Abstract
Objective and Methods
Low bilirubin levels are significantly associated with cardiovascular diseases (CVD). In previous genome-wide linkage studies we identified a major locus on chromosome 2q harboring the candidate gene UDP-glucuronosyltransferase (UGT1A1). The activity of this enzyme is significantly influenced by a TA-repeat polymorphism in the promoter of the gene. In a prospective study individuals with genotype (TA)7/(TA)7 had significantly higher bilirubin levels and approximately one third the risk of CVD as carriers of the wild type (TA)6 allele. In the present study we performed a conditional linkage study to investigate whether this polymorphism explains the observed linkage peak and extended our analysis by a genome-wide association study on bilirubin levels in 1345 individuals.
Results
After adjustment for the bilirubin variance explained by this polymorphism, the LOD score on chromosome 2q dropped from 3.8 to 0.4, demonstrating that this polymorphism explains the previous linkage result. For the genome-wide association study, the closest marker to UGT1A1 was in the top ranking SNPs. The association became even stronger when we considered the TA-repeat polymorphism in the analysis (p=2.68×10−53). Five other SNPs in other regions reached genome-wide significance without obvious connection to bilirubin metabolism.
Conclusions
Our studies suggest that UGT1A1 may be the major gene with strong effects on bilirubin levels and the TA-repeat polymorphism might be the key polymorphism within the gene controlling bilirubin levels. Since this polymorphism has a high frequency and a substantial impact on the development of CVD, the gene might be an important drug target.
Keywords: bilirubin, UGT1A1, linkage analysis, genome-wide association study
Introduction
Many studies have shown a significant association between low serum bilirubin levels and cardiovascular diseases (CVD) (1–8). Lipid oxidation and formation of oxygen radicals are important factors of arterial plaque formation. Bilirubin may serve as a physiological antioxidant providing protection against atherosclerosis and CVD. Segregation analyses in two studies proposed a major gene controlling a substantial amount of serum bilirubin levels (5,9). Two recent independent genome-wide linkage scans identified a major locus on chromosomal 2q telomere controlling serum bilirubin concentrations with maximum multipoint LOD score of 3.8 (10) and 3.2 (11), respectively. The identified chromosomal region harbors the gene of UDP-glucuronosyltransferase (UGT1A1), the major enzyme of bilirubin glucuronidation, which mainly determines bilirubin elimination in humans. The activity of UGT1A1 is significantly influenced by a TA-repeat polymorphism in the promoter region of this gene (12). Individuals homozygous for 7 TA repeats (7/7) were found to have a lower promoter activity and subsequently higher levels of bilirubin than heterozygous (6/7) or wild type homozygous (6/6) (12–14). The (TA)7 allele, also named as UGT1A1*28, is responsible for Gilbert syndrome in Caucasians, a benign, non-hemolytic, mild unconjugated hyperbilirubinemia.
We recently conducted an association study of this TA-repeat polymorphism with bilirubin levels and CVD in the Framingham Heart Study among subjects followed for 24 years. The total variance of bilirubin explained by this polymorphism after adjustment for covariates was 18.6%. Individuals with genotype 7/7 compared to genotypes 6/7 and 6/6 had significantly higher serum bilirubin levels and lower CVD and coronary heart disease (CHD) risk with a hazard ratio of 0.36 and 0.30, respectively (8). Since the frequency of the genotype 7/7 ranges from 12–16% in the Caucasian population, this polymorphism may have a large population impact.
In the present study, we performed 1) a linkage scan conditional on the UGT1A1*28 association analysis to evaluate if the TA-repeat polymorphism in the promoter region of the UGT1A1 can fully explain the linkage peak we identified in our previous linkage studies (10,11) and 2) an Affymetrix 100K SNPs genome-wide association (GWA) study to search for further genes influencing serum bilirubin levels.
Methods
Study subjects
Details on the Framingham Heart Study are described in the Online Supplementary Material. The conditional linkage genome scan was carried out in the 330 largest extended Framingham families used in the previous genome-wide linkage scan (10). The same set of family samples was used for genotyping with the Affymetrix 100K SNP GeneChip for a genome-wide association study with a total number of 1345 genotyped individuals (15). The investigation was in line with the principles outlined in the Declaration of Helsinki.
Laboratory and genotyping methods (see also Online Supplementary Material)
Total serum bilirubin was measured during the first and second examinations of the offspring after a 12 hour fasting period. Details on the genotyping of the 399 microsatellites for genome-wide linkage scan were provided recently (10). Genotyping of the UGT1A1 promoter TA-repeat polymorphism in the TATA box at position −53 was performed using the ABI 3130xl sequencing system as recently described in detail (8). The Affymetrix 100K SNPs provided 112,990 autosomal and 2344 X-chromosomal SNPs in 1345 individuals (16). SNPs with minor allele frequency <10% or a call rate <80% or Hardy-Weinberg equilibrium p-value<0.001 were excluded, leaving 70591 autosomal and 1346 X chromosomal SNPs available for analysis.
