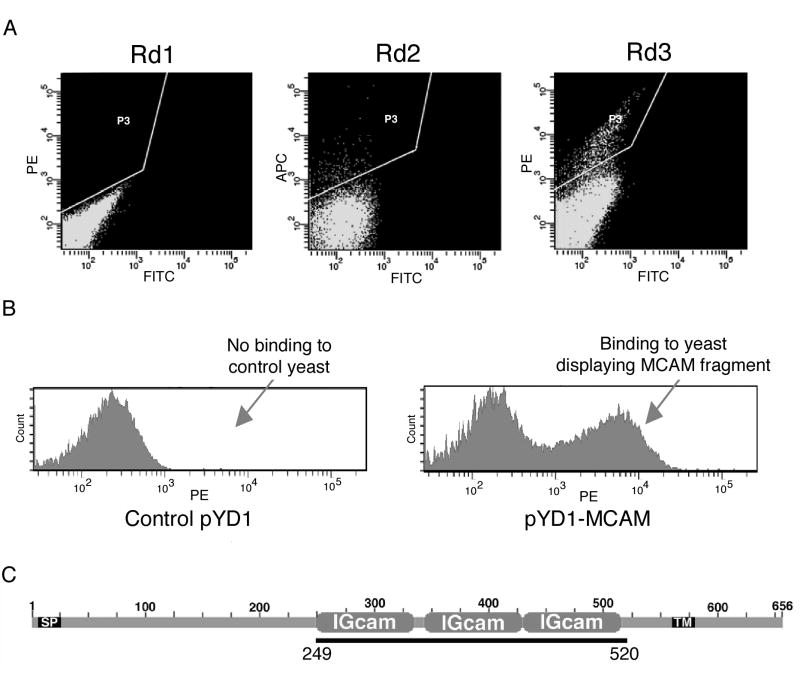
The induced yeast surface display human cDNA library was incubated with biotin-labeled phage antibody, and binding clones were enriched through three rounds of FACS. Very few binding clones (< 0.5%) were present in the initial library population (Figure 2A, Rd1). After two rounds of selection, >15% of the yeast population bound the phage antibody (Figure 2A, Rd3). Individual yeast clones from the third round output population were screened by FACS. Plasmids from M1 phage-binding clones were recovered, retransformed into yeast in order to verify the results of the primary screen, and sequenced to determine the identity of their cDNA inserts. One unique cDNA insert was identified from four clones that bind to the M1 phage antibody (Figure 2B). This cDNA sequence matched perfectly with a portion of the extracellular domain of MCAM (Figure 2C), also known as MUC18 or CD146.
Figure 2.
Yeast surface cDNA display screen identifies MCAM as target antigen of M1 phage antibody. A, Enrichment of yeast clones displaying protein fragments with affinity for the M1 phage antibody through several rounds of FACS. PE and Alexa-647 labeled detection agents were alternated between rounds to reduce the chance of selecting binders to detection agents. The FITC channel is included to indicate autofluorescence. The P3 gate indicates the population selected in each round. B, M1 phage antibody binding to yeast displaying a fragment of the MCAM extracellular domain. Control: yeast transfected with vector pYD1. C, Diagram of the M1 phage-binding MCAM protein fragment. Black bar indicates the region in the MCAM protein corresponding to the recovered cDNA. SP, signal peptide. IGcam: immunoglobulin superfamily cell adhesion molecule domain. TM, transmembrane domain.
