Abstract
Chloride intracellular channel protein 1 (CLIC1) functions as an anion channel in plasma and nuclear membranes when its soluble monomeric form converts to an integral-membrane form. The transmembrane region of CLIC1 is located in its thioredoxin-like domain 1 but the mechanism whereby the protein converts to its membrane conformation has yet to be determined. Since channel formation in membranes is enhanced at low pH (5 to 5.5), a condition that is found at the surface of membranes, the structural dynamics of soluble CLIC1 was studied at pH 7 and at pH 5.5 in the absence of membranes by amide hydrogen-deuterium exchange mass spectrometry (DXMS). Rapid hydrogen exchange data indicate that CLIC1 displays a similar core structure at these pH values. Domain 1 is less stable than the all-helical domain 2 and, while the structure of domain 1 remains intact, its conformational flexibility is further increased in an acidic environment (pH 5.5). In the absence of membrane, an acidic environment appears to prime the solution structure of CLIC1 by destabilising domain 1 in order to lower the activation energy barrier for its conversion to the membrane-insertion conformation. The significantly enhanced H/D-exchange rates at pH 5.5 displayed by two segments (peptides 11-31 and 68-82) could be due to the protonation of acidic residues in salt bridges. One of these segments (peptide 11-31) includes part of the transmembrane region which, in the solution structure, consists of helix α1. This helix is intrinsically stable and is most likely retained in the membrane conformation. Strand β2, another element of the transmembrane region, displays a propensity to form a helical structure and has putative N- and C-capping motifs, suggesting that it too most likely forms a helix in a lipid bilayer.
Keywords: Chloride intracellular channel 1 (CLIC1), hydrogen-deuterium exchange mass spectrometry (DXMS), protein dynamics, glutathione transferase, thioredoxin
Chloride intracellular channel (CLIC) proteins are expressed as soluble proteins without a leader sequence but have been shown to form anion channels in plasma and intracellular membranes (1). Although their physiological roles are uncertain, they have been implicated in a range of processes such as bone resorption (2), regulation of cell motility (3), apoptosis (4,5), tubulogenesis (6) and β-amyloid-induced neurotoxicity (7). The CLIC proteins are unusual for ion channels in that they exist in cells both as soluble and integral membrane forms (8-10). Although high-resolution crystal structures exist for soluble forms of CLICs (11-17), very little is known about the mechanism whereby they convert to their membrane-inserted forms. Further, the structures of these membrane-inserted forms, the stoichiometry required to form a channel and the mechanism of ion conductance are unknown. Soluble CLICs are structural homologues of the canonical cytosolic glutathione transferases (GSTs), notably the class Omega GST (11). Unlike CLICs, GSTs are dimeric and do not insert into membranes. The monomeric CLIC structure is composed of two domains; a thioredoxin-like domain 1 and an all-helical domain 2 (for CLIC1, see Figure 1). The membrane-spanning region is located in the smaller thioredoxin-like domain and there is substantial evidence demonstrating that the amino acid sequence corresponding to the amphipathic α1β2 motif forms the transmembrane region (6,11,12,18-20) (Figure 1). Domain 2 appears not to contribute towards either membrane insertion or channel functioning (20). During the conversion of the soluble precursor to an integral-membrane form, strand β1 and the α1β2 motif are expected to undergo conformational changes that will expose buried hydrophobic regions and allow the replacement of protein-protein interactions with protein-lipid interactions (21).
Figure 1.

Ribbon representation of the crystal structure of soluble CLIC1 (PDB code 1k0m (11)). Domain 1 consists of helices α1, α2 and α3 and strands β1, β2, β3 and β4. Domain 2 consists of helices α4 to α10. The linker region between the domains is indicated. The secondary structural elements α1 and β2, that constitute the transmembrane region, are coloured in black.
Triggers that initiate structural rearrangements in soluble-to-membrane proteins include factors such as redox effects, low pH, lipid charge and membrane composition, the binding of signalling molecules and proteolysis (22-26). Oxidation of CLIC1 in the presence of membranes has been shown to trigger a soluble-to-membrane partitioning of the protein (27). CLIC channel formation and activity have been demonstrated to be highly pH-dependent (10,20,28), in that a low pH (5 to 5.5) significantly enhances the rate of CLIC1 insertion into membranes (28). An acidic pH environment at the surface of membranes results from the attraction of protons by the negatively charged polar head groups in the lipid bilayer (29-31).
Water-soluble proteins that insert into membranes follow multi-step processes that include pH-dependent structural changes that enable them to partition into lipid bilayers (32). Many bacterial pore-forming toxins, for example, have been reported to form acid-induced molten globule-like intermediates for membrane insertion (23, 31, 33-35). Since the molecular mechanism of how the structure of soluble CLIC1 is primed or altered for membrane insertion when it encounters the acidic environment near/at the surface of membranes is unknown, the conformational dynamics of soluble human CLIC1 was investigated as a function of pH (5.5-7.0) in the absence of membranes by amide hydrogen-deuterium exchange mass spectrometry (DXMS) (36-39). Hydrogen-deuterium exchange experiments provide information on the local stability of proteins and we have identified and mapped the sequences most susceptible to pH-induced conformational changes at the level of the crystal structure of soluble CLIC1 (11) when the protein molecule moves from a neutral pH (cytosol) to an acidic pH of 5.5 (near/at membrane surface).
Experimental Procedures
Expression and purification of recombinant CLIC1
Human CLIC1 was expressed as a fusion protein with SjGST from the pGEX-4T-1 plasmid (a gift from Samuel Breit, St. Vincent's Hospital and University of New South Wales, Australia) in Eschericia coli BL-21 cells and the CLIC1 purified as described previously (11,40). Purified CLIC1 was stored in 50 mM sodium phosphate buffer, pH 7, containing 1 mM DTT and 0.02 % sodium azide. A molar extinction coefficient of 17170 M-1cm-1 at 280 nm was used to quantify CLIC1.