Statistical methods (see also Online Supplementary Material)
Linkage analysis conditional on candidate gene association analysis
To evaluate linkage conditional on the −53 TA repeat polymorphism, we used mixed effects variance component models in the SOLAR package to simultaneously incorporate the genotype-specific means of the measured genotype test while performing a variance component-based linkage test (17). As in our previous analysis on the same set of families, we adjusted bilirubin levels for age, sex, height, weight, total cholesterol, hematocrit, albumin, serum glutamic oxaloacetic transaminase, smoking and alcohol (10) and added the −53 TA repeat genotypes as an additional covariate.
100K genome-wide association study
For each SNP, we modeled the log-transformed trait value adjusted for the same covariates as used in the linkage analysis using a linear mixed effects model (LME) in SAS and assuming a recessive, additive, and dominant model. For each genetic model, a p-value was obtained. The smallest of the three p-values was used to rank all SNPs. This was shown to be more robust than using a single p-value based on the association test under an additive model (18,19). Since LME does not take the specific kinship coefficient of each relative pair into account as SOLAR does, we used SOLAR to analyze the 50 LME top-ranked SNPs to compare the results of SOLAR with that of LME. Further details and handling of X-chromosomal SNPs is described in the Online Supplementary Material.
Results
Table 1 presents the characteristics of the study population stratified by −53 TA repeat genotypes from offspring examination 1. Genotype frequencies were 47%, 44% and 9% for genotypes 6/6, 6/7, and 7/7, respectively. No significant differences across genotype groups were found in all characteristics listed in Table 1 except for bilirubin. Mean serum bilirubin was the highest in 7/7 carriers (13.2±5.9 mg/l) and lower in 6/7 (8.2±2.9 mg/l) and 6/6 (6.8±2.2 mg/l) carriers (P<0.01). The characteristics of the study population in the second examination are similar to those of the first examination (data not shown).
Table 1.
General characteristics of the study population stratified by the UGT1A1*28 genotypes.
| Genotype (TA repeats) | 6/6 | 6/7 | 7/7 |
|---|---|---|---|
| Total typed, n | 509 | 485 | 97 |
| Age at entry (years) | 32.3 (10.0) | 31.2 (9.9) | 33.3 (9.4) |
| Height (inches) | 66.2 (4.1) | 66.0 (3.8) | 65.9 (4.2) |
| Weight (pounds) | 156.2 (36.1) | 152.1 (32.7) | 157.9 (32.7) |
| Total cholesterol (mg/dl) | 190.4 (36.7) | 188.4 (35.5) | 188.0 (38.9) |
| Albumin (mg/dl) | 46.9 (3.4) | 47.2 (3.4) | 46.6 (3.0) |
| SGOT (mg/dl) | 43.9 (17.7) | 43.1 (18.2) | 43.4 (18.5) |
| Smoking (# of cigarettes/d) | 11.1 (14.1) | 12.3 (14.6) | 11.7 (16.9) |
| Alcohol (oz/wk) | 3.4 (5.1) | 3.4 (4.2) | 3.3 (3.8) |
| Bilirubin (mg/l)* | 6.8 (2.2) | 8.2 (2.9) | 13.2 (5.9) |
Values are means (SD) as otherwise indicated.
P<0.01
Genome-wide linkage analysis conditional on the UGT1A1*28 association
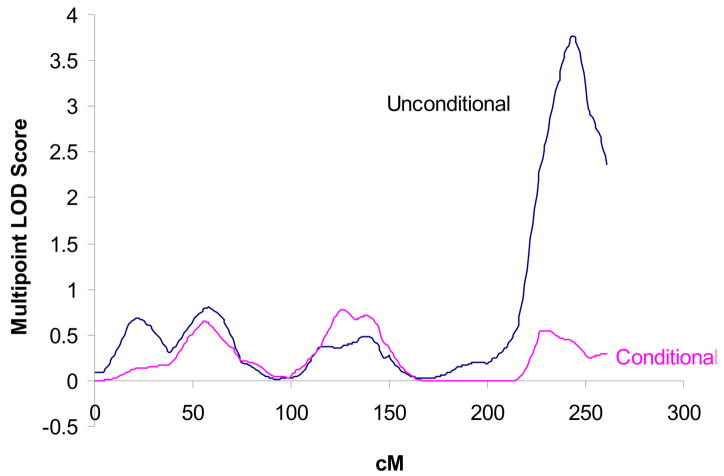
Conditional on the TA-repeat association, the original linkage peak on the 2q telomere dropped from a LOD score of 3.8 to 0.4 (Figure 1). The second linkage peak on chromosome 4 at 96 cM originally described (10) with a LOD score of 1.3 remained unchanged. In two chromosomal regions the LOD scores increased from originally less than 1 to greater than 1: chromosome 7 at 110 cM (increase from 0.51 to 1.65); chromosome 15 at 0 cM (increase from 0.58 to 1.03).
