Abstract
Demand for BRCA1 and BRCA2 mutation screening is increasing, as their identification will impact medical management. However, both the contribution of different mutation types in BRCA1 and BRCA2, and whom should be offered testing for large genomic rearrangements, have not been well established in the United States high risk population. We define the prevalence and spectrum of point mutations and genomic rearrangements in BRCA genes in a large US high-risk clinic population of both non-Ashkenazi and Ashkenazi Jewish descent, using a sample set representative of the US genetic testing population. Two hundred and fifty-one probands ascertained through the University of Pennsylvania high-risk clinic, all with commercial testing for BRCA1 and BRCA2, an estimated prevalence of BRCA mutation ≥ 10% using the Myriad II model and a DNA sample available, were studied. Individuals without deleterious point mutations were screened for genomic rearrangements in BRCA1 and BRCA2. In the 136 non-Ashkenazi Jewish probands, 36 (26%) BRCA point mutations and 8 (6%) genomic rearrangements, seven in BRCA1 and one in BRCA2, were identified. Forty-seven (40%) of the 115 Ashkenazi Jewish probands had point mutations; no genomic rearrangements were identified the group without mutations. In the non-Ashkenazi Jewish probands, genomic rearrangements constituted 18% of all identified BRCA mutations; estimated mutation prevalence (Myriad II model) was not predictive of their presence. While these findings should be confirmed in larger sample sets, our data suggest that genomic rearrangement testing be considered in all non-Ashkenazi Jewish women with an estimated mutation prevalence ≥ 10%.
Keywords: BRCA1, BRCA2, point mutations, genomic rearrangements, US clinic population
INTRODUCTION
A woman born in the United States has an average lifetime risk of 13% of being diagnosed with breast cancer (1). Family history is associated with 10 to 20% percent of breast cancer cases (2), and within that group approximately one-half (5 to 10% of all cases) are strongly hereditary (3). Germline mutations in BRCA1 and BRCA2 are associated with an increased risk of breast and ovarian cancer (4–7). Genetic testing for BRCA1 and BRCA2 mutations has been available for more than a decade (8–10). The usefulness of genetic testing as medical management tool has become increasingly recognized (11), as effective risk reduction procedures, such as prophylactic oophorectomy, and screening measures, such as breast MRI, are available (12–17). Our ability to offer effective management for patients with BRCA1 and BRCA2 mutations has increased the demand for genetic testing and make it imperative that we detect all mutations.
Despite our understanding of the clinical phenotypes most predictive of the presence of BRCA1 and BRCA2 mutations (18–22), a significant number of families with both breast and ovarian cancers do not have identifiable mutations (23). An increase in the rate of mutation detection has resulted from the identification of exon duplication(s) or deletion(s), commonly referred to as genomic rearrangements. Genomic rearrangements are not identified using PCR based methods of mutation screening. Initial studies of genomic rearrangements were limited in various ways – they examined only BRCA1, included small patient populations, were limited to very high risk breast/ovarian cancer families or to one ethnicity (24–27). More recently, highly sensitive DNA-based quantitative techniques have been developed to analyze both BRCA1 and BRCA2 for the presence of genomic rearrangements, and as such are useful in large scale screening. Previous studies, mainly done in homogenous ethnic groups, have suggested that the frequency of genomic rearrangements in BRCA1 in high risk breast cancer families ranges between 1.3–4.4%, accounting for between 8 –19% of the total mutations, with the number dependant on ethnicity and study eligibility criteria (24, 28–35). To date, the prevalence of genomic rearrangements in BRCA2 results lower than that in BRCA1 with the frequency in high risk breast/ovarian cancer families ranging from 0–2.4% with contribution of large genomic rearrangements to all the BRCA2 mutations between 0–11% (29, 32, 33, 36–39). These data have established that genomic rearrangements comprise a significant component of the identifiable mutations in BRCA1 and BRCA2.
While utility of screening for genomic rearrangement in both BRCA1 and BRCA2 is clear, there remain several crucial unanswered questions, such as the prevalence of genomic rearrangements in the US high-risk population, their contribution to mutations in populations of mixed ethnic backgrounds and how patients should be selected for screening. Currently, screening for genomic rearrangements is offered only on a standard basis (i.e. incorporated within initial genetic testing, as opposed to being ordered as a separate test) to women with very high risk personal or family histories (approximated to an estimated mutation prevalence > 30%). The goal of our study was to determine the role of testing for BRCA1 and BRCA2 genomic rearrangements in a US high-risk clinic, representative of the commercial testing population, including women of both Ashkenazi Jewish and non-Ashkenazi Jewish descent.