Spectroscopic measurements
Protein was 5 μM for fluorescence and far-UV CD and 40 μM for near-UV CD measurements in 50 mM sodium phosphate, 1 mM DTT and 0.02 % sodium azide at pH 7.0 or pH 5.5. All measurements were made at 4 and 20 °C. Fluorescence measurements were conducted using a Jasco FP-6300 spectrofluorimeter with excitation at 280 nm and slit widths of 5 nm. Far- and near-UV CD measurements were carried out with a Jasco J-810 spectropolarimeter. Pathlength was 2 mm for far-UV CD and 5 mm for near-UV CD.
Hydrogen-exchange mass spectrometry
Sample preparation
All samples, with the exception of the equilibrium-deuterated control, and buffers were pre-chilled on ice and prepared at 4 °C in the cold room. 15 μl of 0.50 mg/ml CLIC1 (pH 7.0) or 0.34 mg/ml CLIC1 (pH 5.5) in storage buffer (50 mM Na2HPO4, 1 mM DTT, 0.02 % NaN3 pH 7.0 or pH 5.5) were incubated with 45 μl deuteration buffer (10 mM Na2HPO4, 150 mM NaCl in 99.9 % D2O, pH 7.0 or pH 5.5) for varying time periods. As the rate of amide H/D exchange is highly pH dependent; a decrease in pH by 1 unit reduces H/D exchange by approximately 10-fold (41). Since H/D exchange is expected to be about 30-fold slower at pH 5.5 than at pH 7.0, the on-exchange times for CLIC1 at pH 7.0 were 10, 30, 100, 300, 1000 and 3000 seconds, while the on-exchange times at pH 5.5 were 10, 30, 100, 300, 1000, 3000, 9000, 30000 and 90000 seconds. After the allocated time, the hydrogen-deuterium exchange reaction was stopped by adding 90 μl quench buffer, pH 2.3 (0.8 % formic acid, 17 % glycerol, 0.8 M GuHCl for CLIC1 at pH 5.5 or 1.6 M GuHCl for CLIC1 at pH 7.0), and incubating the reaction mixture for 1 min with gentle mixing. In addition to the on-exchange samples a non-deuterated and equilibrium-deuterated controls were also prepared. For the non-deuterated control, 15 μl CLIC1 (pH 7.0) or CLIC1 (pH 5.5) was incubated with a mixture of 45 μl non-deuteration buffer (10 mM Na2HPO4, 150 mM NaCl, pH 7.0) and 90 μl quench buffer. The equilibrium-deuterated control was prepared the night before the rest of the samples where 15 μl CLIC1 (pH 7.0) or CLIC1 (pH 5.5) was incubated with 45 μl equilbrium-deuterated buffer (0.8 % formic acid in 99.9 % D2O) for 16 – 18 hours at 25 °C. After the allocated time, the mixture of CLIC1 (pH 7.0) or CLIC1 (pH 5.5) and equilibrium-deuterated buffer was chillded to 0°C and the exchange reaction was stopped through the addition of 90 ul pre-chilled quench buffer. All samples, including the non- and equilibrium-deuterated controls, were aliquoted in triplicates, snap-frozen on dry ice (immediately after preparation) and express-mailed at dry-ice temperature from the site of preparation (Johannesburg, South Africa) to San Diego CA, USA.
Sample analysis
Sample analysis was performed at the School of Medicine, University of California San Diego (UCSD) employing an apparatus that automatically thawed and proteolysed frozen samples, followed by LC-MS analysis of the resulting peptides. Briefly, frozen protein samples were thawed and, at 0 °C, passed through a solid-phase pepsin column with 0.05 % trifluoroacetic acid at 100 μl/min with 4 min exposure to pepsin. Again at 0 °C, the proteolytic products were passed through a C18 RP-HPLC column (Vydac 50mm ×1mm, 300 A) and eluted with a linear 4 - 36% acetonitrile gradient over thirty minutes. Initial peptide identification was done using collision induced dissociation (CID) on a Finigan LCQ electrospray ion trap mass spectrometer in data-dependent MS2 (tandem MS) mode with capillary temperature at 200 °C. All subsequent samples, including the non-deuterared and equilibrium-deuterated controls as well as 10 – 3000 sec time points, were analysed using the same mass spectrometer in profile mode.
Data processing and peptide validation
The sequence of each peptide was identified using the Sequest software programme (Thermo Finigan Inc) which maps the raw spectral data to the sequence of CLIC1. The resulting peptide pool was quality checked using specialized software (Sierra Analytics, LLC, Modesto, CA) (36-39). Authenticity was validated by a good fit between the theoretical and experimental isotopic envelopes of each peptide. Additionally, parameters such as retention time, m/z range, centroid value and mapping score were scrutinized. The level of deuterium incorporation for each peptide was calculated by subtracting the centroid value of molecular isotope of partially deuterated peptide from the centroid value of non-deuterated peptide as per the method devised by Zhang and Smith (42). The procedure was automated through the use of the specialized software used to quality-check the peptide pool (36-39). Sub-localization of deuterium was performed next, where the partially deuterated peptides were mapped onto the primary sequence of CLIC1. The level of peptide overlap determines the resolution of this step, i.e., multiple overlaping fragmets can narrow the position of deuterium localization. In addition, overlapping fragments that differed by more than two deuterons were identified as outliers and deleted from further analysis.
Corrections for back-exhange, on average ∼30%, were made by employing the methods used by Zhang and Smith (42):
where DO is the average number of deuteriums per peptide at the time of the analysis, m is the average mass of the partially deuterated peptide, m0% is the average mass of the non-deuterated peptide, m100% is the average mass of the equilibrium-deuterated peptide and N is the number of peptide amide linkages in a peptide. Corrections for the final deuterium concentration, 75%, in each sample were also made. The number of peptide amide linkages in a peptide was calculated from
where TN − 2 is the number of residues of the peptide minus the first two amino acids that can not retain deuterium, and TPro is the number of proline residues found in the peptide. It must be noted that the back-exchange equation was shown to introduce an error in DO. For three thousand peptides of random sequence and size this error was calculated to be at an averge of 5.5 % with a standard deviation of 5.5 % (42). Consequently, only differences of 10 % or higher were deemed as significant when comparisons were drawn. Finally, matching peptides for pH 7 and pH 5.5 whose back-exchange values differed by more than 10% were discarded.
