Abstract
Monoamine oxidase A (MAOA) is an enzyme expressed in the brain that metabolizes dopamine, norepinephrine, epinephrine, and serotonin. Abnormalities of serotonin neurotransmission have long been implicated in the psychopathology of autism. A polymorphism exists within the promoter region of the MAOA gene that influences MAOA expression levels so that “low activity” alleles are associated with increased neurotransmitter levels in the brain. Individuals with autism often exhibit elevated serotonin levels. Additional studies indicate that the “low activity” allele may be associated with lower IQ and more severe autistic symptoms. In this study we genotyped the MAOA promoter polymorphism in a group of 29 males (age 2–3 years) with autism and a group of 39 healthy pediatric controls for whom brain MRI data was available. We found a consistent association between the “low activity” allele and larger brain volumes for regions of the cortex in children with autism but not in controls. We did not find evidence for over-transmission of the “low activity” allele in a separate sample of 114 affected sib pairfamilies. Nor did we find any unknown SNPs in yet another sample of 96 probands. Future studies will determine if there is a more severe clinical phenotype associated with both the “low activity” genotype and the larger brain volumes in our sample.
Keywords: autism, MAOA, neuroimaging, genetics
INTRODUCTION
Autism is a behaviorally defined neurodevelopmental disorder with a strong heritable component. Individuals with autism spectrum disorders show marked impairments in development of language, reciprocal social interaction and communication, and excessively rigid and repetitive behaviors. Several lines of evidence from studies of genetics, neurochemistry, and brain morphology have recently converged to suggest that monoamine oxidase A (MAOA) may play a role in the development of autism. We, therefore, studied the effects of a functional MAOA promoter VNTR that potentially unites these findings on brain structure volumes in children with autism.
MAOA is a key regulator of the neurotransmitters serotonin, norepinephrine, and dopamine, abnormalities of all which have been linked to autism [Lam et al., 2006]. These neurotransmitters are metabolized by MAOA and their levels vary in response to levels of MAOA present in the brain [Jonsson et al., 2000; Williams et al., 2003; Zalsman et al., 2005; Ducci et al., 2006]. Serotonin influences multiple aspects of brain development including neurogenesis, cell proliferation, and differentiation [Schumann et al., 2004]. More recent PET studies have demonstrated abnormal serotonin synthesis in brains of individuals with autism [Lotspeich et al., 2004], and serotonin reuptake inhibitor medications (SRIs), which interfere with serotonin transporter (SLC6A4) activity, are effective intreating some aspects of autistic symptomatology. Dopamine has been shown to be involved in a broad range of behaviors from cognitive processing to impulsivity [Claveria et al., 1975; Costa, 1977; Roberts and Sharif, 1978]. With the success of antipsychotics in treating symptoms associated with autism, the role of dopamine in the neurobiology of autism is therefore being more intensively investigated [Anderson and Hoshiono, 1997]. Levels of the catecholamine norepinephrine have also repeatedly been shown to be elevated in children with autism [Lake et al., 1977; Launay et al., 1987; Cook et al., 1990; Leventhal et al., 1990; Leboyer et al., 1992] and finally, deletions and nonsense mutations in MAOA are known to result in Brunner syndrome, which is characterized by low IQ, autistic like symptoms and aggression [Brunner et al., 1993; Brunner, 1996].
Regarding brain morphology, variation in an MAOA promoter VNTR that affects transcriptional efficiency has recently been shown to influence volumes of numerous structures, including the cerebral cortex, in healthy controls [Meyer-Lindenberg and Weinberger, 2006]. A transgenic mouse line in which MAOA is disrupted shows multiple structural brain abnormalities [Cases et al., 1995]. Autism is characterized by consistently replicated morphological abnormalities, including heavier post mortem brain weight, an increased frequency of macrocephaly, and increased total brain volume as determined by MRI. We recently found, in a sample of young autistic children (2 years old), specific enlargement of cerebral cortical gray and white matter [Piven et al., 1992], which is consistent with previous reports of this age group [Lainhart et al., 1997, 2002; Dementieva et al., 2005]. Finally, the MAOA gene is also located in a 20 centiMorgan (cM) region on the X chromosome that has shown modest linkage to autism spectrum disorders (Wassink, unpublished work). Moreover, due to the skewed sex ratio in autism (4 males: 1 female) MAOA has been considered a candidate gene for autism susceptibility based not only on its function, but also on its location on the X chromosome.
The promoter VNTR is 1.2 Kb upstream of exon 1, with a high activity allele (MAOA-H) that is present in 3.5 or 4 repeats and a low activity allele (MAOA-L) present in 3 or 5 repeats [Sabol et al., 1998]. The variant appears to be linked to IQ and symptom severity among autistic children [Yirmiya et al., 2002; Cohen et al., 2003] and has also been associated with panic disorder, anti-social behavior, depressive disorder and responsiveness to selective serotonin reuptake inhibitors (SSRIs) used to treat depression [Deckert et al., 1999; Samochowiec et al., 1999; Yu et al., 2005]. Based on these converging data, we therefore evaluated whether the MAOA promoter VNTR influences autism susceptibility. In this pursuit, rather than being limited to one domain of analysis, we used a comprehensive approach that included: (1) Testing the polymorphism for association with autism, (2) Screening MAOA for rare and potentially deleterious coding sequence variation, and perhaps most novel, (3) Testing for effects of the polymorphism on brain structure volumes in young children with autism.
