Abstract
The vast majority of membrane protein complexes of biological interest cannot be purified to homogeneity, or removed from a physiologically relevant context without loss of function. It is therefore not possible to easily determine the 3D structures of these protein complexes using X-ray crystallography or conventional cryo-electron microscopy. Newly emerging methods that combine cryo-electron tomography with 3D image classification and averaging are, however, beginning to provide unique opportunities for in situ determination of the structures of membrane protein assemblies in intact cells and non-symmetric viruses. Here we review recent progress in this field and assess the potential of these methods to describe the conformation of membrane proteins in their in native environment.
Introduction
Strategies for structure determination using electron crystallography and cryo-electron microscopy rely on the principle of averaging large numbers of images of identical copies of a given molecular complex to determine structural information. For a handful of membrane proteins that can be crystallized in the plane of the lipid bilayer, electron crystallographic methods allow structure determination at atomic resolution by combining information from projection images of thin protein crystals tilted to varying extents relative to the electron beam [1-5]. For protein complexes and viruses with high symmetry that can be purified, single particle cryo-electron microscopic approaches provide powerful tools to combine projection images of complexes in different orientations to progressively build up 3D structures, which in some instances have been obtained at resolutions of 4 Å or better [6-10]. But what if the functional unit of interest is only present in the context of a heterogeneous unit such as a whole cell or virus, and cannot therefore by crystallized or analyzed by the averaging procedures used in single particle cryo-electron microscopy? As discussed in this review, recent advances in cryo-electron tomography, combined with techniques for image averaging and 3D classification [11-13] are beginning to provide powerful tools to describe the molecular structures of membrane proteins without having to isolate them using biochemical purification procedures.
Electron tomography is a well-established method to obtain 3D density distributions of microscopic objects in applications ranging from materials science to biology [14-16]. In biological applications aimed at describing the 3D shapes of “one-of-a-kind” objects under near-native conditions, the images are usually recorded at cryogenic temperatures and at the lowest possible electron doses in order to minimize damage from electron irradiation of the sample. Under these conditions, even in the best cases, the resolutions that can be obtained in a single tomogram of a biological specimen are barely enough to discern molecular shapes. However, by extracting sub-volumes corresponding to specific cellular components such as membrane proteins that are present in multiple copies, 3D averaging can be used to obtain density maps of selected macromolecular complexes at progressively higher signal-to-noise ratios [12,17-22]. A collection of recently reported structures determined using this approach (Figure 1) illustrates that resolutions as high as ∼ 20 Å are within reach [23], and maps of protein complexes at this resolution can already be interpreted in terms of the atomic structures of their individual components determined using X-ray crystallography. Although the experimental and computational approaches are still in an early phase of development, in principle, there are no fundamental barriers to achieving significantly higher resolutions.
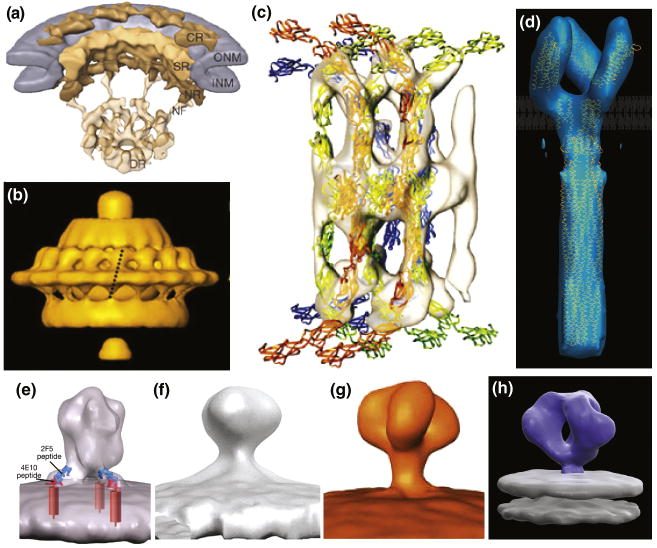
Figure 1.
Representative examples 3D architectures of membrane associated proteins determined using cryo-electron tomography combined with image averaging. (A) Nuclear pore complex [18], (B) Treponema primitia flagellar motor [20], (C) Cadherins in native epidermal desmosomes fitted with coordinates of cadherin molecules obtained by X-ray crystallography [17], (D) Bacterial chemoreceptor for serine (Tsr) fitted with coordinates for the cytoplasmic and ligand-binding domains obtained by X-ray crystallography and the HAMP domain obtained by NMR spectroscopy [19]. (E-H) Four conflicting density maps reported for the SIV/HIV-1 envelope glycoproteins from the work of Zhu et al (E, F) [22,30], Zanetti et al (G) [21] and Liu et al (H) [23].