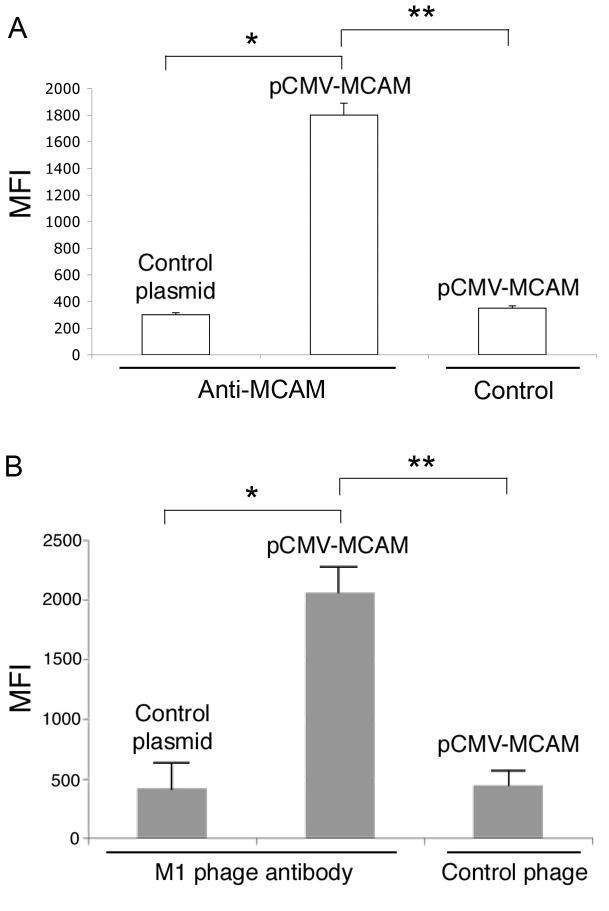
To confirm that MCAM is indeed the antigen bound by the M1 phage antibody, we transiently transfected mammalian cells (BPH-1) that do not express MCAM with a mammalian expression vector containing full-length MCAM cDNA (pCMV-MCAM). After confirming surface expression of MCAM by FACS using an anti-MCAM antibody (Figure 3A), we stained transfected cells with the M1 phage antibody and showed that the M1 phage binds MCAM expressed on the surface of mammalian cells (Figure 3B), confirming that MCAM is the tumor antigen recognized by our M1 phage antibody.
Figure 3.
M1 phage antibody binds to ectopically expressed MCAM. BPH-1 cells were transfected with pCMV-MCAM or pCMV-GLG1 as a negative control and binding of the commercial anti-MCAM antibody and the M1 phage was analyzed by FACS after 72 h. Error bars indicate standard deviations. A, Quality control study of MCAM transfection. The anti-MCAM antibody binds specifically to BPH-1 transfected with pCMV-MCAM but not the control pCMV-GLG1 (*, p < 0.05). Control, secondary antibodies only, no binding is detected compared to the anti-MCAM antibody (**, p < 0.05). B, The M1 scFv binds to ectopically expressed MCAM. The M1 but not the control helper phage bound to BPH-1 cells transfected with the pCMV-MCAM expression plasmid (**, p < 0.05). No binding of M1 phage to BPH-1 cells transfected with the pCMV-GLG1 control plasmid was observed (*, p < 0.05).
MCAM is expressed in mesothelioma tissues
To determine how widely MCAM is expressed by mesothelioma, we performed IHC studies on mesothelioma tissue arrays. MCAM was found to be expressed in > 80% of mesothelioma specimens of all subtypes (epithelioid (28/31), sarcomatoid (8/10) and mixed type (14/14), examples are shown in Figure 4). MCAM is not expressed on normal mesothelium (Figure 4). In addition to tumor cells, MCAM was found to be expressed strongly on tumor-associated blood vessels (Figure 4), consistent with previous reports that MCAM is a marker for angiogenesis (29, 30). These experiments show that MCAM is widely expressed by all subtypes of mesothelioma and by tumor-associated blood vessels, and may thus be an attractive therapeutic target.