METHODS
Samples
For this study, we analyzed data and DNA from four independently ascertained samples: University of North Carolina (UNC) Autism MRI sample, UNC single strand conformation polymorphism (SSCP) screening sample, Collaborative Linkage Study of Autism (CLSA) affected sibling pair sample, and an Iowa Pediatric Normal Control (IPNC) sample.
| Sample | Number of subjects |
Analysis |
|---|---|---|
| UNC autism MRI | 29 | ANCOVA |
| UNC SSCP screening | 94 | SSCP/sequencing |
| CLSA | 513 | FBAT association |
| IPNC | 39 | ANCOVA |
The CLSA sample is comprised of 114 autism affected sibling pair (ASP) families. These families were recruited and assessed through the University of Iowa (UIA) and through the Neurodevelopmental Disorders Research Center (NDRC), University of North Carolina (UNC), Chapel Hill. All affected individuals were at least 3 years old at the time of ascertainment, and diagnosis was confirmed with the Autism Diagnostic Observation Schedule [Lord et al., 1989, 2000] and the Autism Diagnostic Interview—Revised [Le Couteur et al., 1989]. For inclusion, the family was required to have at least two children diagnosed with autism. Affected individuals were excluded if they had fragile X syndrome (based on fragile X DNA testing) or any neurological or medical condition suspected of being associated with autism such as tuberous sclerosis, neurofibromatosis, etc. This sample was used to test for association of the MAOA promoter polymorphism with autism.
The UNC screening sample has also been recruited from the NDRC. Individuals in this sample are from singleton families and were required to have a diagnosis of any Pervasive Developmental Disorder, except for Childhood Disintegrative Disorder, based on a multi-disciplinary clinical evaluation. This sample reflects the high proportion of mental retardation and cognitive impairment in the larger ASD population. Roughly 3/4 of the sample has mental retardation (MR) and ASD, while 1/4 has an ASD but no MR. This sample provided DNA for 94 affected individuals that was used exclusively for mutation screening.
The UNC autism MRI sample is comprised of 51 children with autism between the ages of 18 and 35 months at the time of scan, who are participating in a longitudinal MRI study of the brain (described in detail elsewhere [Piven et al., 1992]). Inclusion and exclusion criteria are as described for the CLSA sample, except that diagnosis was performed clinically at the UNC TEACCH center and verified at repeated intervals over the last 2 years. DNA was available for 29 boys and 5 girls from this sample, but given well established gender differences in brain development and morphology as well as the fact that MAOA is an X chromosome gene, only data from boys were analyzed. All UNC MRI children were Caucasian, and average age at the time of the scan was 2.71 ± 0.30 years.
Lastly, we acquired DNA from a sample of typically developing children (Iowa Pediatric Normal Control [IPNC]) who had undergone an MRI scan at the University of Iowa Hospital and Clinics as part of their involvement in another study. Exclusion criteria for this group included presence of braces, major medical, neurologic, or psychiatric illness, or history of learning disability (information obtained from parents during screening process). Average full scale IQ for this group is 112 (±18). Only genotypes from boys in the IPNC sample were analyzed, resulting in a cohort of 39 Caucasian males with an average age at the time of scan of 12.5 ± 2.21 years. This sample, while still considered pediatric, ranged in age from 7 to 18 years old.
Study approval was acquired from the UNC, Duke, and UIA Institutional Review Boards, and parents or guardians provided written, informed consent for their children after the study had been fully explained to them.
Image Acquisition and Processing
For the UNC MRI sample, imaging was performed at the Duke-UNC Brain Imaging and Analysis Center on a 1.5 Tesla GE Signa MRI scanner. Image acquisition was designed to maximize gray/white tissue contrast for the 18–24 months old child and included: (1) a coronal T1 IR Prepared: T1 300 msec, TR12 msec, TE 5 msec, 20° flip angle, at 1.5 mm thickness with 1 NEX, 20 cm FOV and a 256 × 192 matrix; and (2) a coronal PD/T2 2D dual FSE, TR 7,200 msec, TE 17/75 msec, at 3.0 mm thickness with 1 NEX, 20 cm FOV, and 256 × 160 matrix. Initial image processing to register and align the T1 andPD/T2 scans into a standardized plane was conducted with BRAINS2 developed at the University of Iowa [Andreasen et al., 1992; Magnotta et al., 2002]. Images were processed for tissue segmentation using an adaptation by our lab of the Expectation Maximization Segmentation (EMS) software originally developed at the Catholic University of Leuven [Van Leemput et al., 1999a,b]. We also developed and used a probabilistic atlas for tissue segmentation of the 2-year-old brain. The automated tissue segmentation protocol has been previously described in detail [Hazlett et al., 2005] and was used to generate gray and white matter volumes for the entire cerebral cortex and the frontal, temporal, and combined parietal-occipital lobes.
Images for the IPNC sample were obtained on a 1.5 Tesla GE Signa MR scanner. Three different sequences were acquired for each subject: T1, T2, and Proton Density. Processing of the images after acquisition was done using a locally developed family of software programs called BRAINS (acronym for Brain Research: Analysis of Images, Networks, and Systems). Details of the image analysis are published elsewhere [Andreasen et al., 1992, 1993, 1994; Cohen et al., 1992; Magnotta et al., 2002]. Briefly, The T2 and proton density images were aligned to the spatially normalized T1 image using an automated image coregistration program. A Talairach-based atlas coordinate system was overlaid onto each individual brain, aligning with anatomical landmarks of that brain without normalization to a standardized brain size [Talairach and Tournoux, 1988]. These coordinates were then used to generate automated measurements of frontal, temporal, parietal, and occipital lobes, cerebellum, and subcortical regions. This method permits morphological measurements to be made in non-normalized or “raw” space.