Figure 1.
Multipoint LOD scores for serum bilirubin of the original linkage scan (unconditional) (10) and the conditional (combined linkage/disequilibrium analysis) linkage scan on chromosome 2. After adjusting for bilirubin variance explained by the UGT1A1 TA-repeat polymorphism, the LOD score on chromosome 2q dropped from 3.8 to 0.4 confirming that this polymorphism explains the previous linkage findings. X-axis values are cM. Y-axis values are multipoint LOD scores.
100K genome-wide association study
The results of the top 10 SNPs by LME minimum p-value ranking and then subsequently by SOLAR analyses for the first and second examinations are listed in Table 2. An extended list of the top ranking 50 SNPs is provided in Online Supplementary Tables 1 and 2.
Table 2.
P-values of top 10 SNPs by LME minimum p-value ranking and then subsequently by SOLAR analyses for each of the two examination.
|
LME Analysis |
Solar Analysis |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chro- mosome |
Physical Position |
cM | P recessive |
P additive |
P dominant |
lowest P | P value ranking |
P recessive |
P additive |
P dominant |
lowest P | P value ranking |
| Examination 1 | |||||||||||||
| rs9323037 | 14 | 40501442 | 44.774 | <E-10 | 0.0119618 | 0.02237731 | <E-10 | 1 | 0.34601 | 0.04496 | 0.02305 | 0.02305 | 49 |
| rs1903937 | 12 | 43807754 | 56.322 | 4.191E-05 | 4.214E-07 | 2.9055E-05 | 4.214E-07 | 2 | 0.00023 | 6.36E-06 | 0.0003 | 6.36E-06 | 2 |
| rs10508059 | 13 | 100730832 | 84.627 | 0.0002288 | 1.281E-06 | 2.0363E-05 | 1.281E-06 | 3 | 0.00295 | 3.64E-05 | 0.00025 | 3.64E-05 | 7 |
| rs10516862 | 4 | 91354349 | 100.024 | 0.0131153 | 2.383E-06 | 0.10268287 | 2.383E-06 | 4 | 0.63002 | 0.249626 | 0.20494 | 0.204938 | 50 |
| rs998358 | 8 | 123842133 | 130 | 2.931E-06 | 1.746E-05 | 0.13625395 | 2.931E-06 | 5 | 0.10659 | 0.000192 | 0.00011 | 0.000108 | 21 |
| rs1596516 | 12 | 39490195 | 55.29 | 0.0232866 | 6.309E-05 | 5.8415E-06 | 5.842E-06 | 6 | 3.2E-05 | 0.001041 | 0.17783 | 3.17E-05 | 6 |
| rs281877 | 7 | 131278723 | 136.647 | 6.161E-06 | 0.0166819 | 0.1893402 | 6.161E-06 | 7 | 6.4E-05 | 0.012772 | 0.12514 | 0.000064 | 13 |
| rs1113193 | 2 | 234351137 | 243.308 | 0.0006329 | 8.099E-06 | 0.00012286 | 8.099E-06 | 8 | 0.00271 | 9.33E-06 | 7.7E-05 | 9.33E-06 | 3 |
| rs10492939 | 1 | 3326028 | 3.391 | 9.12E-06 | 0.0032361 | 0.16025649 | 9.12E-06 | 9 | 0.00011 | 0.011897 | 0.27678 | 0.000112 | 22 |
| rs9321588 | 6 | 137487052 | 138.019 | 0.6699534 | 0.0127276 | 9.7611E-06 | 9.761E-06 | 10 | 2.1E-05 | 0.028799 | 0.93903 | 2.11E-05 | 4 |