Figure 4.
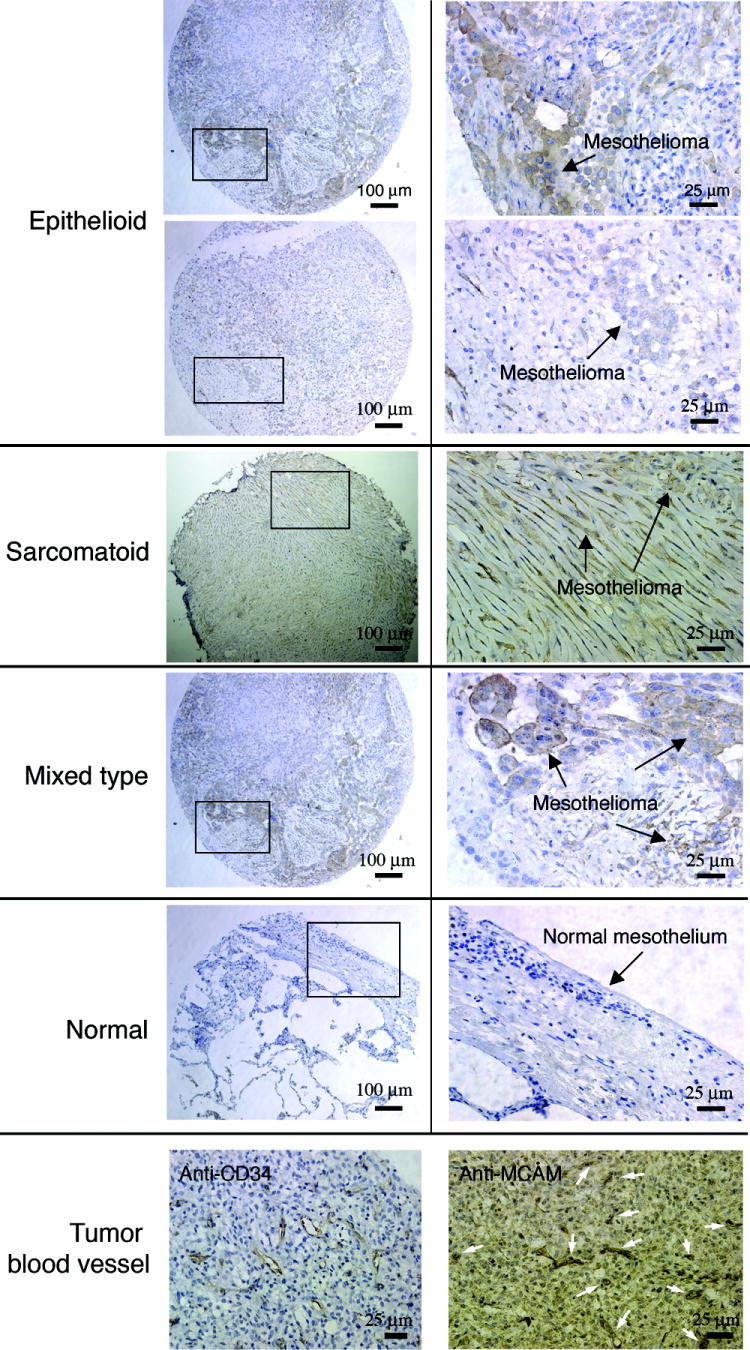
Staining of mesothelioma tissue arrays with anti-MCAM antibody. An anti-MCAM antibody was used to stain mesothelioma tissue arrays containing sarcomatous, epithelioid, and mixed subtypes. Representative images are shown. Boxed regions are shown at higher magnification in right panels. The anti-MCAM antibody stains tumor but not normal lung mesothelium. Two staining examples are shown for epithelioid mesothelioma. The first row shows an example of strong staining. The second row shows an example of moderate staining. The bottom row shows MCAM expression on tumor-associated blood vessels. The anti-CD34 mAb was used to mark blood vessels (arrows) surrounded by mesothelioma cells.
The anti-MCAM scFv targets human mesothelioma cells in ex vivo cultured tumor fragments
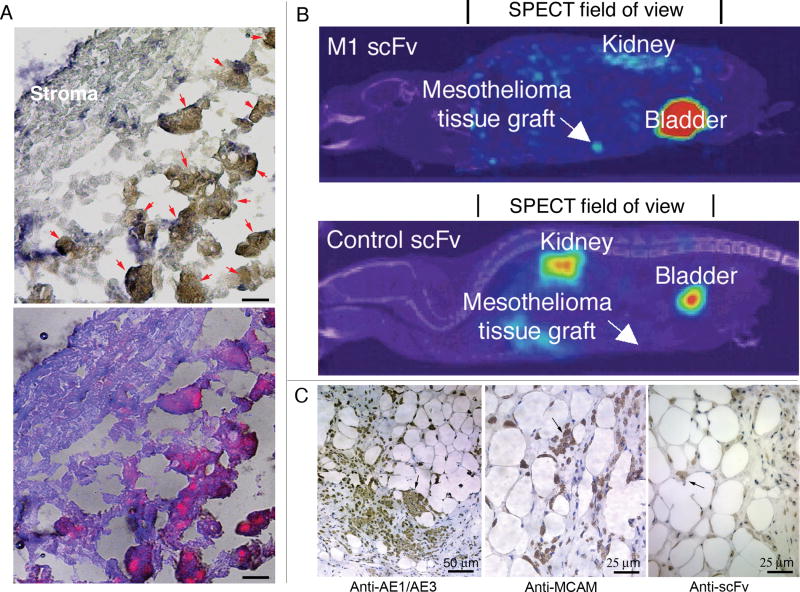
To determine whether the anti-MCAM scFv would target primary human mesothelioma cells, we labeled the anti-MCAM scFv with a near infrared quantum dot (Qdot 705) and incubated the labeled scFv with tumor fragment spheroids grown from mesothelioma obtained from surgical resection (24). After 4 h incubation at 37 °C with labeled anti-MCAM scFv, tumor spheroids were frozen and cryosectioned for viewing by confocal microscopy. In tumor fragments from 5 different mesotheliomas, the anti-MCAM scFv was found to stain tumor cells in all cases (an example is shown in Figure 5A). The cells bound by the anti-MCAM scFv were confirmed to be mesothelioma cells by cytokeratin stain (24) (Figure 5A). The sections incubated with a Qdot 705-labeled control scFv showed no binding. These data show that the anti-MCAM scFv can specifically target primary mesothelioma cells ex vivo in mesothelioma organ culture spheroids.