Genotyping
PCR amplification of the MAOA promoter polymorphism was performed according to a previously described protocol [Ducci et al., 2006]. PCR products were electrophoresed on 6% polyacrylamide gels that were stained with silver and read by two independent raters with discrepancies resolved by re-genotyping.
Mutation Screening
A panel of 96 probands was screened for MAOA mutations using single strand conformation polymorphism (SSCP) screening [Sheffield et al., 1993]. Our screening approach was bioinformatically directed. We utilized TrAPSS, a program developed at the University of Iowa, to first screen MAOA in silico to identify regions of biological interest, such as exons containing secondary structure. We then prioritized and screened these regions in order of biologic importance [Braun et al., 2006]. MAOA is a 90 Kb gene with approximately 4 Kb of coding sequence spanning15 exons. We screened all exons and splice site junctions using amplicons lessthan 250 bp in length. PCR products were electrophoresed on 6% non-denaturing polyacrylamide gels at 20 W for approximately 3 hr at room temperature while being cooled by a fan. The gels were then treated with silver nitrate to visualize the amplified DNA fragments. Any amplicons showing SSCP shifts were then forward and reverse sequenced to determine if a base pair change had occurred. The sequence data were analyzed using the Sequencher gene analysis computer program (Gene Codes, Ann Arbor, MI).
Association in Affected Sib Pair Families
We also genotyped the polymorphism and performed a test of association to determine if any of the alleles were associated with the autism diagnosis in our sample of CLSA affected sib pair (ASP) families. Association analysis was conducted using the commercially available FBAT software package (Golden Helix, Bozeman, MT). The test statistic isa chi-square test (Z2–χ2) that incorporates user-defined covariates and parameters.
Analysis of Genotype Effects on Brain Structure Volumes
Analysis of covariance was used to test for relationships between genotypes and brain structure volumes. Structure volumes were the dependent measures, genotype was the independent measure, and covariates included age at the time of scan acquisition and head circumference. We also tested all interaction terms, which were kept in the model only if significant. The proper genotype grouping for the MAOA promoter VNTR is based on functional expression data which shows that 3 and 5 repeat variants show low enzymatic activity (MAOA-L) while 3.5 and 4 repeats show high activity (MAOA-H) [Sabol et al., 1998]. As MAOA is an X chromosome gene and only males were analyzed, all genotypes were hemizygous.
For any test with a significant F value for genotype we also calculated omega-squared (ω2), an effect size measure which estimates the proportion of variance in a dependent measure accounted for by an independent categorical variable in the population from which the sample was drawn [Kirk, 1982]. Thus we used ω2 to measure the amount of variance in brain morphology accounted for by MAOA genotype in young autistic children. Omega-squared is given by the equation:
where, for our models, SSeffect is the Type III sums of squares for genotype, dfeffect is the number of degrees of freedom for genotype (2 for an additive model, 1 for a dominant model), MSerror is the mean square error for the entire model, and SStotal is the corrected total sums of squares for the entire model.
RESULTS
Association Testing and Mutation Screening
CLSA genotype data was analyzed using PEDCHECK and MERLIN to identify Mendelian inconsistencies and genotyping errors. The test statistic used was a standard univariate FBAT (Golden Helix). The standard null hypothesis was used which stated that no linkage or association was present at this locus. Additionally we conducted the FBAT test under all possible genetic models (additive, dominant, and recessive). Using the FBAT ALL analysis, we found no evidence to support unbalanced transmission of alleles within the CLSA sample (MAOA-L, P = 0.56; MAOA-H P = 0.44).
Similarly, our coding sequence screen did not identify any variants that were not already described in publicly available SNP databases; specifically, we identified no non-sense or missense variants.
Genotype-Brain Structure Volume Analyses
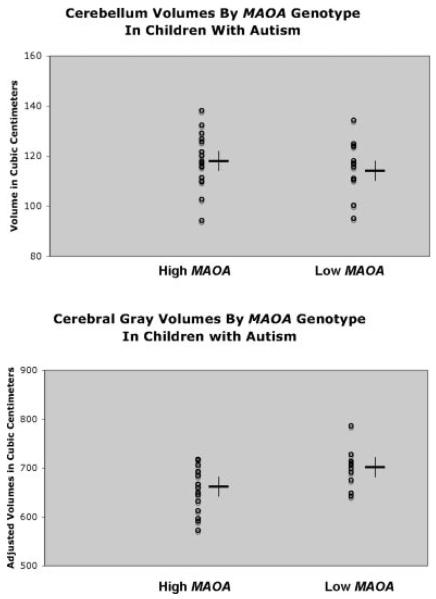
Allele frequencies for the UNC MRI and IPNC samples are shown in Table I. The ANCOVA F test for the 2-genotype MAOA-L/H effect was significant for total cerebral cortical volume (Table II). This led us to perform separate genotype tests for cortical GM and WM, both of which were significant (F = 5.30, P = 0.01 and F = 4.80, P = 0.02, respectively). We then parsed cortical gray and white matter into lobe-based subregions and found significant genotype effects for frontal GM and WM volumes. Volume-genotype relationships in the temporal and parieto-occipital regions showed a similar pattern of relationships, though they did not achieve significance. Genotype omega-squared (ω2) values for structure volumes influenced by genotype ranged from 4% to 9%. We then graphed the adjusted means of the cerebral cortical gray and white matter for each genotype group. Examination of the adjusted means for these structures revealed that the H allele was associated with smaller structure volumes and the L allele with larger volumes (Fig. 1). Genotype did not significantly influence cerebellar GM or WM volumes (Table II) in our sample of children with autism. We also did not find any significant relationships between MAOA-L/H and brain size in typically developing children (Table III). Due to the discrepancy in ages between the two MRI samples, they should not be directly compared. However, the IPNC sample is representative of a healthy pediatric population and the fact remains that the MAOA VNTR alleles produce different effects on brain size in the autism MRI sample and no significant effect on brain size in the IPNC sample.