Structural analysis of receptors in cell membranes
While there is beginning to be major progress in the use of X-ray crystallography to determine the structures of integral membrane proteins, the challenge of determining their structures in the context of an intact cell remains a largely unrealized prospect. Even in cases where atomic models are available for some or all components, knowledge of their intact structures and especially their higher order organization in the plane of the membrane is of great interest. For example, many membrane receptors, especially those involved in mediating signal transduction are often clustered in the cell membranes of prokaryotes and eukaryotes, and receptor conformations in these clusters can reflect functional changes in the cell. An example of the use of cryo-electron tomography to describe structure and conformational changes in the bacterial chemotaxis receptor is discussed below in more detail [19].
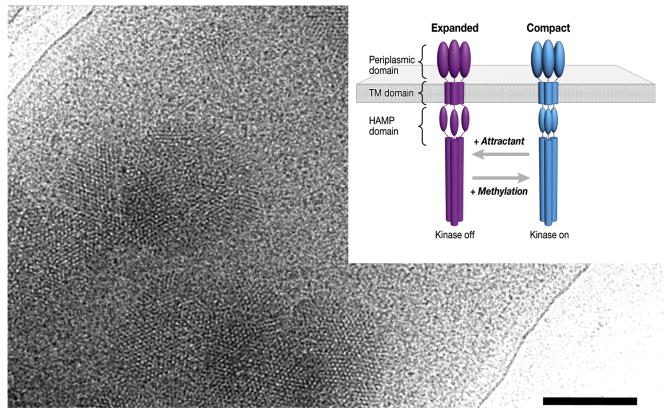
In bacterial chemotaxis, changes in concentration of extracellular ligands influence the conformation of their cognate receptors, resulting in a series of signaling events that regulate rotation of the flagellar motor [24]. Overproduction of the receptors in cells lacking all other chemotaxis components provides a useful way to image the effects of ligand binding in whole cells using cryo-electron tomography. The fact that the receptors are present in small partially ordered clusters (Figure 2) helps locate them in the cell, but is not in any way essential for the process of 3D alignment and classification. By extracting the individual volumes corresponding to each receptor trimer, it was possible to classify and average the dominant class members to deduce that the receptors are organized in two distinct trimer-of-dimer conformations. The two conformations differ in the arrangement of a specific region of the protein (called the HAMP signaling domain) which is on the cytoplasmic side of the membrane (Figure 2 inset). Ligand binding and methylation alter the distribution of chemoreceptors between the two states, with serine binding favoring the expanded state, and chemoreceptor methylation favoring the compact state. These structural analyses, although presently at very modest resolution (∼ 30 Å) nevertheless suggest that HAMP domain rearrangements may play a key structural role in mediating signal transduction across the cell membrane.
Figure 2.
Identification of two distinct receptor conformations of a membrane protein. Projection image of a whole E.coli cell engineered to overproduce the bacterial chemoreceptor for serine. Small patches of the receptor which are present in the cytoplasmic membrane are evident in the electron microscopic image recorded under low-dose illumination. From tomograms of cells such as the one shown, subvolumes corresponding to individual receptor trimers were extracted and classified to obtain two distinct receptor conformations (inset). The relative occupancy of the two states is modulated by changes in the level of serine or the extent of receptor methylation, which have opposing functional effects [24].
Since specimen thickness increases very rapidly at high tilt angles, tomographic data is usually collected over a tilt range spanning from + 70 degrees to − 70 degrees in most cases. This results in a “missing wedge” of data reflecting the incomplete sampling of the full tilt range. When the molecular components being averaged are all in the same orientation (as in the case of the membrane receptors discussed above), the effect of the missing wedge results in anisotropy in resolution of the final density maps, with poorer resolution in the direction of the incident beam, but with minimal effect on image classification. In this case, one can successfully extend to three-dimensions the multivariate statistical analyses employed for classifying 2D projection images in single-particle cryo-electron microscopy [13,25]. However, since these are real space techniques, they will not perform well when there is a broad spread in the orientation of the missing wedge, because the algorithms used will tend to classify volumes according to the orientation of the missing wedge rather than the intrinsic structure. In the general case, where the molecular components are randomly oriented, accounting for the missing wedge in reciprocal space is essential for the proper classification of sub-volumes as illustrated in a recent study by Forster et al [12] aimed at resolving GroEL complexes in a mixture with and without bound GroES. A total of 793 volumes corresponding to individual GroEL molecules with and without GroES attached (592 and 201 volumes, respectively) were each aligned to a 60Å resolution map obtained by filtering the atomic model of the GroEL/ES complex (PDB ID, 1AON). Using the resulting alignments, a strategy for classification using principal component analysis followed by k-means clustering was used to successfully discriminate between volumes corresponding to GroEL alone and those in complex with GroES. However, the success of this approach relies heavily on the availability of an external reference to which all volumes can be aligned and this may limit its practical use. An example of carrying out reference-free alignment and classification with accounting for the missing wedge is presented below.