Figure 5.
Anti-MCAM scFv targets human mesothelioma tissue fragments ex vivo and in vivo. A, Qdot 705-labeled anti-MCAM scFv targets tumor fragments ex vivo. A case of the mixed mesothelioma subtype is shown. Top panel, IHC image. Mesothelioma cells (brown colored; indicated by arrows) are stained with anti-cytokeratin AE1/AE3 mAb; lower panel, fluorescence image showing accumulation of Qdot-labeled scFvs in mesothelioma cells. Scale bar, 10 μm. B, SPECT/CT imaging of the anti-MCAM scFv targeting to human mesothelioma tissues grafted into the peritoneal space of nude mice. For comparison, animals in the control group were injected with the control non-binding scFv (N3M2). Images were taken at 8 h post injection. SPECT and CT scans were performed separately and the images were digitally overlaid. The SPECT field of view (set by the instrument with radius of rotation = 4.01 cm) is indicated. The CT field of view spans the entire animal and is not indicated. C, IHC study on excised xenografts to confirm that grafted tissues contain mesothelioma cells. An anti-human cytokeratin AE1/AE3 mAb was used to stain the mesothelioma cells. The tissue was also stained with an anti-MCAM antibody to confirm the expression of MCAM. Anti-human heavy and light chain antibody was used to stain scFvs. Arrows indicate examples of stained tumor cells (brown color).
The anti-MCAM scFv targets xenografted mesothelioma tissues in vivo
To determine the efficiency of the anti-MCAM scFv in tumor targeting in vivo, we performed molecular imaging studies with technetium (99mTc)-labeled scFv and a combined modality SPECT/CT, which allows simultaneous tomographic imaging of γ– emitting radiopharmaceuticals and anatomic imaging with CT. To increase clinical relevance, we used a novel xenograft model that uses peritoneally implanted fragments of human mesothelioma (24). Mice carrying peritoneally-implanted human mesothelioma tissues were injected with either 99mTc-labeled anti-MCAM M1 scFv or a 99mTc-labeled control scFv and imaged with SPECT/CT. As shown in Figure 5B, peritoneally grafted human mesothelioma tissues were recognized by 99mTc-labeled anti-MCAM scFv but not the control scFv, demonstrating the targeting specificity in vivo. The other organs that showed the greatest contrast were the kidneys and the bladder, consistent with the known route of scFv excretion from the body. After imaging, the tumor fragment spheroids tissues were removed from the mice, sectioned, and stained for human cytokeratin (a mesothelioma marker) to identify the tumor cells and for MCAM to confirm tumor expression of this molecule (Figure 5C). We further used anti-human (heavy and light chains) antibodies to confirm scFvs in the tissue sections (Figure 5C). Next, we performed biodistribution studies using the 99mTc-labeled anti-MCAM and control scFvs. Antibody accumulation in tumor, blood, and major organs was determined at 8h post injection (Figure 6A). The anti-MCAM scFv showed higher tumor accumulation in mice carrying mesothelioma tissue xenografts than the control scFv (Figure 6A), demonstrating targeting specificity of the anti-MCAM scFv. The relative uptake ratios (M1/control) were higher for tumor xenografts compared to other organ sites studied (Figure 6B).
Figure 6.