TABLE I.
MAOA Promoter VNTR Allele Frequencies for MRI Samples
| High activity allele |
Low activity allele |
|||
|---|---|---|---|---|
| Sample | 3.5 repeats |
4 repeats | 3 repeats | 5 repeats |
| UNC-MRI | 0 | 17 | 12 | 0 |
| IPNC | 0 | 28 | 11 | 0 |
TABLE II.
Analysis of Covariance Results for Brain Structure Volumes by Genotype in Children With Autism
| LSM volumes by genotype high activity vs. low activity |
F tests for high vs. low | ||||
|---|---|---|---|---|---|
| (N = 17) | (N = 12) | F | P | ω | |
| ROIa | |||||
| Total brain (standard error) | 1,237.50 (11.69) | 1,314.70 (13.99) | 4.96 | 0.03 | 0.035 |
| Cerebral gray (standard error) | 664.40 (7.03) | 690.10 (8.41) | 5.30 | 0.01 | 0.043 |
| Cerebral white (standard error) | 235.78 (3.56) | 266.18 (4.26) | 4.80 | 0.02 | 0.059 |
| Frontal gray (standard error) | 251.88 (3.05) | 261.67 (3.65) | 4.10 | 0.05 | 0.038 |
| Frontal white (standard error) | 107.23 (1.59) | 114.05 (1.91) | 4.80 | 0.01 | 0.094 |
| Temporal (standard error) | 200.56 (3.22) | 209.96 (3.85) | 3.40 | 0.07 | — |
| Parietal-occipital (standard error) | 358.53 (3.94) | 370.58 (4.71) | 3.73 | 0.06 | — |
| Cerebellum (standard error) | 119.00 (2.47) | 114.60 (2.96) | 1.25 | 0.27 | — |
| Covariatesb | |||||
| Age (years) | 2.67 | 2.76 | 0.58 | 0.44 | — |
| Head circumference (cm) | 50.9 | 51.5 | 1.54 | 0.22 | — |
Region of interest (ROI) mean volumes are in cubic centimeters and are adjusted for the covariates at the time of MRI and head circumference.
Covariates presents raw, unadjusted mean values for the covariates, with the F values from ANOVAs that tested for differences between the covariate means across the genotype groups.
Fig. 1.
Least squares means for each genotype group was calculated using the regional volumes and covariates. Frontal lobe gray and white matter volumes differ significantly between genotype groups in our sample of children with autism. Cerebellum volumes do not differ by MAOA genotype in our sample.
TABLE III.
Analysis of Covariance Results for Brain Structure Volumes by Genotype in a Sample of Healthy Pediatric Controls
| LSM volumes by genotype high activity vs. low activity |
F tests for high vs. low | ||||
|---|---|---|---|---|---|
| (N = 28) | (N = 11) | F | P | ω | |
| ROIa | |||||
| Total brain (standard error) | 1,425.00 (7.17) | 1,357.20 (11.7) | 0.16 | 0.68 | — |
| Cerebral gray (standard error) | 814.68 (4.37) | 798.96 (7.18) | 0.97 | 0.33 | — |
| Cerebral white (standard error) | 422.10 (5.65) | 379.18 (9.29) | 0.04 | 0.83 | — |
| Frontal gray (standard error) | 318.40 (2.42) | 308.30 (3.99) | 1.13 | 0.29 | — |
| Frontal white (standard error) | 165.20 (2.77) | 145.20 (4.56) | 0.01 | 0.92 | — |
| Temporal (standard error) | 245.4 (2.72) | 229.8 (4.47) | 1.44 | 0.23 | — |
| Parietal (standard error) | 274.79 (1.72) | 272.27 (2.83) | 0.93 | 0.34 | — |
| Occipital (standard error) | 133.7 (2.04) | 129.3 (3.36) | 0.02 | 0.86 | — |
| Cerebellum (standard error) | 144.00 (1.68) | 136.9 (2.77) | 1.45 | 0.23 | — |
Region of interest (ROI) mean volumes are in cubic centimeters and are adjusted for the covariates at the time of MRI and head circumference.
DISCUSSION
We found that the low activity MAOA promoter polymorphism allele was associated with increased cerebral cortical volumes in a sample of young male children with autism (Fig. 1). When tissue type and the cerebral lobes were examined separately, the effects were similarly significant for total and frontal gray and white matter volumes, with allelic effect sizes ranging from 3% to 9%. The effects were in the same direction, though not significant, for temporal and parietal-occipital volumes, and no effect was seen in the cerebellum (Fig. 1). Thus the MAOA promoter polymorphism appears to exert a generalized effect on cerebral cortical volumes in young male children with autism. In contrast to the brain structure data, we did not find association of the MAOA promoter polymorphism with autism, nor did we detect any rare disease-causing variants in our mutation screen. These results are consistent with previous studies in which the low activity allele has repeatedly been associated with more severe autism symptomatology while the variant has generally not found to be associated with autism itself [Yirmiya et al., 2002; Cohen et al., 2003].