METHODS
Family Ascertainment
Probands were ascertained from the Cancer Risk Evaluation Program (CREP) at the University of Pennsylvania Health System [Hospital of the University of Pennsylvania (1998–2006) and Pennsylvania Hospital (2001–2006)] (see Eligibility Criteria Flowchart, Supplemental Figure 1). Patients are either self- or physician-referred based on their personal or family history. All participants provided written consent to participate in an institutional review board approved study of the genetics of hereditary breast cancer. More than 99% of patients seen in CREP participate in a research registry. Only one individual from each family was included in this study. If multiple individuals from the same families were enrolled in our study, the individual affected with breast cancer at the earliest age with commercial screening (Myriad Genetics, Salt Lake City, UT) performed was considered the proband. Pathology reports and/or medical records for confirmation for age and type of cancer diagnosis were collected from probands and affected relatives whenever possible. Ethnicity was self reported. The characteristics of the probands are detailed in Table 1.
Table 1.
Characteristics of all probands
| Characteristics | Family N° (%) | |
|---|---|---|
| Ethnicity, All probands (N=251) | ||
| Non-Ashkenazi Jewish | 136 (54) | |
| European | 119 (87) | |
| African American | 16 (12) | |
| Latin American | 1 (1) | |
| Ashkenazi Jewish | 115 (46) | |
|
| ||
| Cancer history | ||
| Non-Ashkenazi Jewish | 136 (100) | |
| Breast only* | 90 (66) | |
| Age of breast diagnosis: | ||
| <30 | 3 (2) | |
| 30–39 | 33 (25) | |
| 40–49 | 46 (33) | |
| 50–59 | 7 (5) | |
| ≥ 60 | 1 (1) | |
| Ovarian only | 13 (9) | |
| Breast and Ovarian | 11 (8) | |
| Bilateral Breast | 14 (11) | |
| Bilateral Breast and Ovarian Cancer | 3 (2) | |
| Male Breast | 5 (4) | |
|
| ||
| Ashkenazi Jewish | 115 (100) | |
| Breast only | 71 (62) | |
| Age of breast diagnosis: | ||
| <30 | 3 (2.5) | |
| 30–39 | 19 (16) | |
| 40–49 | 35 (31) | |
| 50–59 | 10 (9) | |
| ≥ 60 | 4 (3.5) | |
| Ovarian only | 13 (11) | |
| Breast and Ovarian | 6 (5) | |
| Bilateral Breast | 21 (18) | |
| Bilateral Breast and Ovarian Cancer | 2 (2) | |
| Male Breast | 2 (2) | |
Cancer history percentage shown is the percent of total non-Ashkenazi Jewish or Ashkenazi Jewish groups
Study Eligibility
Probands were eligible for the current study if they meet all of the following criteria: 1) were affected with breast or ovarian cancer; 2) had full commercial sequencing or Ashkenazi Jewish founder mutation screening (Myriad Genetics, Salt Lake City, UT); 3) had no prior genetic testing of themselves or any family member; 4) had an estimated mutation prevalence of 10% or higher based on the Myriad II model prevalence tables and 5) had a DNA sample available in the laboratory at the University of Pennsylvania.
Probands were excluded if either they or their relatives had previous genetic testing before being evaluated in CREP for two reasons. First, we wanted to exclude mutation positive patients specifically referred for clinical management, so as to limit bias for mutation positivity. Second, we wanted to exclude individuals were referred to us specifically for research participation after uninformative genetic testing, so as to limit bias for mutation negativity.