Structure of HIV envelope glycoproteins
The envelope glycoproteins (Env) of simian and human immunodeficiency viruses (SIV and HIV, respectively), mediate binding to the cell surface receptor CD4 on target cells in order to initiate infection. In a recent study, three conformational states of Env displayed on the surface of intact viruses were reported: an unliganded state, a complex of Env with the broadly neutralizing antibody b12 and a ternary complex of Env with CD4 and the antibody 17b [23]. In this case, the three distinct conformational states were analyzed by preparing separate specimens of the virus in the unbound or antibody/ligand-bound conformations. While these studies demonstrate that distinct conformational states of the Env trimer can be distinguished, an interesting question is whether they could be distinguished if they were present simultaneously in the same preparation. If this were possible, it would represent an important step forward towards using these methods for analyzing conformational heterogeneity in membrane proteins.
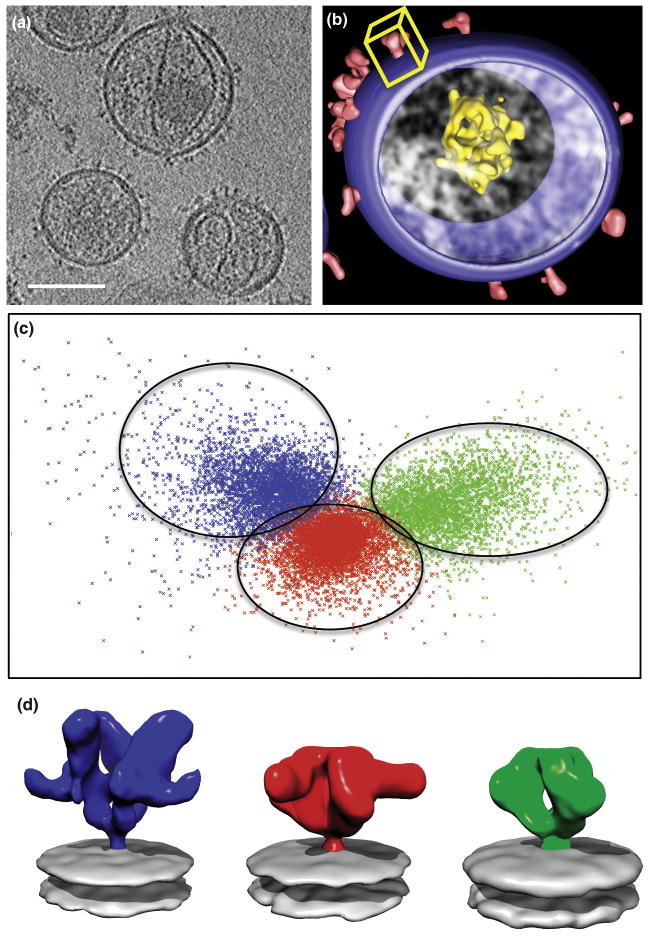
To test this idea, we merged the aligned 3D volumes from the three datasets and assessed whether the classification techniques we have developed would be capable of clearly resolving the distinct structures, given that we can compare the structures derived from the classes with the “ground truth” version obtained before shuffling of the three datasets. The total data set included 13964 individual spike volumes, with roughly equal numbers of contributing images from unliganded (4741), b12-bound (4323) and CD4/17b-bound (4900) Env trimers. As illustrated in Figure 3, the classification methods are very robust, and the combined data set can be cleanly separated into three classes that contain ∼ 89%, 88% and 97% of the individual input datasets. The success of this computational exercise indicates that the use of proper 3D classification strategies can be a powerful tool to assess conformational heterogeneity even at the modest resolutions presently achieved with cryo-electron tomography.
Figure 3.
Structural heterogeneity of HIV-1 envelope glycoprotein spikes studied by image classification. (A) Tomographic slice showing Env spikes on the surface of individual HIV-1 viruses [23]. (B) Segmented rendering of a single virion highlighting the viral membrane, core and spikes; the yellow box represents the subvolume corresponding to a single spike extracted for further analysis. (C) Two-dimensional factor map representation of the three sets of spikes that were combined, shuffled and subjected to classification into 3 groups. Class 1 (green) contained 89% of volumes corresponding to unliganded Env, Class 2 (red) contained 88% of volumes corresponding to b12-bound spikes, and Class 3 (blue) contained 97% of volumes corresponding to 17b-CD4 bound spikes. (D) Corresponding class averages demonstrating the ability of image classification to successfully separate the different conformations of the HIV-1 Env trimer in this experiment (color code as before).