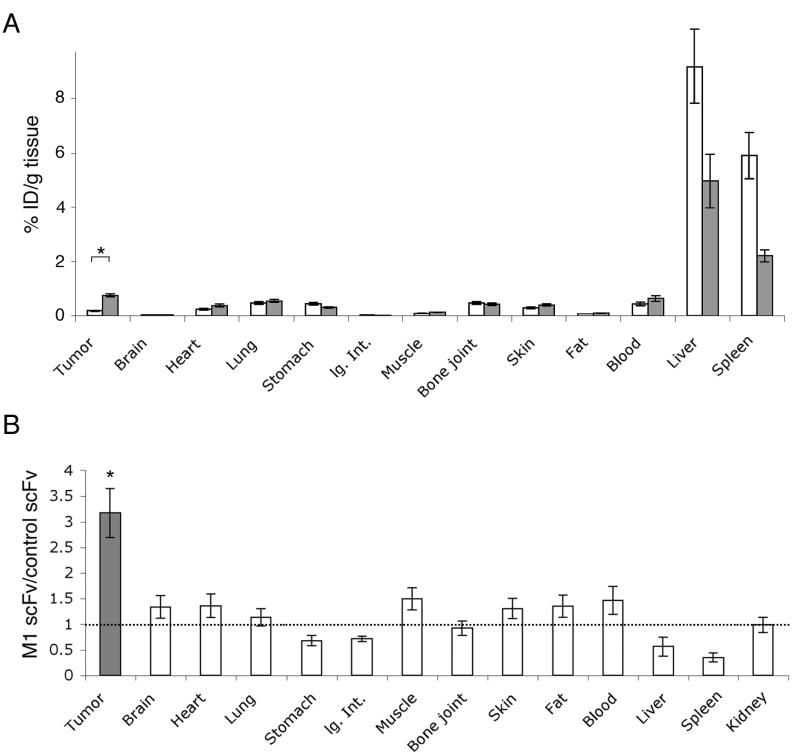
Biodistribution study of intravenuously injected anti-MCAM M1 scFv. Tissues were collected 8h post-injection. A, The average %ID/g tissue of the anti-MCAM scFv (n = 10, grey bars) and the control scFv (n = 10, white bars) was plotted for tumor, blood and other organs/tissues. The values for kidney (not plotted) are 52.4 %ID/g tissue for the anti-MCAM scFv and 53.0 %ID/g tissue for the control scFv. Standard deviations are indicated. lg.Int., large intestine. *, there is a significant difference (p < 0.05) between %ID/g of the M1 scFv and that of the control scFv. B, The ratio of %ID/g tissue (anti-MCAM over control scFv) was plotted for tumor, blood and other organs/tissues. The dashed line indicates ratio = 1. Standard deviations are indicated. *, there is a significant difference (p < 0.05) between tumor and each of the normal organ sites.
DISCUSSION
We have previously selected a panel of human scFvs from a phage antibody library that bind to clinically represented, internalizing epitopes on the mesothelioma cell surface (19). We have further shown that these scFvs can mediate tumor-specific intracellular delivery of small molecule drugs, which selectively kill mesothelioma cells in vitro (19). In this study, we sought to identify the target antigen bound by one of these antibodies, the M1 scFv. We focused our initial identification efforts on the M1 scFv because it has demonstrated payload delivery function and binds to several tumor cell lines in addition to mesothelioma cell lines, suggesting that it may be broadly useful as a tumor-targeting agent (19). We used a novel expression cloning strategy based on yeast surface display of human protein fragments (20, 21) to identify MCAM as the antigen bound by the M1 phage antibody. MCAM is a transmembrane glycoprotein that belongs to the immunoglobulin superfamily (31, 32) and functions as a Ca2+-independent adhesion molecule. It was originally described as a marker for advanced melanoma (33-36). In immunohistochemistry studies using a large panel of tissues, MCAM expression was observed in a relatively limited spectrum (9/42) of normal human adult tissues (endothelium, smooth muscle, Schwann cells, ganglion cells, myofibroblasts, cerebellar cortex, breast, hair follicles, and dendritic cells) (37). Notably, CD146 expression was not observed in normal mesothelium nor any of the endocrine tissues tested and was only present in 1/12 epithelial tissues tested (breast) (37). The expression on endothelium is limited to certain tissues. Among the 14 normal tissues studied, MCAM expression was found in 5 endothelium (stomach, colon, breast, ovary and lung) (30).
The discovery of MCAM expression in mesothelioma tissues is significant for therapeutic development against this disease for several reasons. First, our study showed that MCAM is expressed by all subtypes of mesothelioma. In contrast, mesothelin, a currently used marker for mesothelioma, recognizes the epithelioid but not the sarcomatous subtype of mesothelioma (16), a particularly recalcitrant form of this disease. Second, consistent with previous reports of MCAM expression on blood vessels (29, 30), our study using mesothelioma tissue microarrays showed that MCAM is expressed on both mesothelioma cells and tumor-associated blood vessels, making MCAM a potentially attractive target for a combined anti-tumor and anti-angiogenesis therapy (38). Finally, our results show that overlapping sets of cell surface antigens exist between tumors of diverse tissue origins. While the etiology of mesothelioma may be unique, it nevertheless shares characteristics with other commonly occurring tumors such as melanoma. Treatment of mesothelioma may thus benefit from ongoing therapeutic development for other oncological indications.