The estimated prevalence of BRCA mutations was generated using the Myriad II model, which consists of two mutation prevalence tables, stratified by Ashkenazi Jewish ethnicity3. The tables were developed following the guidelines initially published by Frank et al. (21) and provide a composite mutation prevalence for BRCA1 and BRCA2. The Myriad II model has been demonstrated to perform similarly to BRCAPRO and BOADICA, and be the most sensitive in predicting mutations in non-Ashkenazi Jewish families (40). All models use only first and second degree relatives. The 507 probands that met above criteria 1–3 were evaluated using the Myriad II model. The predicted prevalence of BRCA mutations was calculated for both the maternal and paternal lineages; the lineage with the highest prevalence was used for analytical purposes. Twelve probands had one parent each of Ashkenazi Jewish and non-Ashkenazi Jewish origin. These probands were grouped with the lineage with the highest estimated mutation prevalence – six Ashkenazi Jewish and six non-Ashkenazi Jewish. Sixty-two probands (40 Ashkenazi Jewish and 22 non-Ashkenazi Jewish) had an estimated mutation prevalence over 10%, but did not provide samples. A comparison between the sample and non-sampled groups is included in the Results section. Two hundred and fifty-one probands had both an estimated BRCA mutation prevalence of over 10% in either the maternal or paternal lineage and a sample available in the laboratory and thus were included in the study.
Point Mutation Analysis
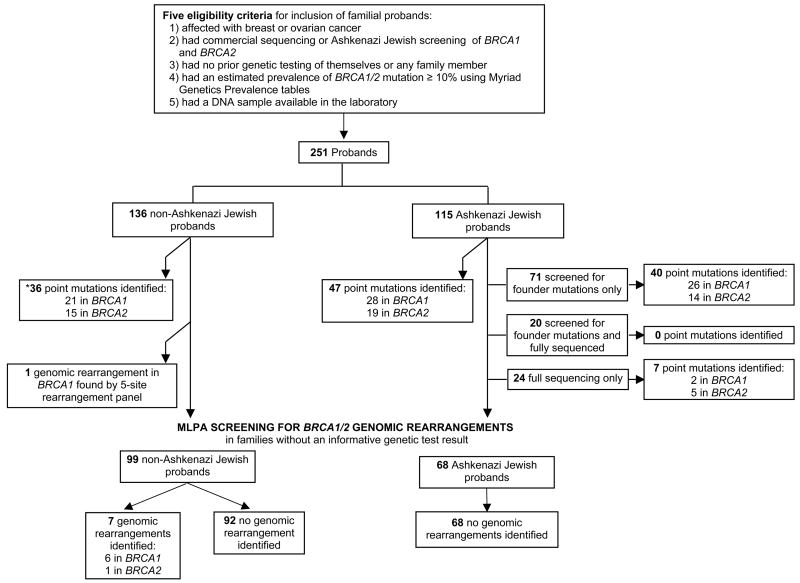
DNA was extracted from peripheral blood mononuclear cells by using standard procedures. Either blood or DNA samples were provided to Myriad Genetics Laboratory (Salt Lake City, UT) for mutation detection. After analysis, all patients were classified as having a deleterious mutation, a variant of uncertain significance, or no mutation (see Supplemental Tables 1 and 2). The classification of mutations was performed as previously described (21). As detailed in Figure 1, full sequencing in probands with Ashkenazi Jewish ancestry was done in a total of 44 women, either because genetic testing was done prior to the availability of the Ashkenazi Jewish founder mutation panel, mixed heritage (probands with non-Ashkenazi Jewish ancestors), high prior probability or based on patient request.
Figure 1.
Analytical strategy of the study and numbers of patients identified in each group. *Mutations are listed in Supplementary Table 1
Screening for Genomic Rearrangements
Ninety-four probands (72 non-Ashkenazi Jewish and 22 Ashkenazi Jewish) had testing for the presence of five BRCA1 genomic rearrangements included in the commercial screening since 2001 at Myriad Genetics Laboratory. The rearrangement panel includes five specific mutations in BRCA1: 7.1-kb deletion of exons 8–9, 3.4-kb deletion of exon 13, 6-kb duplication of exon 13, 26-kb deletion of exons 14–20 and 510-bp deletion of exon 22 (24, 26, 41–43). Multiplex ligation dependent probe amplification (MLPA) was used to screen for genomic rearrangements in BRCA1 and BRCA2 in all the mutation negative families. MLPA was performed according to the instructions provided by the manufacturer (MRC-Holland, Amsterdam, Netherlands). The probe mixes MLPA P002 and P087 were used to screen BRCA1. The probe mix MLPA P045 kit was used to screen BRCA2 and for the CHEK2 1100delC mutation. The fragments were analyzed on ABI 3100 capillary sequencer (Applied Biosystems Inc., Foster City, CA, USA) using Genescan software. Variation in peak height was evaluated by comparing each test sample to the normal control present in the same experiment and by cumulative comparison of four to six samples always from the same experiment. Any sample with variation in peak height was repeated a total of three times.