Results
Global structure of soluble CLIC1
Fluorescence and CD spectroscopy were used to characterize the global structure of CLIC1 at low temperature (4 °C) and at 20 °C. The data shown in Figure 2 are consistent with the features of the crystal structure of CLIC1 (11,43), and indicate that the protein's conformation is not significantly affected by pH between 7 and 5.5 and by temperature between 4 and 20 °C. It is, therefore, unlikely that the labeling of CLIC1 with deuterium at low temperature involves an artifactual low temperature conformation of the protein.
Figure 2.
Far-UV CD spectra (A), near-UV CD spectra (B), and fluorescence emission spectra (C) for native CLIC1 at pH 7 (black) and pH 5.5 (grey). Spectra were recorded at 20 °C (solid line) and at 4 °C (dotted line). There are no significant changes in the secondary and tertiary structures of CLIC1 over this temperature range.
CLIC1 Sequence and Pepsin Digestion
The CLIC1 sequence used in the H/D exchange study differs from that reported for the crystal structure, in that it has 2 additional residues at the N-terminus due to the thrombin cleave site, and two mutations, E63Q and G151E (Figure 3). The numbering of all peptides reported in this study excludes the additional two residues at the N-terminus and corresponds to that for the structure. Pepsin digestion of CLIC1 in 1 M GuHCL and 0.8 % (v/v) formic acid for a duration of 4 min over immobilized pepsin, yields peptides that cover the entire sequence of CLIC1 at both pH 7 and pH 5.5. Of these peptides, 75 were well resolved and common to CLIC1 at pH 7 and pH 5.5 covering 95% of the sequence (Figure 3). The only region not covered is residues 42-54. The number of deuterons incorporated was measured for all 75 peptides, including peptides with multiple charge states, and of these 21 peptides were chosen to generate the deuterium exchange data shown in the graphics. However, all 75 peptides were analysed to ensure that the exchange data agreed with those of the 21 selected peptides. Of the 21 peptides, 7 come from the thioredoxin-like domain, domain 1, and 13 come from the all-helical domain 2 (Figure 3).
Figure 3.
Sequence and peptic fragments of CLIC1. The sequence used in the DXMS study is shown above while that reported for the crystal structure of CLIC1 (PDB code 1k0m (11)) is shown below. Numbering is based on the structure's sequence. The asterisks indicate the differences between the sequences, as described under Results. The broken and dotted lines under the protein sequence indicate peptic fragments that are common to CLIC1 at pH 7 and pH 5.5. Peptides chosen for this study are shown by the broken lines. The only region not covered is residues 42-54. The regions corresponding to the two domains and the linker are indicated by the solid lines above the sequence.
Hydrogen-Deuterium Exchange at pH 7 and 5.5
As the rate of amide H/D exchange is highly pH dependent and H/D exchange is expected to be about 30-fold slower at pH 5.5 than at pH 7.0 (41), the on exchange time points for pH 5.5 are 30-fold longer (300, 3000, 9000, 30000 and 90000 sec) than the corresponding time points for pH 7 (10, 100, 300, 1000 and 3000 sec). Differences in deuteration of CLIC1-derived peptides at pH 5.5 and pH 7.0 and at the corresponding on-exchange time points were revealed by overlaying isotopic profiles of the individual peptides. For example, the changes in the isotopic profiles for peptides 11-31 and 68-82 indicate a more rapid shift toward a higher mass at pH 5.5 than at pH 7.0 (see Supporting Information Figure S1).
The extent of deuteration at pH 7 for 10 sec and at pH 5.5 for 300 sec are similar and do not differ by more than 10% (Figure 4), indicating comparable core structures of the protein under these conditions. Further, the intensity of deuterium incorporation of soluble CLIC1 correlates well with structural parameters calculated for the crystal structure (PDB code 1k0m (11)), i.e., secondary structure, hydrogen bonding and solvent accessible surface area of the backbone (Figure 4), in agreement with the above spectroscopic data. The high levels of rapidly exchanging protons for the N-terminus comprising strand β1 and for peptide 152-172 comprising helix α6 are explained by their highly dynamic behavior in CLIC1, as indicated by a lack of well-defined electron density in some crystal structures (11).
Figure 4.
Exchange of amide protons of peptides from CLIC1 at pH 7 and at pH 5.5 for 10 sec and 300 sec on-exchange times, respectively. The longer on-exchange time at pH 5.5 is to account for the 30-fold slower exchange rate at this pH (41). The percentages of deuterium incorporation are indicated by the colour bar. The single colour bars are for the levels of deuteration at both pH 7 and 5.5 except for segment 203-213 which has two colour bars to show the difference in % deuteration at pH 7 (light blue) and pH 5.5 (dark blue). The solvent accessible surface area (SASA) of the backbone in CLIC1 (PDB code 1k0m (11)) was determined by GetArea (http://www.scsb.utmb.edu/getarea) and is shown by the broken line plot. The positions of α-helical (α) and β-strand (β) regions are shown by solid horizontal bars, prolines are indicated by diamonds, and backbone hydrogen atoms involved in hydrogen bonding interactions are labeled with vertical lines. TMR indicates the sequence (residues 25-46) that corresponds to the transmembrane region.