| Examination 2 | |||||||||||||
| rs1113193 | 2 | 234351137 | 243.31 | 0.0345571 | 1.377E-08 | 7.485E-07 | 1.377E-08 | 1 | 0.084459 | 2.48E-05 | 1.68E-05 | 1.68E-05 | 2 |
| rs2269337 | 9 | 130602238 | 144.16 | no converge | 7.242E-08 | 0.0011683 | 7.242E-08 | 2 | 0.012702 | 0.002836 | 0.013068 | 0.002836 | 45 |
| rs618171 | 1 | 215537693 | 239.06 | 1.997E-07 | 0.0493516 | 0.5044529 | 1.997E-07 | 3 | 0.01397 | 0.126232 | 0.583849 | 0.01397 | 48 |
| rs434310 | 6 | 95061282 | 102.81 | 7.412E-05 | 6.592E-07 | 2.328E-05 | 6.592E-07 | 4 | 0.002277 | 1.63E-05 | 0.000132 | 1.63E-05 | 1 |
| rs233818 | 4 | 103561912 | 107.95 | 0.1167418 | 0.0078786 | 8.359E-07 | 8.359E-07 | 5 | 0.035446 | 0.071176 | 0.241546 | 0.035446 | 50 |
| rs9304554 | 18 | 26099175 | 56.71 | 0.0017456 | 1.054E-06 | 1.938E-06 | 1.054E-06 | 6 | 0.002777 | 0.000215 | 0.00168 | 0.000215 | 30 |
| rs1477627 | 11 | 97005057 | 94.029 | 1.406E-06 | 8.673E-05 | 0.4790098 | 1.406E-06 | 7 | 0.771445 | 0.009052 | 0.000352 | 0.000352 | 36 |
| rs1413754 | 6 | 131192319 | 128.93 | 1.452E-06 | 5.857E-05 | 0.0139882 | 1.452E-06 | 8 | 2.32E-05 | 0.000486 | 0.039586 | 2.32E-05 | 5 |
| rs4849360 | 2 | 111532102 | 121.99 | 0.6357593 | 0.1239215 | 1.743E-06 | 1.743E-06 | 9 | 0.005472 | 0.303775 | 0.782526 | 0.005472 | 47 |
| rs756418 | 4 | 11224236 | 23.177 | 0.2779108 | 4.158E-06 | 1.841E-06 | 1.841E-06 | 10 | 0.322777 | 3.72E-05 | 2.46E-05 | 2.46E-05 | 6 |
An extended list with the top 50 SNPs is provided in the Supplementary Tables 1 and 2.
Grey shaded p-values are those with genome-wide significance (p < 0.05/71937 quality-controlled SNPs = <6.95×10−7).
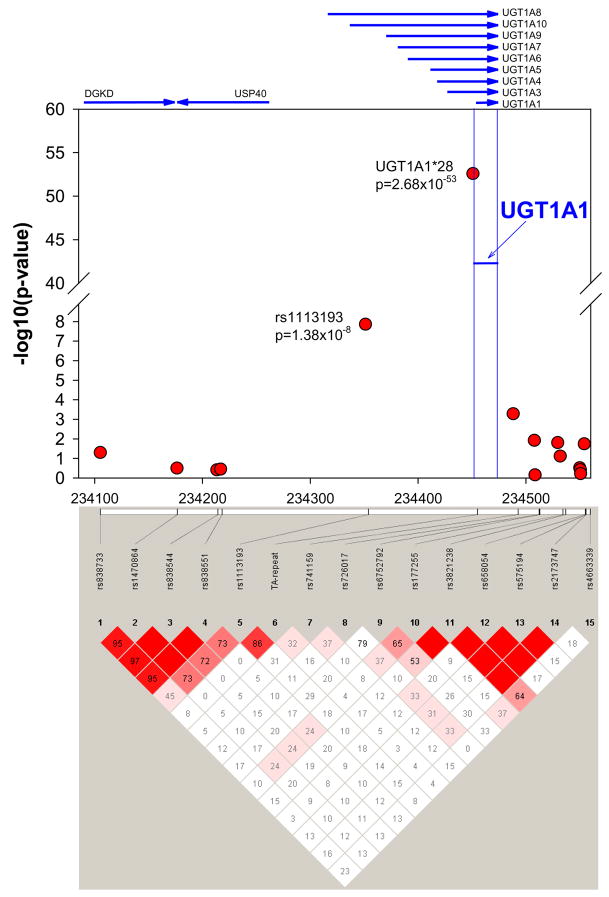
For analyses of bilirubin from both examinations, there were six SNPs that reached genome-wide significance in the LME analysis: two were associated with bilirubin levels measured from the first examination (rs9323037 and rs1903937) and four were from the second examination (rs1113193, rs2269337, rs618171 and rs434310). The top ranked SNP from the second examination, rs1113193, is about 100kb upstream of the UGT1A1 gene, the closest SNP to the gene and is in significant LD with the UGT1A1*28 (TA-repeat) polymorphism, with D′=0.86 (Figure 2). This SNP was ranked number 2 in the subsequent SOLAR analysis. For the first examination, this SNP was ranked number 8 in LME and number 3 in SOLAR by the minimum p-value method. Except for this SNP, the rest of the 49 SNPs in each examination did not overlap (Table 2 and Supplementary Tables 1 and 2).