Quantitative Analysis of MLPA
We used GeneMarker, Version 1.4 (Softgenetics LLC) to perform data normalization and analysis. GeneMarker normalizes the peak height (fluorescence intensity) of each MLPA fragment with either the internal control probes or the entire population (all the fragments). Samples for which we obtained a height ratio (normalized fluorescence intensity of each individual probe of a patient sample to a control DNA sample) less than 0.7 (for deletion) and greater than 1.2 (for duplication) had MLPA repeated. Controls with known genomic deletions were studied and had height ratios of ~1.5, 1.0, 0.5, or 0.0 for regions with duplication, the wild-type sequence, a deletion, or absence of the sequence, in that order.
Identification and Confirmation of Rearrangements
For the rearrangements in BRCA1 and BRCA2, we confirmed the presence of the genomic deletions using both MLPA kits for BRCA1 and real-time quantitative PCR for the mutations in BRCA1 and BRCA2. Real-time PCR was performed using the LightCycler (Roche Applied Science, V.3.5.3) and SYBR Green I chemistry (LightCycler FastStart DNA Master SYBR Green I). Primers and probes for exons within BRCA1 and BRCA2 were designed as described in Barrois et al (44). We selected one test exon within the deleted region of interest. As an endogenous control, we normalized each sample to ALB (encoding albumin at 4q11-q13). We determined heterozygosity for a deletion when the ratio of ALB:test exon was 2:1. Each assay was run in triplicate; two normal controls (ratio 1:1) and a non-template control were included. PCR reactions were set up in a total volume of 20 μl by distributing aliquots of 18 μl of master mix into the capillaries, followed by 2 μl of DNA adjusted to 10 ng/μl. The reaction mixture contained 0.4 μl of each primer, 2 μl of FastStart DNA kit SYBR Green I and contained 5mM MgCl2. The thermal cycling conditions were: an initial denaturation step at 95°C for 10 min, 45 cycles at 95°C for 10 sec, primer annealing temperature for 5 sec and extension at 72° C for 6 sec. (Primers and annealing temperatures are available on request.) A melting curve was produced after each run for every sample. Quantification data were analyzed using the fit point method of the LightCycler software (Supplemental Figures 2 and 3). In the fit point method for the Lightcycler (Roche, Inc), the relative expression ratio is calculated from the real-time efficiencies and the crossing point (where fluorescence rises above the threshold) deviation of an unknown sample as compared to a control.
All genomic rearrangements identified by MLPA, a sub-set of which were confirmed by real-time PCR, were sent for the BRACAnalysis® Rearrangement Test (BART) (Myriad Genetics, Salt Lake City, UT). All mutations confirmed by real-time PCR also were identified by BART. No mutations unconfirmed by real-time PCR were identified by BART. In three families, additional affected family members were available and co-segregation of the mutation with disease was observed.
Statistical Methods
Comparisons of discrete and quantitative variables were made by using Fisher’s exact tests and Kruskal-Wallis chi-square tests, respectively (STATA v. 9, StataCorp, Texas). The latter was used to to test the null hypothesis that there was no difference in the median ages of cancer diagnosis in the mutation and non-mutation carrying groups and between the estimated mutation prevalences (Myriad II model) in the sampled and non-sampled groups. Chi-square tests comparing observed vs. expected mutation distributions also were computed. Logistic regression was used to evaluate whether the estimated Myriad prevalences predicted the presence of genomic rearrangement. Null hypotheses were rejected at a p-value of p<0.05, and all comparisons were made using two-sided tests.