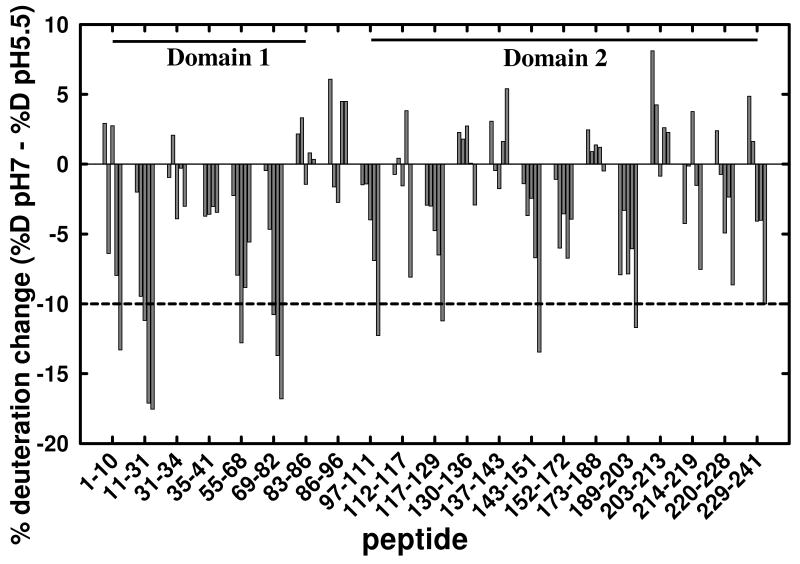
Figure 5 shows deuterium incorporation profiles of the 21 chosen peptides at the five on-exchange times for each pH condition. No common peptides were obtained for the region covered by residues 42-54. To determine which peptides display significant changes in deuteration, we calculated the differences between pH 7 and pH 5.5 at the corresponding time points (Figure 6). Of the 21 peptides, 13 do not show significant differences in their levels of deuterium incorporation (i.e., differences are < 10%) (Figure 6).
Figure 5.
Amide hydrogen-deuterium exchange analysis of the effect of pH on CLIC1. Each pH condition (7.0 and 5.5) is divided into rows of blocks (peptides) that, from top to bottom, correspond to five on-exchange time points from 10 s to 3000 s for pH 7, and from 300 s to 90000 s for pH 5.5, as indicated. The longer on-exchange times at pH 5.5 are to account for the 30-fold slower exchange rate at this pH (41). The deuteration levels for each block (peptide) at each time point are shown by different colours as indicated by the colour bar. The CLIC1 sequence and the α-helical (α) and β-strand (β) regions are shown above. TMR indicates the sequence (residues 25-46) that corresponds to the transmembrane region.
Figure 6.
Differences in the level of deuteration of CLIC1 peptides upon lowering the on-exchange pH from 7 to 5.5. The % deuteration change (%D pH 7 - %D pH 5.5) for each peptide (sequence indicated on x-axis) was calculated by subtracting the % of deuteration at pH 5.5 (%D pH 5.5) from the % of deuteration at pH 7 (%D pH7). Positive differences represent decreased levels of deuteration at pH 5.5, while negative differences represent increased deuteration at pH 5.5. Each peptide (sequence indicated on x-axis), is represented by the difference calculated between pH 7 and pH 5.5 for the five on-exchange time points (vertical bars). The time points for pH 7 were 10, 100, 300, 1000 and 3000 sec, whereas, given the 30-fold slower exchange rate at pH 5.5 (41), the corresponding time points at pH 5.5 were 300, 3000, 9000, 30000 and 90000 sec. Differences of greater than 10% (broken line) are considered to be significant (see Materials and Methods for further explanation). No data were obtained for residues 42-54 due to the absence of common peptides in this region. The peptides corresponding to the two domains are indicated by the two solid horizontal lines.
The 8 peptides that display exchange differences of >10% (see Figure 6) are mapped onto the structure of CLIC1 together with their H/D exchange kinetics, as shown in Figure 7A. The data indicate that the protection of amide protons in much of domain 1 is affected by lowering the pH from 7 to 5.5 resulting in enhanced levels of H/D exchange in segments comprising portions of β1 (peptide 1-10), β1α1 (peptide 11-31), α2β3 (peptide 55-68) and β4α3 (peptide 68-82). Helix α1 forms part of the transmembrane region (TMR, residues 25-46 (6,11,18-20)) of membrane-inserted CLIC1 (Figure 1). The biggest increases in deuterium incorporation at pH 5.5 occur in peptides 11-31 and 68-82. Unfortunately, there were no common peptides for residues 42-46 (strand β2) which also form part of the TMR (Figure 1). However, nearly all amide protons in a peptide 42-48 obtained only at pH 5.5 are rapidly exchanged within 300 secs, indicative of a highly flexible segment at least at this pH (Figure 7A). Four segments in domain 2 also display enhanced levels of H/D exchange: the N- and C-terminal regions of α4 (peptides 97-111 and 117-129, respectively), the N- and C-terminal regions of α5 (peptides 117-129 and 143-151, respectively) and the C-terminal region of α7 and the loop connecting α7 to α8 (Figure 7A).
Figure 7.