Figure 2.
P-values of 14 genotyped SNPs from the genome-wide association study and the TA-repeat (UGT1A1*28) polymorphism within the promoter of UGT1A1. P-values for the genome-wide association study SNPs were calculated using a linear mixed effects model (LME). The figure illustrates the lowest p-value from the two examinations. Chromosomal position were taken from HapMap (NCBI build 36, dbSNP b126). The blue lines show the boundaries of the UGT1A1 region. The lower panel provides the linkage disequilibrium pattern (D′, Haploview 4.0).
There were no genes located within 100–200kb of four other top ranked SNPs (rs9323037, rs1903937, rs618171 and rs434310). rs2269337 was located within exosome component 2 (EXOSC2) which is involved in various mRNA decay pathways (20). The wider chromosomal area hosts a few genes involved in the development of cancer and leukemia and no obvious genes involved in bilirubin metabolism.
The results of the top 10 SNPs from the X chromosome using LME analysis are listed in Supplementary Table 3. No SNP reached significance after correction for multiple comparisons and none of the top 10 SNPs from the first and second examinations overlapped.
Association analysis including the TA-repeat polymorphism to the GWA SNP analysis
Using SOLAR to analyze the association between the −53 TA-repeat polymorphism and bilirubin levels from the first examination, the p-values were 3.06×10−39, 2.68×10−53, and 2.81×10−31 for the dominant, additive, and recessive models, respectively. Figure 2 shows the p-values of 14 SNPs genotyped in the GWA study and of the UGT1A1*28 TA-repeat polymorphism in the chromosomal region of UGT1A1 including the upstream 350 kb and downstream 100kb.
Discussion
Most knowledge regarding the relationship of the UGT1A1 gene and serum bilirubin has been derived from association studies using polymorphisms within this gene in relatively small samples of unrelated individuals. Although these studies provided important evidence for the genetic effect of this gene, they are limited to this known candidate gene and are susceptible to type I error due to possible hidden population stratification. Our study is the first study using a large familial sample to measure the effect of the −53 TA-repeat (UGT1A1*28) polymorphism and to explore the possibility of other loci controlling serum bilirubin levels through genome-wide linkage, genome-wide linkage/disequilibrium (conditional linkage), and genome-wide association studies. Our findings support an important continued role for linkage analysis and the collection of family data in the age of GWA studies (21,22).
UGT1A1 as the major gene influencing bilirubin levels
Our study extends the previous observation that individuals with the UGT1A1*28 allele have significantly higher serum bilirubin levels. In conditional linkage analysis, when an association between the UGT1A1*28 and bilirubin levels was allowed simultaneously, the evidence for linkage on the 2q telomere was eliminated. This indicates that the TA repeat polymorphism accounts for the genetic linkage signal we observed earlier (10,11). The conclusion is supported by the results of the 100K SNPs GWA study. Of the SNPs screened in our 100K scan, rs1113193 is the SNP that is physically close to and in significant linkage disequilibrium with UGT1A1*28. It is the only SNP to be consistently ranked in the top 10 SNPs with different analytical approaches, different ranking methods, and two different bilirubin measurements at the two Framingham examinations. This SNP reaches genome-wide significance and is the top-hit in the second examination by LME analysis.
Although several other mutations within this gene have been identified, their allelic frequencies are low in Caucasians (23), and the UGT1A1*28 was thought to be the main cause of Gilbert syndrome in Caucasians due to its functional influence on the promoter activity (12,24). This is supported by a recent association study on 13 polymorphisms within the UGT1A1 gene and serum bilirubin levels, that conditional on the TA repeat no other variant was shown to be significantly associated with bilirubin in both genotype and haplotype analyses (25). Recently, another polymorphism in the phenobarbital responsive enhancer module of this gene was found to reduce transcriptional activity (26). However, this polymorphism was in complete LD with the TA-repeat (27,28). Therefore, association studies can not distinguish their effect.