RESULTS
Family Characteristics
Probands from 251 families met the eligibility criteria for inclusion, 244 females and 7 males (Table 1). Of the 251 high risk families, 115 (46%) were of Ashkenazi Jewish descent and 136 (54%) were of non-Ashkenazi Jewish descent. The proband characteristics of the non-Ashkenazi Jewish and Ashkenazi Jewish families were not significantly different. The range of estimated prevalence of point mutations in BRCA1 and BRCA2 was from 11.2 to 79.3% with a median of 17.6%; in the non-Ashkenazi Jewish probands, from 11.2 to 79.3%, with a median of 22.9%; and in the Ashkenazi Jewish probands from 11.6 to 75%, with a median of 16%. The characteristics of the non-Ashkenazi Jewish (Table 2) and Ashkenazi Jewish families were very similar: over 95% of families contained women with unilateral breast cancers, 45% at least one woman with ovarian cancer, 15% at least one woman with breast and ovarian cancers, 5–7% at least one male breast cancer case and 20% at least one man with prostate cancer. The familial characteristics differed significantly between non-Ashkenazi Jewish and Ashkenazi Jewish probands only in the percentage of families with women with breast cancer diagnosed under age 50, 96% and 86%, respectively (p=0.01). We identified 62 probands who met the study eligibility requirements but did not provide samples; of these 40 were Ashkenazi Jewish and 22 were non-Ashkenazi Jewish.
Table 2.
Characteristics of non-Ashkenazi Jewish families*
| All Families (n = 136) | BRCA1 Point Mutation Positive Families (n = 21) | BRCA2 Point Mutation Positive Families (n = 15) | BRCA1 Genomic Rearrangement Positive Families (n = 7) | BRCA2 Genomic Rearrangement Positive Families (n = 1) | All Mutation Positive Families (n = 44) | All Mutation Negative Families (n = 92) | p value† | |
|---|---|---|---|---|---|---|---|---|
| Familial Characteristics | ||||||||
| Any Breast Cancer (%) | 134 (99) | 21 (100) | 15 (100) | 7 (100) | 1 (100) | 44 (100) | 90 (98) | 0.55 |
| Breast Cancer <50 y (%) | 130 (96) | 20 (95) | 15 (100) | 7 (100) | 1 (100) | 43 (98) | 87 (95) | 0.55 |
| Br/Ov Cancer in the same individual (%) | 22 (16) | 6 (29) | 4 (27) | 1 (14) | 0 | 11 (25) | 11 (12) | 0.05 |
| Any Ovarian Cancer (%) | 63 (46) | 14 (67) | 8 (53) | 4 (57) | 1 (100) | 27 (61) | 36 (39) | 0.02 |
| Male Breast Cancer (%) | 9 (7) | 1 (5) | 1 (6) | 0 | 1 (100) | 3 (7) | 6 (7) | >0.99 |
| Pancreatic Cancer (%) | 9 (7) | 1 (5) | 5 (33) | 0 | 0 | 6 (14) | 3 (3) | 0.03 |
| Prostate Cancer (%) | 26 (19) | 7 (33) | 3 (9) | 2 (29) | 1 (100) | 13 (29) | 13 (14) | 0.04 |
| Proband Characteristics | ||||||||
| Breast Cancer Only (%) | 90 (66) | 9 (43) | 10 (67) | 4 (57) | 0 | 23 (52) | 67 (73) | 0.02 |
| Bilateral Breast Cancer (%) | 15 (11) | 4 (19) | 1 (7) | 2 (29) | 1 (100) | 8 (18) | 7 (8) | 0.08 |
| Ovarian Cancer Only (%) | 13 (10) | 3 (14) | 1 (7) | 1 (14) | 0 | 5 (11) | 8 (9) | 0.75 |
| Br/Ov Cancer in the same individual (%) | 13 (10) | 4 (19) | 2 (13) | 0 | 0 | 6 (14) | 7 (8) | 0.34 |
| Male Breast Cancer (%) | 5 (4) | 1 (5) | 1 (7) | 0 | 0 | 2 (5) | 3 (3) | >0.99 |
| Breast Cancer Average Age Dx | 43.1 | 40.4 | 43.5 | 35.7 | --- | 40.8 | 44.0 | 0.14‡ |
| Ovarian Cancer Average Age Dx | 57.5 | 51 | 59 | --- | --- | 53.7 | 60.3 | 0.57 |
includes up to fourth degree relatives
comparison of all mutation positive and negative families using two-tailed Fisher Exact test
comparison of average age of diagnoses using Krushal Wallis test
Point mutations and variants of uncertain significance in BRCA1 and BRCA2
In total, 83 (33%) of the 251 total probands studied were found to have deleterious point mutations, 49 (59%) in BRCA1 and 34 (41%) in BRCA2, respectively (Figure 1). The 83 mutations are comprised of 22 and 15 distinct mutations in BRCA1 and BRCA2, respectively. All mutations are listed in Supplementary Data Table 1. The three Ashkenazi Jewish founder mutations accounted for 47 (56%) of the mutations detected. While a higher rate of point mutations was found in the Ashkenazi Jewish group (40%) than in the non-Ashkenazi Jewish group (26%) (p=0.02), the proportion of mutations identified in BRCA1 (60%) and BRCA2 (40%) was identical in both groups.