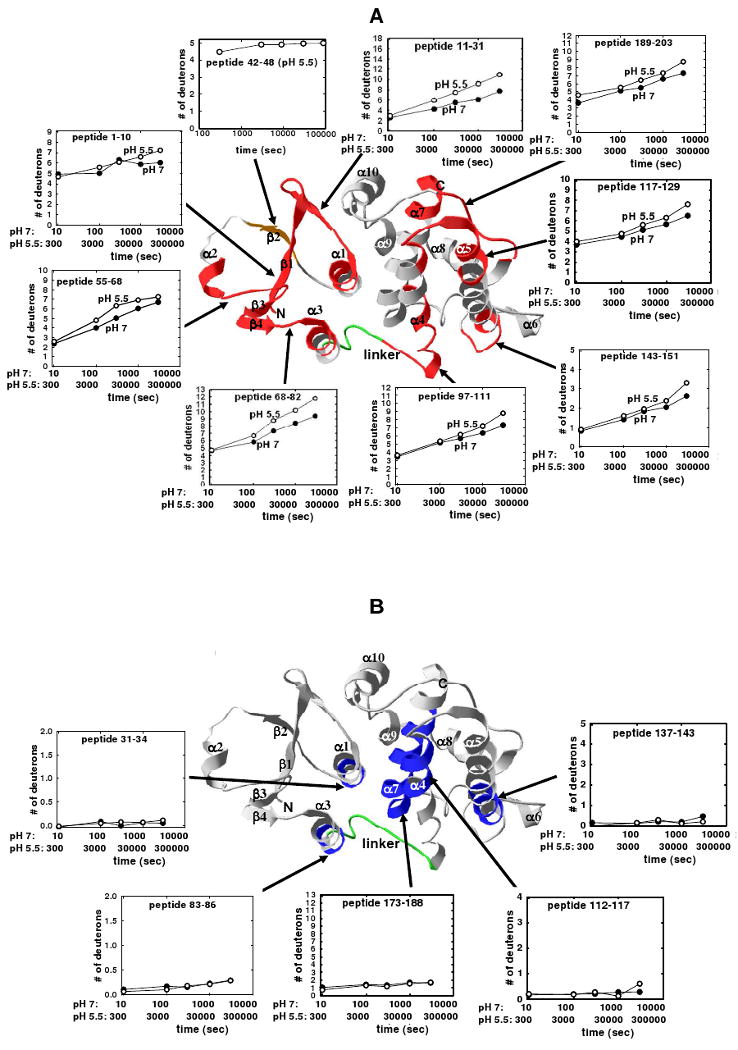
H/D exchange kinetics for CLIC1 segments mapped on the crystal structure of CLIC1 (PDB code 1k0m (11)). A. The red ribbons represent the eight regions for which the difference in deuteration at pH 7 and pH 5.5 at least at one time point was > 10% (see Figure 6), while the orange ribbon represents the exchange data for a peptide (residues 42-48), in the transmembrane region, that was only obtained at pH 5.5. B. The blue ribbons represent the five regions that display little or no deuterium incorporation at pH 7 and pH 5.5 (see Figure 5). α-Helices, β-strands, and the N- and C-termini are labeled. The linker region connecting the two domains is in green. The time axes for the H/D exchange kinetics at pH 7 and at pH 5.5 are shown and account for the 30-fold slower exchange rate at pH 5.5 (41). H/D exchange kinetics at pH 7 are in filled circles and the kinetics at pH 5.5 are in open circles.
Figure 7B shows the peptides mapped on the structure of CLIC1 that display very little H/D exchange at pH 7 and 5.5 throughout the experiment. They include the C-terminal regions of α1 and α3 (peptides 31-34 and 83-86, respectively), both in domain 1, and the central-to-C-terminal region of α4 (peptide 112-117), the central-to-C-terminal region of α5 (peptide 137-143) and most of α7 (peptide 173-188), in domain 2. The stable and highly protected segments 31-34, 112-117 and 173-188 in CLIC1 correspond to regions in the structurally related rGST M1-1 that display no increase in amide H/D exchange (44). The H/D exchange kinetics data for several peptides from CLIC1 are shown in Figure S2 of the Supporting Information.
Overall, the amide H/D exchange results for soluble CLIC1 show that the all-helical domain 2 is more stable and better protected than the thioredoxin-like domain 1 when the pH is lowered from 7 to 5.5. The proximity of the stable segments 112-117, 137-143 and 173-188 in a bundle formed by helices 4, 5 and 7 (Figure 7B), indicates an important structural motif for the stability of the core of domain 2. The stability of helix 7 (equivalent to helix 6 in GSTs) is consistent with its putative role in the nucleation of GST folding (45).
Discussion
Low pH conditions facilitate the appearance and activity of CLIC1 anion channels in membranes, although the mechanism involved remains to be determined (10,20,28). An acidic pH at the surface of a membrane (29,31) can function to prime or alter the structure of a soluble protein to achieve a lower activation energy barrier for its conversion to a membrane-insertion conformation (31,46). In contrast to other membrane-insertable proteins (23,31,47-51), CLIC1 does not undergo significant acid-induced structural changes in the absence of membranes (this study and ref 43). Recently, it has been shown that the changes needed for the soluble anti-apoptotic protein Bcl-xL to convert to its membrane conformation also requires both acidic conditions (pH 4.9) and the presence of membranes (52).
The H/D exchange data reported in this study indicate that the thioredoxin-like domain 1 of CLIC1 is less stable than domain 2, and that, while the structure of domain 1 remains intact, its stability is further reduced in an acidic environment (pH 5.5), a condition shown by others to facilitate the protein's insertion into membranes (28). The conformational stability of soluble CLIC1 has been shown to diminish substantially at pH 5.5 resulting in the formation of a partially unfolded intermediate not observed at pH 7 (43). Unfolding of the native protein to the non-molten globule-like intermediate involves helix α1 the structural element in domain 1 that makes the majority of contacts with domain 2. Substantial evidence exists indicating that the amino acid sequence corresponding to the amphipathic α1β2 motif in domain 1 forms the transmembrane region that spans membranes (6,11,18-20). For this motif to function as a transmembrane region, it will have to become detached from domain 2 then extend and refold into its membrane-insertable conformation. It is, therefore, possible that, as soluble CLIC1 encounters an acidic environment (e.g., pH 5.5) near the surface of a membrane, the enhanced flexibility and diminished stability of domain 1 allows it to loosen up and become primed for the structural changes needed for it to insert into membranes. While the conformation of the membrane-inserted transmembrane region is unknown, it is most likely to be helical. Helix α1 in soluble CLIC1 has a highly stable central region, as indicated by the absence of H/D exchange in peptide 31-34 at pH 7 and 5.5 (Figure 7B). When compared to most members of both the thioredoxin and GST superfamilies, the sequences of helix 1 in CLIC proteins display the highest propensities to form a helix according to the AGADIR helix-coil algorithm (53). Furthermore, helix 1 has an N-capping motif (Ser27) and a C-capping motif (Lys37), both of which can stabilise helical structures substantially (54). GST Kappa, a non-canonical GST, and calsequestrin, a member of the thioredoxin family, both of which are known to interact with membranes (55,56), also display high helix propensities for their corresponding helix 1 sequences. However, the sequence for helix 1 in class Omega GST, the closest structural homologue of CLIC1 (11), does not display as high a helical propensity. Since membrane spanning structures are typically helical, it is reasonable to assume that helix 1 is retained in membrane-inserted CLIC1. Although residues 42-46 in the trans-membrane region form strand β2 in the crystal structure, this highly dynamic segment at pH 5.5 (Figure 7A) also exhibits a propensity to form a helix with potential N- and C-capping motifs.