The second aim of our study was to further explore, whether there are other genes with large effects contributing to the variation of bilirubin levels. However, from our previous genome scans (10,11) and the conditional linkage genome scan in this study, we observe no additional linkage peaks with a LOD score above 2 in the rest of the genome. The results are in line with our GWA study which showed that except for rs1113193, no other SNP was consistently ranked in the top list. This implies that UGT1A1 might be one of the most important genes with a large effect controlling bilirubin levels in a Caucasian population as proposed by earlier segregation analyses (5,9). However, one has to bear in mind that neither our linkage scan nor our 100K GWA study adequately covered the genome in a high enough resolution which leaves some room for the identification of further genes in the future. Furthermore, the total variance of bilirubin explained by the UGT1A1 TA-repeat polymorphism was 18.6% (8) which is very high compared to other phenotypes. Recent GWA studies on e.g. lipid levels found that several genes combined explained only up to 4.8% of variance in lipids (29) which was at the best about 1.4% for a singular gene (30). The effect of the UGT1A1 TA-repeat polymorphism on bilirubin levels can therefore be considered as exceptionally high which might be explained by the fact that the TA-repeat polymorphism was already shown to have a strong functional influence (12–14). Therefore, future large GWA studies including meta-analyses using microarrays with a higher coverage and HapMap-imputed genotyes might be able to find further genes with small- and medium-sized effects on bilirubin levels for which our present study does not have the power.
Why was UGT1A1 not identified by GWA studies on CVD?
One may question why this locus was not identified by recent GWA studies on CVD. Table 3 lists all previous studies which investigated the association between the UGT1A1 TA-repeat polymorphisms and both, bilirubin levels and atherosclerosis outcomes (6–8,31,32). Although these studies found clear associations of this polymorphism with bilirubin levels and most studies found an association of bilirubin levels with atherosclerosis outcomes, only the prospective Framingham Heart Study (8) found an association of the polymorphism with CVD endpoints. The major difference between our previous Framingham Study (8) and these retrospective studies and GWA studies on CVD is that the Framingham Study was a prospective population-based cohort study followed for 24 years with an average age of 36 years at baseline, whereas the other studies were cross-sectional case-control studies with much higher mean ages at recruitment, mostly 60 years or older. First of all, a survival bias due to older ages at recruitment is possible, because up to half of the incident cases do not survive the first half year after a major CHD event and thus are not available for inclusion into the studies. CVD in those individuals could be due to stronger genetic effects and thus be more severe. Furthermore, the further survival after the first event is influenced by genetic factors (33). Second, genetic heterogeneity at this locus in those GWA studies can not be excluded. All those GWA studies included a large study population and some with several independent samples. This increased the possibility to include people with different genetic backgrounds.
Table 3.
Previous studies investigating the association of UGT1A1 TA-repeat polymorphism with bilirubin levels and cardiovascular endpoints.
| Genotype 7/7 versus 6/6 TA-repeats |
||||||
|---|---|---|---|---|---|---|
| Study | Study design | CVD endpoint | Number of cases/controls | bilirubin levels % changea | CVD endpoint OR (95%CI) | Association of bilirubin with CVD endpoint OR (95%CI) |
| Rotterdam Study (31) | retrospective | MI | 185/255 | +90%/+53% | 1.3 (0.8–2.2) | 0.96 (0.90–1.03) per 1 μmol/L b |
| ECTIM Study (32) | retrospective | MI | 366/314 | n.d. | 1.9 (1.1–3.4) | n.d. |
| Lingenhel et al. (7) | retrospective | CAD | men: 365/397 | +102%/+94% | 0.97 (0.61–1.54) | 0.90 (0.86–0.94) per 0.1 mg/dL |
| women: 112/222 | +121%/+15% | 0.93 (0.45–1.93) | 0.77 (0.68–0.87) per 0.1 mg/dL | |||
| Rantner et al. (6) | retrospective | PAD | 255/255 | +117%/+52% | n.s. | 0.89 (0.84–0.93) per 0.1 mg/dL |
| Framingham Heart Study (8) | prospective 24 years | CVD | 156/1583 | + 81% | 0.36 (0.18–0.74) | 0.90 (0.84–0.96) per 0.1 mg/dL |
MI, myocardial infarction; CAD, coronary artery disease; PAD, peripheral arterial disease; CVD, cardiovascular disease (includes fatal and nonfatal myocardial infarction, angina pectoris, coronary insufficiency, stroke, transient ischemic attack, intermittent claudication, congestive heart failure, or death from coronary heart disease).
Relative difference of bilirubin levels in carriers of 7/7 TA-repeats compared to wildtype 6/6 TA-repeats.
Serum bilirubin concentrations were available only for 114 cases and 162 controls.
Strengths and limitations of the study
One lesson we have learned from recent GWA studies is that due to multi-comparison issues, power may be low and it is not easy to reach genome-wide significance even for associated SNPs with moderate to large effect unless the sample size is large. Therefore, it may be appealing to use different analytical methods, ranking methods and repeated measurements to identify those SNPs that consistently rank in top position. This was shown to be more robust than using a single p-value based on the association test under an additive model (18,19). The probability that those SNPs are truly associated SNPs might be high. This might be the reason why rs1113193 did not reach genome-wide significance and rank in top position in the earlier 100K GWA scan on bilirubin measured on the Framingham Offspring second examination using a less extended analysis method and different covariates (15). Our study demonstrates that GWA study results can still be robust in the case where a SNP is in linkage disequilibrium with a functional variant with an effect size which is not too small, even if the marker density is low as with the 100K gene chip, and SNPs related to the phenotype are far apart with distances of 100kb and more. However, due to type I error, some SNPs that are not truly associated could have small p-values in one analytical method, or using one ranking method, or on one examination, but have larger p-values otherwise.