In the 115 Ashkenazi Jewish probands, 47 mutations (40% of families) were identified, 28 (24%) in BRCA1 and 19 (16%) in BRCA2, respectively. Of the identified mutations, forty-five were Ashkenazi Jewish founder mutations (BRCA1 185delAG - 22, 5382insC – 5; BRCA2 – 6174delT – 18) and two were non-founder mutations (1135insA in BRCA1 and 5466insT in BRCA2). BRCA1 1135insA, has been described as an ancient founder mutation in the Norwegian population and previously has been reported in an Ashkenazi Jewish family of Norwegian descent, as our family is (45, 46).
After full sequencing in the probands from the 136 non-Ashkenazi Jewish families, 36 point mutations (26% of families) were identified. Twenty-one mutations (15%) and 15 (11%) were identified in BRCA1 and in BRCA2, respectively. The only mutations identified more than once were both in BRCA1, 5832insC in two probands of Polish descent and IVS16+6T>6 in two African American probands. Five of 16 (31%) African Americans had identified mutations as compared to 31/120 (25.8%) European Americans. The estimated mutation prevalence for the African American probands ranged from 16.3 to 73.7% with a median of 26.3%. Twenty variants of uncertain significance (VUS) were identified in the 251 probands (see Supplementary Data Table 2). Four VUS were found in patients who also had deleterious mutations. Eight of the VUS were identified in BRCA1 and 12 in BRCA2. Of these, BRCA1 V1688del may be deleterious, but it is not yet formally classified as such (47). Eight of the 20 (40%) VUS were found in probands of African American ancestry.
Comparison of sampled and non-sampled probands
In order to assess any bias based on sample collection, we evaluated in 62 probands who met eligibility criteria but did not provide a sample. Comparing the sampled (251) and non-sampled (62) probands, there was no significant difference in the estimated mutation prevalence (p=0.4, Ashkenazi Jewish and p=0.12, non-Ashkenazi Jewish). In the 40 Ashkenazi Jewish probands, 12 (29%) had mutations, not significantly different from the 115 Ashkenazi Jewish probands with samples (p=0.12). None of the 22 non-Ashkenazi Jewish probands carried mutations, significantly different than the sampled group (p=0.002). However, the inclusion of the non-sampled probands did not significantly changed the rate of point mutations, which decreased from 26 to 23% (p=0.49). Eighty-six percent of the potential non-Ashkenazi Jewish probands were included in the study.
Comparison of Testing Population with Myriad Genetics Testing Population
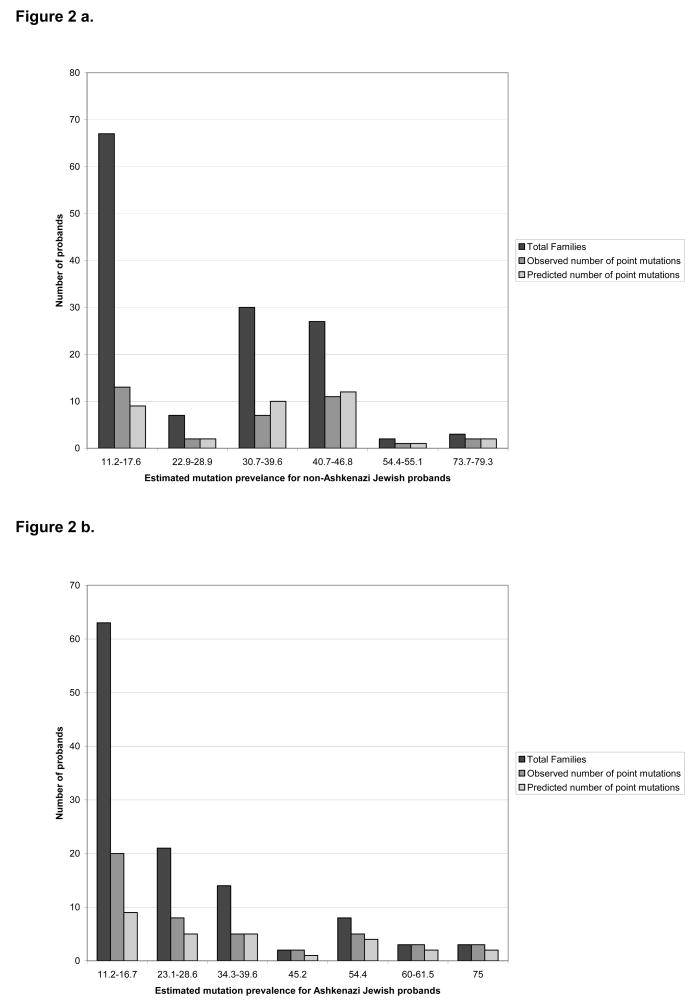
In order to determine whether we could extrapolate our findings to the larger commercial testing population, we compared the expected number to the observed number of point mutations based on the Myriad II model. The majority of probands had an estimated mutation prevalence between 10 and 20%, comprising 49% and 55% of those studied in the non-Ashkenazi Jewish and Ashkenazi Jewish groups, respectively. In the non-Ashkenazi Jewish group, there was no significant difference between the number of observed and expected mutations (Figure 2a). In the entire Ashkenazi Jewish