Since membrane insertion of CLIC1 is favoured at low pH (5 - 5.5), the involvement of protonation and electrostatic effects in bringing about structural changes in the protein are suggested. Within the pH range of 7 to 5.5, decreased stability of domain 1 might be associated with a decrease in the level of electrostatic attraction that occurs upon protonation of acidic residues. CLIC1 has three histidines, 12 aspartates and 21 glutamates. Inspection of the crystal structure (PDB code 1k0m), indicates that none of the histidine residues are directly involved in side chain-side chain interactions with other residues but are involved in water-mediated hydrogen bonding. At this stage it is not clear if and how protonation of these histidines might impact the conformation of CLIC1. It is interesting to note that a histidine residue (His74) is located in a segment (peptide 68-82) that displays enhanced H/D exchange when the pH is lowered from 7 to 5.5 (Figure 7A). Domain 1 has two salt bridges (Arg29-Glu81 and Lys37-Glu85), while one exists across the domain interface (Lys20-Asp225). Interestingly, these salt bridges involve residues located in the two segments (peptide 11-31 and peptide 68-82) that display significantly enhanced H/D-exchange rates at pH 5.5 (Figures 7A). Although the pKa values of the acidic partners are unknown, it is tempting to speculate that at a low pH they may become protonated and neutral (at least partly at pH 5.5). Since Arg29 and Lys37 are buried at the domain interface, their positively charged side chains would diminish the conformational stability of the respective regions making them more mobile, in accordance with the exchange data. Furthermore, disruption of the Arg29-Glu81 salt bridge between helices 1 and 3 could also destabilise the interdomain interaction at Met32. Once domain 1 is destabilised and primed within the acidic environment at the surface of a membrane, the presence of an acidic lipid bilayer may induce the domain to unfold, exposing the transmembrane region which is then driven to undergo conformational changes and insert into the membrane.
Supplementary Material
Abbreviations
- CLIC1
chloride intracellular channel protein 1
- DXMS
hydrogen-deuterium exchange mass spectrometry
- GuHCl
guanidine hydrochloride
- H/D
hydrogen/deuterium
- GST
glutathione transferase
- TMR
transmembrane region
Footnotes
This work was supported by the University of the Witwatersrand, the South African National Research Foundation (Grant 205359), the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation (Grant 64788), grants from the NIH Innovative Technologies for the Molecular Analysis of Cancer (IMAT) program (CA099835 and CA118595, VLW), NIH grant AI076961 (VLW), and a grant from the University of California Industry-University Cooperative Research Program (UC10591), BiogenIDEC corporate sponsor (VLW).
Supporting Information Available: Two figures as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ashley RH. Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins. Mol Membr Biol. 2003;20:1–11. doi: 10.1080/09687680210042746. [DOI] [PubMed] [Google Scholar]
- 2.Schlesinger PH, Blair HC, Teitelbaum SL, Edwards JC. Characterization of the osteoclast ruffled border chloride channel and its role in bone resorption. J Biol Chem. 1997;272:18636–18643. doi: 10.1074/jbc.272.30.18636. [DOI] [PubMed] [Google Scholar]
- 3.Ronnov-Jessen L, Villadsen R, Edwards JC, Petersen OW. Differential expression of a chloride intracellular channel gene, CLIC4, in transforming growth factor-beta1-mediated conversion of fibroblasts to myofibroblasts. Am J Pathol. 2002;161:471–480. doi: 10.1016/s0002-9440(10)64203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandes-Salas E, Suh KS, Speransky VV, Bowers WL, Levy JM, Adams T, et al. mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol Cell Biol. 2002;22:3610–3620. doi: 10.1128/MCB.22.11.3610-3620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suh KS, Mutoh M, Gerdes M, Crutchley JM, Mutoh T, Edwards LE, et al. Antisense suppression of the chloride intracellular channel family induces apoptosis, enhances tumor necrosis factor {alpha}-induced apoptosis, and inhibits tumor growth. Cancer Res. 2005;65:562–571. [PubMed] [Google Scholar]
- 6.Berry KL, Bülow HE, Hall DH, Hobert OA. A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science. 2003;302:2134–2137. doi: 10.1126/science.1087667. [DOI] [PubMed] [Google Scholar]
- 7.Novarino G, Fabrizi C, Tonini R, Denti MA, Malchiodi-Albedi F, Lauro GM, Sacchetti B, Paradisi S, Ferroni A, Curmi PM, Breit SN, Mazzanti M. Involvement of the intracellular ion channel CLIC1 in microglia-mediated beta-amyloid-induced neurotoxicity. J Neurosci. 2004;24:5322–5330. doi: 10.1523/JNEUROSCI.1170-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonini R, Ferroni A, Valenzuela SM, Warton K, Campbell TJ, Breit SN, Mazzanti M. Functional characterisation of the NCC27 nuclear protein in stable transfected CHO-K1 cells. FASEB J. 2000;14:1171–1178. doi: 10.1096/fasebj.14.9.1171. [DOI] [PubMed] [Google Scholar]
- 9.Tulk BM, Schlesinger PH, Kapadia SA, Edwards JC. CLIC-1 functions as a chloride channel when expressed and purified from bacteria. J Biol Chem. 2000;275:26986–26998. doi: 10.1074/jbc.M004301200. [DOI] [PubMed] [Google Scholar]