The other SNPs identified with genome-wide significance but less consistent results with the various analysis methods clearly need further investigation in other cohorts. Four of the other five SNPs identified with genome-wide significance were far outside of genes. Since long-range effects of regulatory regions can not be excluded (30), these regions might be of interest for future studies.
Conclusions
Our studies suggest that UGT1A1 may be the only major gene with strong effects on bilirubin levels and the TA-repeat polymorphism might be the key polymorphism within the gene controlling serum bilirubin levels in the Caucasian population. With an allele frequency of 30%–40% of the (TA)7 repeats and a frequency of homozygous individuals of 10%–16% (6–8,12,24), the gene might have an important effect on preventing the development of CHD and CVD. Therefore, future research to examine possible therapeutic prevention targeting of this genetic variant and its influence on bilirubin levels may have a large population impact (34).
Supplementary Material
Acknowledgments
We are very grateful to Robert C. Elston and Nancy L. Geller for their helpful comments on the data analysis and critical review of the manuscript.
This work was supported by the National Heart, Lung, and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195). This work was also supported by grants to Florian Kronenberg from the “Tiroler Wissenschaftsfonds” (Project 404/505), the “Austrian Nationalbank” (Project 9331) and the “Genomics of Lipid-associated Disorders – GOLD” of the “Austrian Genome Research Programme GEN-AU”, Schoepfstr. 41, A-6020 Innsbruck.
Footnotes
Conflict of interest: None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schwertner HA, Jackson WG, Tolan G. Association of low serum concentration of bilirubin with increased risk of coronary artery disease. Clin Chem. 1994;40:18–23. [PubMed] [Google Scholar]
- 2.Breimer LH, Wannamethee G, Ebrahim S, Shaper AG. Serum bilirubin and risk of ischemic heart disease in middle-aged British men. Clin Chem. 1995;41:1504–1508. [PubMed] [Google Scholar]
- 3.Hopkins PN, Wu LL, Hunt SC, et al. Higher serum bilirubin is associated with decreased risk for early familial coronary artery disease. Arterioscler Thromb Vasc Biol. 1996;16:250–255. doi: 10.1161/01.atv.16.2.250. [DOI] [PubMed] [Google Scholar]
- 4.Levinson SS. Relationship between bilirubin, apolipoprotein B, and coronary artery disease. Ann Clin Lab Sci. 1997;27:185–192. [PubMed] [Google Scholar]
- 5.Hunt SC, Kronenberg F, Eckfeldt JH, et al. Association of plasma bilirubin with coronary heart disease and segregation of bilirubin as a major gene trait: The NHLBI Family Heart Study. Atherosclerosis. 2001;154:747–754. doi: 10.1016/s0021-9150(00)00420-2. [DOI] [PubMed] [Google Scholar]
- 6.Rantner B, Kollerits B, Anderwald-Stadler M, et al. Association between the UGT1A1 TA-repeat polymorphism and bilirubin concentration in patients with intermittent claudication: results from the CAVASIC Study. Clin. Chem. 2008;54:851–857. doi: 10.1373/clinchem.2007.102046. [DOI] [PubMed] [Google Scholar]
- 7.Lingenhel A, Kollerits B, Schwaiger JP, et al. Serum bilirubin levels, UGT1A1 polymorphisms and risk for coronary artery disease. Exp. Gerontol. 2008;43:1102–1107. doi: 10.1016/j.exger.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J-P, O’Donnell CJ, Schwaiger JP, et al. Association between the UGT1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation. 2006;114:1476–1481. doi: 10.1161/CIRCULATIONAHA.106.633206. [DOI] [PubMed] [Google Scholar]
- 9.Hunt SC, Wu LL, Hopkins PN, Williams RR. Evidence for a major gene elevating serum bilirubin concentration in Utah pedigrees. Arterioscler Thromb Vasc Biol. 1996;16:912–917. doi: 10.1161/01.atv.16.8.912. [DOI] [PubMed] [Google Scholar]
- 10.Lin JP, Cupples LA, Wilson PW, Heard-Costa N, O’Donnell CJ. Evidence for a gene influencing serum bilirubin on chromosome 2q telomere: a genomewide scan in the Framingham study. Am J Hum Genet. 2003;72:1029–1034. doi: 10.1086/373964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronenberg F, Coon H, Gutin A, et al. A genome scan for loci influencing anti-atherogenic serum bilirubin levels. Eur J Hum Genet. 2002;10:539–546. doi: 10.1038/sj.ejhg.5200842. [DOI] [PubMed] [Google Scholar]