Using human tumor fragments cultured ex vivo, we showed that the anti-MCAM scFv penetrates the tumor fragments and homes specifically to primary mesothelioma cells. To be useful for targeted therapy, antibodies or antibody fragments must be able to accumulate in tumor tissues in vivo after systemic administration. The in vivo biodistribution of the anti-MCAM M1 scFv was evaluated in a novel mesothelioma organotypic xenograft model using SPECT/CT. SPECT/CT combines functional imaging (SPECT) and structural imaging (CT) to achieve accurate and sensitive tumor detection in vivo. We found that the anti-MCAM M1 scFv, but not the control scFv, preferentially accumulated in mesothelioma xenografts compared to surrounding soft tissues, demonstrating its potential in noninvasive imaging and targeted immunotherapy. This result is most impressive since the organotypic xenograft model is more clinically relevant compared to models based on cell lines (24).
We have selected scFvs on live mesothelioma cells to identify those that target novel internalizing epitopes. These scFv-targeted epitopes are in their native conformation as opposed to MHC-presented ones. As such, these scFvs are well suited for targeting live tumor cells ex vivo and in vivo, as we have demonstrated in this study, but may have limitations in detecting denatured epitopes such as those in paraffin-embedded tissues. For example, the M1 scFv binds to live mesothelioma cells and mesothelioma cells in situ in frozen tissues as we have demonstrated previously (19), but does not stain paraffin-embedded tissues. As such, we have used the commercial anti-MCAM antibody to stain the paraffin-embedded mesothelioma tissue arrays.
We used a novel, FACS-based expression cloning strategy based on yeast surface cDNA display to identify the target antigen. The yeast display technology was originally developed by Wittrup and colleagues to study eukaryotic protein functions (39-41). We have previously adapted this technology for human proteome display, and constructed a large yeast surface display human cDNA fragment library (20, 21). We screened the library by FACS to identify cellular proteins binding to post-translational modifications (20) and small molecules (21). A major advantage of this cloning system is that the “bait” can be of diverse chemical and molecular composition, as long as it can be fluorescently detected (20, 21). In this study, we used phage particles displaying the M1 scFv as the bait, greatly simplifying the identification process. Since 50,000-70,000 cells can be sorted per second, the FACS-based method allows the full diversity of large libraries to be practically screened. The combination of phage antibody library selection on the surface of living tumor cells and rapid target antigen identification by screening the yeast surface-displayed human proteome could be a powerful method for mapping the tumor cell surface epitope space.
Acknowledgments
This work is supported by grants from the National Institute of Health (R01 CA118919 to BL, and CA95671 to VCB) and the Mesothelioma Applied Research Foundation (to BL and VCB). We thank Dr. Stephen L. Nishimura for help with histopathology study, and Dr. Shannon Wilson for help with tumor fragment spheroid experiments.