- 10.Tulk BM, Kapadia S, Edwards JC. CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am J Physiol Cell Physiol. 2002;282:C1103–C1112. doi: 10.1152/ajpcell.00402.2001. [DOI] [PubMed] [Google Scholar]
- 11.Harrop SJ, De Maere MZ, Fairlie WD, Rezstsova T, Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Jankova L, Warton K, Bauskin AR, Wu WM, Pankhurst S, Campbell TJ, Breit SN, Curmi PMG. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4Å resolution. J Biol Chem. 2001;276:44993–45000. doi: 10.1074/jbc.M107804200. [DOI] [PubMed] [Google Scholar]
- 12.Littler DR, Harrop SJ, Fairlie D, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, Mazzanti M, Breit SN, Curmi PMG. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004;279:9298–9305. doi: 10.1074/jbc.M308444200. [DOI] [PubMed] [Google Scholar]
- 13.Li YF, Li DF, Zeng ZH, Wang DC. Trimeric structure of the wild soluble chloride intracellular ion channel CLIC4 observed in crystals. Biochem Biophys Res Commun. 2006;343:1272–1278. doi: 10.1016/j.bbrc.2006.03.099. [DOI] [PubMed] [Google Scholar]
- 14.Littler DR, Harrop SJ, Brown LJ, Pankhurst GJ, Mynott AV, Luciani P, Mandyam RA, Mazzanti M, Tanda S, Berryman MA, Breit SN, Curmi PMG. Comparison of vertebrate and invertebrate CLIC proteins: The crystal structures of Caenorhabditis elegans EXC-4 and Drosophila melanogaster DmCLIC. Proteins. 2007;71:364–378. doi: 10.1002/prot.21704. [DOI] [PubMed] [Google Scholar]
- 15.Littler DR, Assaad NN, Harrop SJ, Brown LJ, Pankhurst GJ, Luciani P, Aguilar MI, Mazzanti M, Berryman MA, Breit SN, Curmi PMG. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J. 2005;272:4996–5007. doi: 10.1111/j.1742-4658.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- 16.Mi W, Liang YH, Li L, Su XD. The crystal structure of human chloride intracellular channel protein 2: A disulfide bond with functional implications. Proteins. 2008;71:509–513. doi: 10.1002/prot.21922. [DOI] [PubMed] [Google Scholar]
- 17.Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Littler DR, Brown LJ, Mazzanti M, Breit SN, Curmi PMG, Dulhunty AF, Board PG, Parker MW. Structure of the Janus Protein Human CLIC2. J Mol Biol. 2007;374:719–731. doi: 10.1016/j.jmb.2007.09.041. [DOI] [PubMed] [Google Scholar]
- 18.Duncan RR, Westwood PK, Boyd A, Ashley RH. Rat brain p64H1, expression of a new member of the p64 chloride channel protein family in endoplasmic reticulum. J Biol Chem. 1997;272:23880–23886. doi: 10.1074/jbc.272.38.23880. [DOI] [PubMed] [Google Scholar]
- 19.Singh H, Ashley RH. Redox regulation of CLIC1 by cysteine residues associated with the putative channel pore. Biophys J. 2006;90:1628–1638. doi: 10.1529/biophysj.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry KL, Hobert O. Mapping functional domains of chloride intracellular channel (CLIC) proteins in vivo. J Mol Biol. 2006;359:1316–1333. doi: 10.1016/j.jmb.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 21.von Heijne G, Blomberg C. Trans-membrane translocation of proteins. The direct transfer model. Eur J Biochem. 1979;97:175–181. doi: 10.1111/j.1432-1033.1979.tb13100.x. [DOI] [PubMed] [Google Scholar]
- 22.Epand RM. Lipid polymorphism and protein-lipid interactions. Biochim Biophys Acta. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 23.Chenal A, Savarin P, Nizard P, Guillain F, Gillet D, Forge V. Membrane protein insertion regulated by bringing electrostatic and hydrophobic interactions into play. A case study with the translocation domain of diphtheria toxin. J Biol Chem. 2002;277:43425–43432. doi: 10.1074/jbc.M204148200. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JE, Xie M, Singh LM, Edge R, Cornell RB. Both acidic and basic amino acids in an amphitropic enzyme, CTP:phosphocholine cytidyltransferase, dictate its selectivity for anionic membranes. J Biol Chem. 2003;278:514–522. doi: 10.1074/jbc.M206072200. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida Y, Kinuta M, Abe T, Liang S, Araki K, Cremona O. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J. 2004;23:3483–3491. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chenal A, Vernier G, Savarin P, Bushmarina NA, Gèze A, Guillain F, Gillet D, Forge V. Conformational states and thermodynamics of α-lactalbumin bound to membranes: a case study of the effects of pH, calcium, lipid membrane curvature and charge. J Mol Biol. 2005;349:890–905. doi: 10.1016/j.jmb.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 27.Goodchild SC, Howell MW, Cordina NM, Littler DR, Breit SN, Curmi PMG, Brown LJ. Oxidation promotes insertion of the CLIC1 chloride intracellular channel into the membrane. 2009 doi: 10.1007/s00249-009-0450-0. in press. [DOI] [PubMed] [Google Scholar]
- 28.Warton K, Tonini R, Fairlie WD, Mathews JM, Valenzuela SM, Qiu MR, Wu WM, Pankhurst S, Bauskin AR, Harrop SJ, Campbell TJ, Curmi PMG, Breit SN, Mazzanti M. Recombinant CLIC1 (NCC27) assembles in lipid bilayers via a pH-dependent two-state process to form chloride ion channels with identical characteristics to those observed in Chinese hamster ovary cells expressing CLIC1. J Biol Chem. 2002;277:26003–26011. doi: 10.1074/jbc.M203666200. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin S. The electrostatic properties of membranes. Annu Rev Biophys Biophys Chem. 1989;18:113–136. doi: 10.1146/annurev.bb.18.060189.000553. [DOI] [PubMed] [Google Scholar]