group, we found a significant difference between the number of observed and expected point mutations (p=0.008); this result was entirely due to the considerable difference between number of observed (20) and expected (9) mutations in the group with the estimated mutation prevalence between 11.2–16.7% (Figure 2b). In the 52 Ashkenazi Jewish probands with an estimated mutation prevalence over 20%, there was no significant difference between the number of observed and expected point mutations.
Figure 2.
Figure 2a. The total number of non-Ashkenazi Jewish probands in each estimated mutation prevalence, group showing the estimated and observed number of point mutations
Figure 2b: The total number of Ashkenazi Jewish probands in each mutation prevalence group, showing the estimated and observed number of point mutation
Prevalence of Genomic Rearrangements in BRCA1 and BRCA2
In total, we screened 167 probands for genomic rearrangements in BRCA1 and BRCA2 using MLPA. Probands were included if they had negative result after full sequencing, the Ashkenazi Jewish founder mutation screen (if Ashkenazi Jewish) or a variant of uncertain significance. Using MLPA in the 68 Ashkenazi Jewish probands, four samples with decreases in peak height as compared to controls were identified, all of BRCA2 exons 1–2. None of the peak height decreases were confirmed as true deletions by real time PCR of exon 2 of BRCA2 or by BART. In summary, no genomic rearrangements were identified in the 68 Ashkenazi Jewish probands studied. Twenty-five (37%) had an estimated mutation prevalence over 10%, which is the cutoff used for non-Ashkenazi Jewish women.
In the 100 non-Ashkenazi Jewish probands without deleterious point mutations, eight genomic rearrangements were detected - seven in BRCA1 and one in BRCA2, all deletions (Table 3). One large genomic rearrangement (deletion of BRCA1 exons 14–20) was identified by the 5-site rearrangement panel (Myriad Genetics, Salt Lake City, UT). The additional five in BRCA1 and one in BRCA2 were identified by MLPA and confirmed by real-time PCR of one or more of the relevant exon(s) of BRCA1 or BRCA2 (Supplemental Figures 2 and 3) and BART. A benign polymorphism, BRCA2 2192C→ G (P655R), known to be found in African Americans, within the MLPA BRCA2 exon 11 probe binding site (probe 2279-L1770) created a false positive in two samples, which was not confirmed by real time PCR or BART. Five of these genomic rearrangements previously have been reported (33, 43, 48, 49). We identified two novel large genomic deletions - the deletion of exons 11–12 in BRCA1 and of exons 1–13 in BRCA2. We identified a deletion of BRCA1 exons 21, 22, 23 and 24 in two independent families. The deletion of exons 8–9 in the African American family presumably is different than the previously reported deletion in women of European origin, (42) as it was not detected by the commercially available 5-site genomic rearrangement panel. Walsh and colleagues also detected a deletion of exons 8 and 9 in an African American family so it is possible that it is a founder mutation in this population (48). In our family, the deletion was in linkage disequilibrium with BRCA1 A102G.