The MAOA promoter VNTR has been studied in numerous psychiatric phenotypes, including antisocial behavior [Samochowiec et al., 1999; Sjoberg et al., 2007], aggression [Brunner et al., 1993; Manuck et al., 2000; Newman et al., 2005; Meyer-Lindenberg and Weinberger, 2006], attention deficit hyperactivity disorder [Lawson et al., 2003; Li et al., 2007], bipolar disorder [Preisig et al., 2000; Gutierrez et al., 2004], anorexia nervosa [Urwin et al., 2003; Urwin and Nunn, 2005], autism [Yirmiya et al., 2002; Cohen, 2004], and depression [Yu et al., 2005]. MAOA has most consistently been shown to play a role in aggression, as in Brunner Syndrome, a disorder caused by a nonsense mutation of MAOA and characterized by mental retardation, aggression, autistic-like symptoms, and poor impulse control [Brunner et al., 1993; Brunner, 1996; Lenders et al., 1996]. Mouse knockout models of MAOA show a dramatic increase in aggressive activity and elevated levels of cortical serotonin and dopamine [Cases et al., 1995]. More recent studies have shown that not only deletion of the MAOA gene, but also low activity MAOA may confer susceptibility to impulsivity and aggression, particularly in the context of environmental factors such as previous child abuse [Caspi et al., 2002; Foley et al., 2004; Kim-Cohen et al., 2006; Nilsson et al., 2006;Widom and Brzustowicz, 2006]. Meyer-Lindenberg et al. (2006) found that psychiatrically normal males with the low activity MAOA promoter VNTR allele had volume reductions in the amygdala, poorer impulse control, and increased reactivity to negatively charged emotional stimuli as compared to males with the high activity variant.
Similar effects of MAOA have also been found in autism, with two separate studies finding the low activity allele to be associated with lower IQ and more severe symptoms [Yirmiya et al., 2002; Cohen et al., 2003]. The findings from our study complement these others nicely, as we show that the low activity MAOA allele is associated with increased white and gray matter volumes of the cerebral cortex, but not the cerebellum, in young males with autism. Early cortical enlargement is considered a hallmark of autism pathology. Individuals with autism, when compared to controls, have an increased frequency of macrocephaly and heavier post mortem brain weight and, from an early age, increased MRI volumes of a number of brain structures. Our University of North Carolina (UNC) autism research group recently found generalized enlargement of cerebral cortical gray and white matter, but not of the cerebellum, in the same group of subjects analyzed in this report. Thus our data extends information suggesting a pathological role for the low activity MAOA VNTR allele in autism and other behavioral disorders. This assertion is further supported by the lack of association between the MAOA VNTR and brain structure volumes in the Iowa pediatric normal control sample.
The influence of MAOA on brain structure is most likely mediated through its effects on serotonin and dopamine [Caspi et al., 2002; Yirmiya et al., 2002; Cohen, 2004; Foley et al., 2004; Kim-Cohen et al., 2006; Nilsson et al., 2006; Widom and Brzustowicz, 2006]. It is possible, for example, that other factors in the genetic background of an individual with autism, such as variants in serotonin or dopamine transporters, act in concert and facilitate the effect of the MAOA gene on brain morphology. MAOA functions to maintain low levels of monoamine neurotransmitters in CNS neurons, but the low activity promoter polymorphism allele has been associated with higher levels of serotonin and dopamine metabolites [Zalsman et al., 2005; Ducci et al., 2006], and both serotonin and dopamine as well as their precursors are known to affect brain morphology [Sodhi and Sanders-Bush, 2004; Prakash and Wurst, 2006a,b]. Also of interest, MAOA is expressed early in development, during the presumed period of risk for autism [Vitalis et al., 2002]. This early MAOA expression is strongest in the cortex, thalamus, and hypothalamus and weakest in the cerebellum and brainstem, where MAOB expression is strongest [Jahng et al., 1997]. This expression pattern mirrors nicely our volume findings in the autism sample.
These assertions are made, however, in the context of certain limitations. First, our sample, though large for this type of study in autism, is nonetheless by objective standards small, and we cannot rule out the possibility of false positive findings. Second, the MAOA promoter polymorphism is likely not the only MAOA variant that has an effect on brain structure volumes. We have not yet exhaustively genotyped all MAOA variants, but as we do, we may be able to delineate genotype-phenotype relationships with more precision than our current data permits. Lastly, because we analyzed only male children, we cannot determine whether the observed relationships are specific to males or whether they would also apply to females. Taken together with existing data on the MAOA promoter polymorphism, our results, which suggest a contribution of MAOA to the brain enlargement seen in young autistic children, provide further evidence for the involvement of the low activity allele in impairments of social cognition. The small effect size of the MAOA VNTR is most likely due to the underlying genetic and phenotypic complexity of brain development in the context of pathology. Further studies on the function of MAOA in brain development and behavior will clarify the full role of MAOA in autism spectrum disorders.
ACKNOWLEDGMENTS
This research was supported by NIH grants MH61696 (JP), HD03110 (JP), and MH066418 (JP and THW).
Grant sponsor: NIH; Grant numbers: MH61696, HD03110, MH066418.