- 30.Menestrina G, Forti S, Gambale F. Interaction of tetanus toxin with lipid vesicles. Effects of pH, surface charge, and transmembrane potential on the kinetics of channel formation. Biophys J. 1989;55:393–405. doi: 10.1016/S0006-3495(89)82833-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Goot FG, González-Manãs JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 32.Parker MW, Feil SC. Pore-forming protein toxin: from structure to function. Prog Biophys Mol Biol. 2005;88:91–142. doi: 10.1016/j.pbiomolbio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 33.London E. Diphtheria toxin: membrane interaction and membrane translocation. Biochim Biophys Acta. 1992;1113:25–51. doi: 10.1016/0304-4157(92)90033-7. [DOI] [PubMed] [Google Scholar]
- 34.Lacy DB, Stevens RC. Unraveling the structures and modes of action of bacterial toxins. Curr Opin Struct Biol. 1998;8:778–784. doi: 10.1016/s0959-440x(98)80098-5. [DOI] [PubMed] [Google Scholar]
- 35.Zakharov SD, Cramer WA. Colicin crystal structures: pathways and mechanisms for colicin insertion into membranes. Biochim Biophys Acta. 2002;1565:333–346. doi: 10.1016/s0005-2736(02)00579-5. [DOI] [PubMed] [Google Scholar]
- 36.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. [Google Scholar]
- 37.Brudler R, Gessner CR, Li S, Tyndall S, Getzoff ED, Woods VL., Jr PAS domain allostery and light-induced conformational changes in photoactive yellow protein upon I2 intermediate formation, probed with enhanced hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2006;363:148–160. doi: 10.1016/j.jmb.2006.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL., Jr Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14:2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci U S A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valenzuela SM, Martin DK, Por SB, Robbins JM, Warton K, Bootcov MR, Schofield PR, Campbell TJ, Breit SN. Molecular cloning and expression of a chloride ion channel of cell nuclei. J Biol Chem. 1997;272:12575–12582. doi: 10.1074/jbc.272.19.12575. [DOI] [PubMed] [Google Scholar]
- 41.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins Struct Funct Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fanucchi S, Adamson RJ, Dirr HW. Formation of an unfolding intermediate state of soluble chloride intracellular channel protein CLIC1 at acidic pH. Biochemistry. 2008;47:11674–11681. doi: 10.1021/bi801147r. [DOI] [PubMed] [Google Scholar]
- 44.Thompson LC, Walters J, Burke J, Parsons JF, Armstrong RN, Dirr HW. Double mutation at the subunit interface of glutathione transferase rGSTM1-1 results in a stable, folded monomer. Biochemistry. 2006;45:2267–2273. doi: 10.1021/bi0519506. [DOI] [PubMed] [Google Scholar]
- 45.Stenberg G, Dragani B, Cocco R, Mannervik B, Aceto A. A conserved “hydrophobic staple motif” plays a crucial role in the refolding of human glutathione transferase P1-1. J Biol Chem. 2000;275:10421–10426. doi: 10.1074/jbc.275.14.10421. [DOI] [PubMed] [Google Scholar]
- 46.Manceva SD, Pusztai-Carey M, Butco P. Effect of pH and ionic strength on the cytolytic toxin Cyt1A: a fluorescence spectroscopy study. Biochim Biophys Acta. 2004;1699:123–130. doi: 10.1016/j.bbapap.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Parker MW, Tucker AD, Tsernoglou D, Pattus F. Insights into membrane insertion based on studies of colicins. Trends Biochem Sci. 1990;15:126–129. doi: 10.1016/0968-0004(90)90205-p. [DOI] [PubMed] [Google Scholar]
- 48.Blewitt MG, Chung LA, London E. Effect of pH on the conformation of diphtheria toxin and its implications for membrane penetration. Biochemistry. 1985;24:5458–5464. doi: 10.1021/bi00341a027. [DOI] [PubMed] [Google Scholar]
- 49.Bychkova VE, Dujsekina AE, Klenin SI, Tiktopulo EI, Uversky VN, Ptitsyn OB. Molten globule-like state of cytochrome c under conditions simulating those near the membrane surface. Biochemistry. 1996;35:6058–6063. doi: 10.1021/bi9522460. [DOI] [PubMed] [Google Scholar]
- 50.Song M, Shao H, Mujeeb A, James TL, Miller WL. Molten–globule structure and membrane binding of the N-terminal protease-resistant domain (63–193) of the steroidogenic acute regulatory protein (StAR) Biochem J. 2001;356:151–158. doi: 10.1042/0264-6021:3560151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nam GH, Choi KY. Association of human tumor necrosis factor-related apoptosis inducing ligand with membrane upon acidification. Eur J Biochem. 2002;269:5280–5287. doi: 10.1046/j.1432-1033.2002.03242.x. [DOI] [PubMed] [Google Scholar]
- 52.Thuduppathy GR, Terrones O, Craig JW, Basañez G, Hill RB. The N-terminal domain of Bcl-XL reversibly binds membranes in a pH-dependent manner. Biochemistry. 2006;45:14533–14542. doi: 10.1021/bi0616652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of a-helices: local motifs, long-range electrostatics, ionic strength dependence and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 54.Scholtz JM, Baldwin RL. Annu Rev Biophys Biomol Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 55.Ladner JE, Parsons JF, Rife CL, Gilliand GL, Armstrong RN. Parallel evolutionary pathways for glutathione transferases: structure and mechanism of the mitochondria class kappa enzyme rGST K1-1. Biochemistry. 2004;43:352–361. doi: 10.1021/bi035832z. [DOI] [PubMed] [Google Scholar]
- 56.Wang S, Trumble WR, Liao H, Wesson CR, Dunker AK, Kang C. Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nature Struct Biol. 1998;5:476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.