Abbreviations
- MCAM
melanoma cell adhesion molecule
- ScFv
single-chain variable domain fragment
- mAb
monoclonal antibody
- SPECT/CT
single-photon emission computed tomography/computed tomography
- IHC
immunohistochemistry
- HRP
horseradish peroxidase
- OCT
optimal cutting temperature
- MFI
mean fluorescence intensity
- IGcam
immunoglobulin superfamily cell adhesion molecule domain
- TM
transmembrane domain
References
- 1.Bertazzi PA. Descriptive epidemiology of malignant mesothelioma. Med Lav. 2005;96:287–303. [PubMed] [Google Scholar]
- 2.Dunleavey R. Malignant mesothelioma: risk factors and current management. Nurs Times. 2004;100:40–3. [PubMed] [Google Scholar]
- 3.Corson JM. Pathology of mesothelioma. Thorac Surg Clin. 2004;14:447–60. doi: 10.1016/j.thorsurg.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Scherpereel A, Lee YC. Biomarkers for mesothelioma. Curr Opin Pulm Med. 2007;13:339–443. doi: 10.1097/MCP.0b013e32812144bb. [DOI] [PubMed] [Google Scholar]
- 5.Lucas DR, Pass HI, Madan SK, et al. Sarcomatoid mesothelioma and its histological mimics: a comparative immunohistochemical study. Histopathology. 2003;42:270–9. doi: 10.1046/j.1365-2559.2003.01583.x. [DOI] [PubMed] [Google Scholar]
- 6.Pass HI, Kranda K, Temeck BK, Feuerstein I, Steinberg SM. Surgically debulked malignant pleural mesothelioma: results and prognostic factors. Ann Surg Oncol. 1997;4:215–22. doi: 10.1007/BF02306613. [DOI] [PubMed] [Google Scholar]
- 7.Tomek S, Manegold C. Chemotherapy for malignant pleural mesothelioma: past results and recent developments. Lung Cancer. 2004;45(Suppl 1):S103–19. doi: 10.1016/j.lungcan.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Kindler HL. Moving beyond chemotherapy: novel cytostatic agents for malignant mesothelioma. Lung Cancer. 2004;45(Suppl 1):S125–7. doi: 10.1016/j.lungcan.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen UB, Kirpotin DB, Pickering EM, et al. Therapeutic efficacy of anti-ErbB2 immunoliposomes targeted by a phage antibody selected for cellular endocytosis. Biochim Biophys Acta. 2002;1591:109–18. doi: 10.1016/s0167-4889(02)00256-2. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Stuckert P, Bosch I, Marks JD, Marasco WA. Single-chain antibody-mediated gene delivery into ErbB2-positive human breast cancer cells. Cancer Gene Ther. 2001;8:555–65. doi: 10.1038/sj.cgt.7700337. [DOI] [PubMed] [Google Scholar]
- 11.King GD, Curtin JF, Candolfi M, et al. Gene therapy and targeted toxins for glioma. Curr Gene Ther. 2005;5:535–57. doi: 10.2174/156652305774964631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng G, Chen J, Li H, Glickson JD. Rerouting lipoprotein nanoparticles to selected alternate receptors for the targeted delivery of cancer diagnostic and therapeutic agents. Proc Natl Acad Sci U S A. 2005;102:17757–62. doi: 10.1073/pnas.0508677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnett M. Targeted drug conjugates: principles and progress. Adv Drug Deliv Rev. 2001;53:171–216. doi: 10.1016/s0169-409x(01)00227-7. [DOI] [PubMed] [Google Scholar]
- 14.Zeng L, Fleury-Feith J, Monnet I, et al. Immunocytochemical characterization of cell lines from human malignant mesothelioma: characterization of human mesothelioma cell lines by immunocytochemistry with a panel of monoclonal antibodies. Hum Pathol. 1994;25:227–34. doi: 10.1016/0046-8177(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 15.Hassan R, Bera T, Pastan I. Mesothelin: a new target for immunotherapy. Clin Cancer Res. 2004;10:3937–42. doi: 10.1158/1078-0432.CCR-03-0801. [DOI] [PubMed] [Google Scholar]
- 16.Ordonez NG. Immunohistochemical diagnosis of epithelioid mesothelioma: an update. Arch Pathol Lab Med. 2005;129:1407–14. doi: 10.5858/2005-129-1407-IDOEMA. [DOI] [PubMed] [Google Scholar]
- 17.Roth A, Drummond DC, Conrad F, et al. Anti-CD166 Single Chain Antibody-Mediated Intracellular Delivery of Liposomal Drugs to Prostate Cancer Cells. Mol Cancer Ther. 2007;6:2737–46. doi: 10.1158/1535-7163.MCT-07-0140. [DOI] [PubMed] [Google Scholar]
- 18.Mamot C, Drummond DC, Noble CO, et al. Epidermal growth factor receptor-targeted immunoliposomes significantly enhance the efficacy of multiple anticancer drugs in vivo. Cancer Res. 2005;65:11631–8. doi: 10.1158/0008-5472.CAN-05-1093. [DOI] [PubMed] [Google Scholar]