REFERENCES
- Anderson GM, Hoshiono Y. Neurochemical studies of autism. In: Cohen DJ, Volkmar FR, editors. Handbook of autism and pervasive developmental disorders. Wiley; New York: 1997. pp. 325–343. [Google Scholar]
- Andreasen NC, Cohen G, Harris G, Cizadlo T, Parkkinen J, Rezai K, et al. Image processing for the study of brain structure and function: Problems and programs. J Neuropsychiatry Clin Neurosci. 1992;4:125–133. doi: 10.1176/jnp.4.2.125. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Cizadlo T, Harris G, Swayze V, 2nd, O'Leary DS, Cohen G, et al. Voxel processing techniques for the antemortem study of neuroanatomy and neuropathology using magnetic resonance imaging. J Neuropsychiatry Clin Neurosci. 1993;5:121–130. doi: 10.1176/jnp.5.2.121. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Harris G, Cizadlo T, Arndt S, O'Leary DS, Swayze V, et al. Techniques for measuring sulcal/gyral patterns in the brain as visualized through magnetic resonance scanning: BRAINPLOT and BRAINMAP. Proc Natl Acad Sci USA. 1994;91:93–97. doi: 10.1073/pnas.91.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun TA, Shankar SP, Davis S, O'Leary B, Scheetz TE, Clark AF, et al. Prioritizing regions of candidate genes for efficient mutation screening. Hum Mutat. 2006;27:195–200. doi: 10.1002/humu.20247. [DOI] [PubMed] [Google Scholar]
- Brunner HG. MAOA deficiency and abnormalbehaviour: Perspectives on an association. Ciba Found Symp. 1996;194:155–164. doi: 10.1002/9780470514825.ch9. discussion 164–167. [DOI] [PubMed] [Google Scholar]
- Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262:578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science. 1995;268:1763–1766. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Claveria LE, Teychenne PF, Calne DB, Petrie A, Bassendine MF. Dopaminergic agonists in Parkinsonism. Adv Neurol. 1975;9:383–392. [PubMed] [Google Scholar]
- Cohen G, Andreasen NC, Alliger R, Arndt S, Kuan J, Yuh WT, et al. Segmentation techniques for the classification of brain tissue using magnetic resonance imaging. Psychiatry Res. 1992;45:33–51. doi: 10.1016/0925-4927(92)90012-s. [DOI] [PubMed] [Google Scholar]
- Cohen IL, Liu X, Schutz C, White BN, Jenkins EC, Brown WT, et al. Association of autism severity with a monoamine oxidase A functional polymorphism. Clin Genet. 2003;64:190–197. doi: 10.1034/j.1399-0004.2003.00115.x. [DOI] [PubMed] [Google Scholar]
- Cohen BI. Rationale for further investigation of chromosome 16p13.3, a region implicated for autism. Autism. 2004;8:445–447. doi: 10.1177/1362361304047226. [DOI] [PubMed] [Google Scholar]
- Cook EH, Jr, Leventhal BL, Heller W, Metz J, Wainwright M, Freedman DX. Autistic children and their first-degree relatives: Relationships between serotonin and norepinephrine levels and intelligence. J Neuropsychiatry Clin Neurosci. 1990;2:268–274. doi: 10.1176/jnp.2.3.268. [DOI] [PubMed] [Google Scholar]
- Costa E. Nonstriatal dopaminergic neurons: Section X. Morphine, amphetamine, and noncataleptogenic neuroleptics: Introduction: Morphine, amphetamine, and noncataleptogenic neuroleptics. Adv Biochem Psychopharmacol. 1977;16:557–563. [PubMed] [Google Scholar]
- Deckert J, Catalano M, Syagailo YV, Bosi M, Okladnova O, Di Bella D, et al. Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet. 1999;8:621–624. doi: 10.1093/hmg/8.4.621. [DOI] [PubMed] [Google Scholar]
- Dementieva YA, Vance DD, Donnelly SL, Elston LA, Wolpert CM, Ravan SA, et al. Accelerated head growth in early development of individuals with autism. Pediatr Neurol. 2005;32:102–108. doi: 10.1016/j.pediatrneurol.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Ducci F, Newman TK, Funt S, Brown GL, Virkkunen M, Goldman D, et al. A functional polymorphism in the MAOA gene promoter (MAOA-LPR) predicts central dopamine function and body mass index. Mol Psychiatry. 2006;11:858–866. doi: 10.1038/sj.mp.4001856. [DOI] [PubMed] [Google Scholar]
- Foley DL, Eaves LJ, Wormley B, Silberg JL, Maes HH, Kuhn J, et al. Childhood adversity, monoamine oxidase a genotype, and risk for conduct disorder. Arch Gen Psychiatry. 2004;61:738–744. doi: 10.1001/archpsyc.61.7.738. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Arias B, Gasto C, Catalan R, Papiol S, Pintor L, et al. Association analysis between a functional polymorphism in the monoamine oxidase A gene promoter and severe mood disorders. Psychiatr Genet. 2004;14:203–208. doi: 10.1097/00041444-200412000-00007. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe M, Gerig G, Smith RG, Provenzale J, Ross A, et al. Magnetic resonance imaging and head circumference studyof brain size in autism: Birth through age 2 years. Arch Gen Psychiatry. 2005;62:1366–1376. doi: 10.1001/archpsyc.62.12.1366. [DOI] [PubMed] [Google Scholar]
- Jahng JW, Houpt TA, Wessel TC, Chen K, Shih JC, Joh TH. Localization of monoamine oxidase A and B mRNA in the rat brain by in situ hybridization. Synapse. 1997;25:30–36. doi: 10.1002/(SICI)1098-2396(199701)25:1<30::AID-SYN4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Norton N, Gustavsson JP, Oreland L, Owen MJ, Sedvall GC. A promoter polymorphism in the monoamine oxidase A gene and its relationships to monoamine metabolite concentrations in CSF of healthy volunteers. J Psychiatr Res. 2000;34:239–244. doi: 10.1016/s0022-3956(00)00013-3. [DOI] [PubMed] [Google Scholar]
- Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, Craig IW, et al. MAOA, maltreatment, and gene-environment interaction predicting children's mental health: New evidence and a meta-analysis. Mol Psychiatry. 2006;11:903–913. doi: 10.1038/sj.mp.4001851. [DOI] [PubMed] [Google Scholar]
- Kirk R. Experimental design: Procedures for the behavioral sciences. 2nd edition Brooks/Cole; Belmont, CA: 1982. [Google Scholar]
- Lainhart JE, Piven J, Wzorek M, Landa R, Santangelo SL, Coon H, et al. Macrocephaly in children and adults with autism. J Am Acad Child Adolesc Psychiatry. 1997;36:282–290. doi: 10.1097/00004583-199702000-00019. [DOI] [PubMed] [Google Scholar]
- Lainhart JE, Ozonoff S, Coon H, Krasny L, Dinh E, Nice J, et al. Autism, regression, and the broader autism phenotype. Am J Med Genet. 2002;113:231–237. doi: 10.1002/ajmg.10615. [DOI] [PubMed] [Google Scholar]
- Lake CR, Ziegler MG, Murphy DL. Increased norepinephrine levels and decreased dopamine-beta-hydroxylase activity in primary autism. Arch Gen Psychiatry. 1977;34:553–556. doi: 10.1001/archpsyc.1977.01770170063005. [DOI] [PubMed] [Google Scholar]
- Lam KS, Aman MG, Arnold LE. Neurochemical correlates of autistic disorder: A review of the literature. Res Dev Disabil. 2006;27:254–289. doi: 10.1016/j.ridd.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Launay JM, Bursztejn C, Ferrari P, Dreux C, Braconnier A, Zarifian E, et al. Catecholamines metabolism in infantile autism: A controlled study of 22 autistic children. J Autism Dev Disord. 1987;17:333–347. doi: 10.1007/BF01487064. [DOI] [PubMed] [Google Scholar]
- Lawson DC, Turic D, Langley K, Pay HM, Govan CF, Norton N, et al. Association analysis of monoamine oxidase A and attention deficit hyperactivity disorder. Am J Med Genet Part B. 2003;116B:84–89. doi: 10.1002/ajmg.b.10002. [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Rutter M, Lord C, Rios P, Robertson S, Holdgrafer M, et al. Autism diagnostic interview: A standardized investigator-based instrument. J Autism Dev Disord. 1989;19:363–387. doi: 10.1007/BF02212936. [DOI] [PubMed] [Google Scholar]
- Leboyer M, Bouvard MP, Launay JM, Tabuteau F, Waller D, Dugas M, et al. Brief report: A double-blind study of naltrexone in infantile autism. J Autism Dev Disord. 1992;22:309–319. doi: 10.1007/BF01058158. [DOI] [PubMed] [Google Scholar]
- Lenders JW, Eisenhofer G, Abeling NG, Berger W, Murphy DL, Konings CH, et al. Specific genetic deficiencies of the A and B isoenzymes of monoamine oxidase are characterized by distinct neurochemical and clinical phenotypes. J Clin Invest. 1996;97:1010–1019. doi: 10.1172/JCI118492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal BL, Cook EH, Jr, Morford M, Ravitz A, Freedman DX. Relationships of whole blood serotonin and plasma norepinephrine within families. J Autism Dev Disord. 1990;20:499–511. doi: 10.1007/BF02216055. [DOI] [PubMed] [Google Scholar]
- Li J, Kang C, Zhang H, Wang Y, Zhou R, Wang B, et al. Monoamine oxidase A gene polymorphism predicts adolescent outcome of attention-deficit/hyperactivity disorder. Am J Med Genet Part B. 2007;144:430–433. doi: 10.1002/ajmg.b.30421. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, et al. Autism diagnostic observation schedule: A standardized observation of communicative and social behavior. J Autism Dev Disord. 1989;19:185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr, Leventhal BL, DiLavore PC, et al. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- Lotspeich LJ, Kwon H, Schumann CM, Fryer SL, Goodlin-Jones BL, Buonocore MH, et al. Investigation of neuroanatomical differences between autism and Asperger syndrome. Arch Gen Psychiatry. 2004;61:291–298. doi: 10.1001/archpsyc.61.3.291. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Harris G, Andreasen NC, O'Leary DS, Yuh WT, Heckel D. Structural MR image processing using the BRAINS2 toolbox. Comput Med Imaging Graph. 2002;26:251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Manuck SB, Flory JD, Ferrell RE, Mann JJ, Muldoon MF. A regulatory polymorphism of the monoamine oxidase-A gene may be associated with variability in aggression, impulsivity, and central nervous system serotonergic responsivity. Psychiatry Res. 2000;95:9–23. doi: 10.1016/s0165-1781(00)00162-1. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Buckholtz JW, Kolachana B, R Hariri A, Pezawas L, Blasi G, et al. Neural Mechanisms of Genetic Risk for Impulsivity and Violence in humans. Proc Natl Acad Sci. 2006;103:6269–6974. doi: 10.1073/pnas.0511311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Newman TK, Syagailo YV, Barr CS, Wendland JR, Champoux M, Graessle M, et al. Monoamine oxidase A gene promoter variation and rearing experience influences aggressive behavior in rhesus monkeys. Biol Psychiatry. 2005;57:167–172. doi: 10.1016/j.biopsych.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Nilsson KW, Sjoberg RL, Damberg M, Leppert J, Ohrvik J, Alm PO, et al. Role of monoamine oxidase A genotype and psychosocial factors in male adolescent criminal activity. Biol Psychiatry. 2006;59:121–127. doi: 10.1016/j.biopsych.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Piven J, Nehme E, Simon J, Barta P, Pearlson G, Folstein SE. Magnetic resonance imaging inautism: Measurement of the cerebellum, pons, and fourth ventricle. Biol Psychiatry. 1992;31:491–504. doi: 10.1016/0006-3223(92)90260-7. [DOI] [PubMed] [Google Scholar]