- 12.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP- glucuronosyltransferase 1 in Gilbert’s syndrome. N Engl J Med. 1995;333:1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 13.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet. 1996;347:578–581. doi: 10.1016/s0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- 14.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci USA. 1998;95:8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cupples LA, Arruda HT, Benjamin EJ, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8 (Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 17.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 19.Zheng G, Joo J, Lin JP, et al. Robust ranks of true associations in genome-wide case-control association studies. BMC Proc. 2007;1 (Suppl 1):S165. doi: 10.1186/1753-6561-1-s1-s165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clerget-Darpoux F, Elston RC. Are linkage analysis and the collection of family data dead? Prospects for family studies in the age of genome-wide association. Hum Hered. 2007;64:91–96. doi: 10.1159/000101960. [DOI] [PubMed] [Google Scholar]
- 22.Visscher PM, Andrew T, Nyholt DR. Genome-wide association studies of quantitative traits with related individuals: little (power) lost but much to be gained. Eur J Hum Genet. 2008;16:387–390. doi: 10.1038/sj.ejhg.5201990. [DOI] [PubMed] [Google Scholar]
- 23.Maitland ML, Grimsley C, Kuttab-Boulos H, et al. Comparative genomics analysis of human sequence variation in the UGT1A gene cluster. Pharmacogenomics J. 2006;6:52–62. doi: 10.1038/sj.tpj.6500351. [DOI] [PubMed] [Google Scholar]
- 24.Borlak J, Thum T, Landt O, Erb K, Hermann R. Molecular diagnosis of a familial nonhemolytic hyperbilirubinemia (Gilbert’s syndrome) in healthy subjects. Hepatology. 2000;32:792–795. doi: 10.1053/jhep.2000.18193. [DOI] [PubMed] [Google Scholar]
- 25.Hong AL, Huo D, Kim HJ, et al. UDP-Glucuronosyltransferase 1A1 Gene Polymorphisms and Total Bilirubin Levels in an Ethnically Diverse Cohort of Women. Drug Metab Dispos. 2007;35:1254–1261. doi: 10.1124/dmd.106.014183. [DOI] [PubMed] [Google Scholar]
- 26.Sugatani J, Yamakawa K, Yoshinari K, et al. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun. 2002;292:492–497. doi: 10.1006/bbrc.2002.6683. [DOI] [PubMed] [Google Scholar]
- 27.Maruo Y, D’Addario C, Mori A, et al. Two linked polymorphic mutations (A(TA)7TAA and T-3279G) of UGT1A1 as the principal cause of Gilbert syndrome. Hum Genet. 2004;115:525–526. doi: 10.1007/s00439-004-1183-x. [DOI] [PubMed] [Google Scholar]
- 28.Lin Z, Fontaine J, Watchko JF. Coexpression of gene polymorphisms involved in bilirubin production and metabolism. Pediatrics. 2008;122:e156–e162. doi: 10.1542/peds.2007-3249. [DOI] [PubMed] [Google Scholar]
- 29.Aulchenko YS, Ripatti S, Lindquist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heid IM, Boes E, Müller A, et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA Study sheds new light on intergenic regions. Circ Cardiovasc Genetics. 2008;1:10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- 31.Bosma PJ, van dM I, Bakker CT, et al. UGT1A1*28 allele and coronary heart disease: the Rotterdam Study. Clin Chem. 2003;49:1180–1181. doi: 10.1373/49.7.1180. [DOI] [PubMed] [Google Scholar]
- 32.Gajdos V, Petit FM, Perret C, et al. Further evidence that the UGT1A1*28 allele is not associated with coronary heart disease: The ECTIM Study. Clin Chem. 2006;52:2313–2314. doi: 10.1373/clinchem.2006.078667. [DOI] [PubMed] [Google Scholar]
- 33.Marin F, Gonzalez-Conejero R, Lee KW, et al. A pharmacogenetic effect of factor XIII valine 34 leucine polymorphism on fibrinolytic therapy for acute myocardial infarction. J Am Coll Cardiol. 2005;45:25–29. doi: 10.1016/j.jacc.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 34.Schwertner HA, Vitek L. Gilbert syndrome, UGT1A1*28 allele, and cardiovascular disease risk: possible protective effects and therapeutic applications of bilirubin. Atherosclerosis. 2008;198:1–11. doi: 10.1016/j.atherosclerosis.2008.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.