- 19.An F, Drummond DC, Wilson S, et al. Targeted drug delivery to mesothelioma cells using functionally selected internalizing human single-chain antibodies. Mol Cancer Ther. 2008;7:569–78. doi: 10.1158/1535-7163.MCT-07-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidlingmaier S, Liu B. Construction and application of a yeast surface-displayed human cDNA library to identify post-translational modification-dependent protein-protein interactions. Mol Cell Proteomics. 2006;5:533–40. doi: 10.1074/mcp.M500309-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Bidlingmaier S, Liu B. Interrogating Yeast Surface-displayed Human Proteome to Identify Small Molecule-binding Proteins. Mol Cell Proteomics. 2007;6:2012–20. doi: 10.1074/mcp.M700223-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Battifora H. The multitumor (sausage) tissue block: novel method for immunohistochemical antibody testing. Lab Invest. 1986;55:244–8. [PubMed] [Google Scholar]
- 23.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 24.Kim KU, Wilson SM, Abayasiriwardana KS, et al. A novel in vitro model of human mesothelioma for studying tumor biology and apoptotic resistance. Am J Respir Cell Mol Biol. 2005;33:541–8. doi: 10.1165/rcmb.2004-0355OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schier R, Marks JD, Wolf EJ, et al. In vitro and in vivo characterization of a human anti-c-erbB-2 single-chain Fv isolated from a filamentous phage antibody library. Immunotechnology. 1995;1:73–81. doi: 10.1016/1380-2933(95)00007-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu B, Conrad F, Cooperberg MR, Kirpotin DB, Marks JD. Mapping tumor epitope space by direct selection of single-chain Fv antibody libraries on prostate cancer cells. Cancer Res. 2004;64:704–10. doi: 10.1158/0008-5472.can-03-2732. [DOI] [PubMed] [Google Scholar]
- 27.Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301:1149–61. doi: 10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- 28.Waibel R, Alberto R, Willuda J, et al. Stable one-step technetium-99m labeling of His-tagged recombinant proteins with a novel Tc(I)-carbonyl complex. Nat Biotechnol. 1999;17:897–901. doi: 10.1038/12890. [DOI] [PubMed] [Google Scholar]
- 29.Sers C, Riethmuller G, Johnson JP. MUC18, a melanoma-progression associated molecule, and its potential role in tumor vascularization and hematogenous spread. Cancer Res. 1994;54:5689–94. [PubMed] [Google Scholar]
- 30.Yan X, Lin Y, Yang D, et al. A novel anti-CD146 monoclonal antibody, AA98, inhibits angiogenesis and tumor growth. Blood. 2003;102:184–91. doi: 10.1182/blood-2002-04-1004. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann JM, Riethmuller G, Johnson JP. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc Natl Acad Sci U S A. 1989;86:9891–5. doi: 10.1073/pnas.86.24.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sers C, Kirsch K, Rothbacher U, Riethmuller G, Johnson JP. Genomic organization of the melanoma-associated glycoprotein MUC18: implications for the evolution of the immunoglobulin domains. Proc Natl Acad Sci U S A. 1993;90:8514–8. doi: 10.1073/pnas.90.18.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denton KJ, Stretch JR, Gatter KC, Harris AL. A study of adhesion molecules as markers of progression in malignant melanoma. J Pathol. 1992;167:187–91. doi: 10.1002/path.1711670205. [DOI] [PubMed] [Google Scholar]
- 34.Kraus A, Masat L, Johnson JP. Analysis of the expression of intercellular adhesion molecule-1 and MUC18 on benign and malignant melanocytic lesions using monoclonal antibodies directed against distinct epitopes and recognizing denatured, non-glycosylated antigen. Melanoma Res. 1997;7(Suppl 2):S75–81. [PubMed] [Google Scholar]
- 35.Shih IM, Elder DE, Hsu MY, Herlyn M. Regulation of Mel-CAM/MUC18 expression on melanocytes of different stages of tumor progression by normal keratinocytes. Am J Pathol. 1994;145:837–45. [PMC free article] [PubMed] [Google Scholar]
- 36.Pacifico MD, Grover R, Richman PI, et al. Development of a tissue array for primary melanoma with long-term follow-up: discovering melanoma cell adhesion molecule as an important prognostic marker. Plast Reconstr Surg. 2005;115:367–75. doi: 10.1097/01.prs.0000148417.86768.c9. [DOI] [PubMed] [Google Scholar]
- 37.Shih IM, Nesbit M, Herlyn M, Kurman RJ. A new Mel-CAM (CD146)-specific monoclonal antibody, MN-4, on paraffin-embedded tissue. Mod Pathol. 1998;11:1098–106. [PubMed] [Google Scholar]
- 38.Mills L, Tellez C, Huang S, et al. Fully human antibodies to MCAM/MUC18 inhibit tumor growth and metastasis of human melanoma. Cancer Res. 2002;62:5106–14. [PubMed] [Google Scholar]
- 39.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–7. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 40.Feldhaus MJ, Siegel RW, Opresko LK, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–70. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 41.Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat Biotechnol. 2000;18:754–9. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]