- Prakash N, Wurst W. Development of dopaminergic neurons in the mammalian brain. Cell Mol Life Sci. 2006a;63:187–206. doi: 10.1007/s00018-005-5387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Wurst W. Genetic networks controlling the development of midbrain dopaminergic neurons. J Physiol. 2006b;575:403–410. doi: 10.1113/jphysiol.2006.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig M, Bellivier F, Fenton BT, Baud P, Berney A, Courtet P, et al. Association between bipolar disorder and monoamine oxidase A gene polymorphisms: Results of a multicenter study. Am J Psychiatry. 2000;157:948–955. doi: 10.1176/appi.ajp.157.6.948. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Sharif NA. Effects of l-glutamate and related amino acids upon the release of [3H]dopamine from rat striatal slices. Brain Res. 1978;157:391–395. doi: 10.1016/0006-8993(78)90048-3. [DOI] [PubMed] [Google Scholar]
- Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–279. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- Samochowiec J, Lesch KP, Rottmann M, Smolka M, Syagailo YV, Okladnova O, et al. Association of a regulatory polymorphism in the promoter region of the monoamine oxidase A gene with antisocial alcoholism. Psychiatry Res. 1999;86:67–72. doi: 10.1016/s0165-1781(99)00020-7. [DOI] [PubMed] [Google Scholar]
- Schumann CM, Hamstra J, Goodlin-Jones BL, Lotspeich LJ, Kwon H, Buonocore MH, et al. The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J Neurosci. 2004;24:6392–6401. doi: 10.1523/JNEUROSCI.1297-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Beck JS, Kwitek AE, Sandstrom DW, Stone EM. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics. 1993;16:325–332. doi: 10.1006/geno.1993.1193. [DOI] [PubMed] [Google Scholar]
- Sjoberg RL, Ducci F, Barr CS, Newman TK, Dell'osso L, Virkkunen M, Goldman D, et al. A non-additive interaction of a functional MAO-A VNTR and testosterone predicts antisocial behavior. Neuropsycho-pharmacology. 2007;32:425–430. doi: 10.1038/sj.npp.1301417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi MS, Sanders-Bush E. Serotonin and brain development. Int Rev Neurobiol. 2004;59:111–174. doi: 10.1016/S0074-7742(04)59006-2. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. Thieme Medical Publishers; New York: 1988. [Google Scholar]
- Urwin RE, Bennetts BH, Wilcken B, Lampropoulos B, Beumont PJ, Russell JD, et al. Gene-gene interaction between the monoamine oxidase A gene and solute carrier family 6 (neurotransmitter transporter, noradrenalin) member 2 gene in anorexia nervosa (restrictive subtype) Eur J Hum Genet. 2003;11:945–950. doi: 10.1038/sj.ejhg.5201077. [DOI] [PubMed] [Google Scholar]
- Urwin RE, Nunn KP. Epistatic interaction between the monoamine oxidase A and serotonin transporter genes in anorexia nervosa. Eur J Hum Genet. 2005;13:370–375. doi: 10.1038/sj.ejhg.5201328. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imaging. 1999a;18:897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Trans Med Imaging. 1999b;18:885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- Vitalis T, Fouquet C, Alvarez C, Seif I, Price D, Gaspar P, et al. Developmental expression of monoamine oxidases A and B in the central and peripheral nervous systems of the mouse. J Comp Neurol. 2002;442:331–347. doi: 10.1002/cne.10093. [DOI] [PubMed] [Google Scholar]
- Widom CS, Brzustowicz LM. MAOA and the “cycle of violence:” childhood abuse and neglect, MAOA genotype, and risk for violent and antisocial behavior. Biol Psychiatry. 2006;60:684–689. doi: 10.1016/j.biopsych.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Williams RB, Marchuk DA, Gadde KM, Barefoot JC, Grichnik K, Helms MJ, et al. Serotonin-related gene polymorphisms and central nervous system serotonin function. Neuropsychopharmacology. 2003;28:533–541. doi: 10.1038/sj.npp.1300054. [DOI] [PubMed] [Google Scholar]
- Yirmiya N, Pilowsky T, Tidhar S, Nemanov L, Altmark L, Ebstein RP. Family-based and population study of a functional promoter-region monoamine oxidase A polymorphism in autism: Possible association with IQ. Am J Med Genet. 2002;114:284–287. doi: 10.1002/ajmg.10189. [DOI] [PubMed] [Google Scholar]
- Yu YW, Tsai SJ, Hong CJ, Chen TJ, Chen MC, Yang CW. Association study of a monoamine oxidase a gene promoter polymorphism with major depressive disorder and antidepressant response. Neuropsychopharmacology. 2005;30:1719–1723. doi: 10.1038/sj.npp.1300785. [DOI] [PubMed] [Google Scholar]
- Zalsman G, Huang YY, Harkavy-Friedman JM, Oquendo MA, Ellis SP, Mann JJ. Relationship of MAO-A promoter (u-VNTR) and COMT (V158M) gene polymorphisms to CSF monoamine metabolites levels in a psychiatric sample of caucasians: A preliminary report. Am J Med Genet Part B. 2005;132B:100–103. doi: 10.1002/ajmg.b.30094. [DOI] [PubMed] [Google Scholar]