Analyzing conformational variability
While it is encouraging that structures with large differences such as unliganded and liganded Env spikes can be successfully classified and separated, there is much room for further assessment and development of the computational arsenal needed to tackle subtler differences in conformational space. For example, given the intrinsic flexibility of large macromolecular complexes, it will be particularly important to find ways characterize the presence and extent of conformational variability within single viral or cell preparations. Thus, in addition to obtaining averaged structures, classification of the 3D images to resolve the different conformations of the same molecular species is therefore an important tool for describing structural variability. Further, it will be important to assess to what extent effective class separation depends on alignment of individual noisy 3D images to their respective averages as the iterative classification is carried out.
It is also not clear at this time what resolution barriers need to be crossed before subtler conformational changes that do not involve mass differences can be resolved for complexes such as the Env trimer. The significance of this type of analysis has been recognized previously in the context of single particle reconstruction involving classification of 2D projection images [26,27], but similar methods in cryo-electron tomography are yet to be developed. In part, this is a result of lower signal-to-noise ratios in the data, the presence of the missing wedge and the intrinsic structural variability that is a hallmark of biological specimens in their native habitat. Correction of the contrast transfer function will also undoubtedly be an essential part of any effort to improve resolution, and some recent reports have begun to address this problem [28,29]. With continued advances on both experimental and computational fronts, there is the exciting hope that it will be possible in the future to determine structures and structural changes of some of these protein complexes at near-atomic resolution in situ.
References
- 1.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 3.Raunser S, Walz T. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys. 2009;38:89–105. doi: 10.1146/annurev.biophys.050708.133649. [DOI] [PubMed] [Google Scholar]
- 4.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 5.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Cong Y, Zhang Q, Woolford D, Schweikardt T, Khant H, Dougherty M, Ludtke SJ, Chiu W, Decker H. Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy. Structure. 2009;17:749–758. doi: 10.1016/j.str.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc Natl Acad Sci U S A. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryoelectron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that single particle cryo-electron microscopy can be used to obtain density maps of icosahedral viruses at near atomic resolution.
- 11*.Bartesaghi A, Sprechmann P, Liu J, Randall G, Sapiro G, Subramaniam S. Classification and 3D averaging with missing wedge correction in biological electron tomography. J Struct Biol. 2008;162:436–450. doi: 10.1016/j.jsb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Computational framework for alignment, classification, and averaging of subvolumes derived by electron tomography with emphasis on computational efficiency and accuracy that accounts for the effects of the missing wedge and does not require external references.
- 12.Forster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron sub-tomograms using constrained correlation. J Struct Biol. 2008;161:276–286. doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Barnard JS, Sharp J, Tong JR, Midgley PA. High-resolution three-dimensional imaging of dislocations. Science. 2006;313:319. doi: 10.1126/science.1125783. [DOI] [PubMed] [Google Scholar]
- 15.Leis A, Rockel B, Andrees L, Baumeister W. Visualizing cells at the nanoscale. Trends Biochem Sci. 2009;34:60–70. doi: 10.1016/j.tibs.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 16.McIntosh JR. In: Cellular Electron Microscopy. McIntosh JR, editor. Vol. 79. San Diego: Academic Press; 2007. [Google Scholar]
- 17.Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- 18.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 19*.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]; First report of structure determination of a membrane protein in a whole cell, and the description of two closely related protein conformations present in the cell membrane whose relative proportions are altered by ligand binding and receptor methylation.
- 20.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 21.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 23*.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]; Determination of the 3D structures of envelope glycoprotein trimers on infectious HIV-1 particles using cryo-electron tomography combined with 3D classification and averaging, and discovery of the large CD4-induced change in quaternary structure.
- 24.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walz J, Typke D, Nitsch M, Koster AJ, Hegerl R, Baumeister W. Electron Tomography of Single Ice-Embedded Macromolecules: Three-Dimensional Alignment and Classification. J Struct Biol. 1997;120:387–395. doi: 10.1006/jsbi.1997.3934. [DOI] [PubMed] [Google Scholar]
- 26.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Kimmel M, Spahn CM, Penczek PA. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure. 2008;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Fernandez JJ, Li S, Crowther RA. CTF determination and correction in electron cryotomography. Ultramicroscopy. 2006;106:587–596. doi: 10.1016/j.ultramic.2006.02.004. [DOI] [PubMed] [Google Scholar]; A method for CTF detection and defocus determination of cryo electron microscopy tilt-series based on strip-based periodogram averaging that overcomes the low contrast conditions found in cryo electron tomography.
- 29.Winkler H, Taylor KA. Focus gradient correction applied to tilt series image data used in electron tomography. J Struct Biol. 2003;143:24–32. doi: 10.1016/s1047-8477(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhu P, Winkler H, Chertova E, Taylor KA, Roux KH. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS Pathog. 2008;4:e1000203. doi: 10.1371/journal.ppat.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]