Table 3.
Description of genomic rearrangements
| Gene | Exon(s) | Type of genomic change |
Myriad II model estimated mutation prevalence† |
Phenotype proband | Family history (# of cases)‡ | Ethnicity | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Primary Cancer(s) (Age of Dx) | Br Ca | Br Ca Dx < 50 |
Male Br Ca |
Ov Ca | Br/Ov Ca |
|||||
| BRCA1 | 1–2 | Deletion | 17.6 | Breast ca (45) | 1 | 1 | --- | 1 | --- | White (Irish/English/German) |
| BRCA1 | 3 | Deletion | 46.8 | Ovarian ca (54) | 2 | 1 | --- | --- | 1 | White (Italian) |
| BRCA1 | 8–9 | Deletion | 17.6 | Breast ca (29) | 1 | 1 | --- | 1 | --- | African-American |
| BRCA1 | part of 11–12 | Deletion | 17.6 | Breast ca (39) | 1 | 1 | --- | 1 | --- | White (Western Europe) |
| BRCA1 | 14–20* | Deletion | 16.3 | Bilateral breast ca (27, 33) | 2 | 2 | --- | --- | --- | White (British) |
| BRCA1 | 21–24 | Deletion | 16.3 | Bilateral breast ca (45, 47) | 3 | 2 | --- | --- | --- | White (Irish/German) White |
| BRCA1 | 21–24 | Deletion | 30.7 | Breast ca (29) | 6 | 5 | --- | --- | --- | (German/British/Italian/Czech) White |
| BRCA2 | 1–13 | Deletion | 11.2 | Bilateral breast ca (55, 55) | 6 | 4 | 1 | 1 | --- | (Scottish/English/German/Norwegian) |
Identified by the Myriad Genetics 5-site rearrangement screen
Using 1st and 2nd degree relatives
Up to 4th degree relatives included in family history
The frequency of genomic rearrangements in all non-Ashkenazi Jewish probands from our clinic based population was 6% (8/136), increasing to 8% (8/100) in those without identified point mutations. Together the genomic rearrangements constituted 18% (8/44) of all mutations identified in the non-Ashkenazi Jewish group; 29% (8/28) and 6% (1/16) of those in BRCA1 and BRCA2, respectively. One of the genomic rearrangements was found in an African American individual; in this population, it constituted 1/6 (16%) of all mutations identified. The estimated mutation prevalences using the Myriad Genetics tables were not predictive of identifying genomic rearrangements in BRCA1 or BRCA2 in the non-Ashkenazi Jewish group (p=0.27). The median estimated mutation prevalence in the non-Ashkenazi Jewish probands was 22.9%.
In families without identified point mutations in BRCA1 or BRCA2, the median estimated mutation prevalence was 17.9%; in the eight of these families subsequently identified with a genomic rearrangement the median estimated mutation prevalence was 17.6%, supporting a family history comparable to those families that did not have identified mutations. While the number of individuals identified with genomic rearrangements was small, those with rearrangements in BRCA1 were more likely to have bilateral breast cancer (37.5% vs. 8%, p=0.03) than the mutation negative probands. There were no other significant differences between the personal or family history of the probands with genomic rearrangements as compared to the mutation negative probands.
DISCUSSION
While the presence of genomic rearrangements in addition to point mutations in both BRCA1 and BRCA2 has been well established, the proportion of women carrying mutations and their relative contribution to the total mutations in a United States based high risk clinic representative of the commercial testing population has not been determined. In our study, 6% of non-Ashkenazi Jewish women with an estimated mutation prevalence over 10% had a genomic rearrangement, comprising 8% of those without an identified point mutation. Of importance, genomic rearrangements accounted for 18% of the mutations identified and their presence did not correlate with the estimated prevalence of BRCA mutation. Specifically, six of the eight women with genomic rearrangements had an estimated mutation prevalence under 30%, and thus would not have been eligible for standard screening for genomic rearrangements, which is offered only to women with an estimated mutation prevalence over 30% (approximated from the Myriad Genetics criteria). While it is likely that the phenotypic predictors for genomic rearrangements in BRCA1 and BRCA2 are the same as those for point mutations, we did not identify enough rearrangements to fully evaluate mutation predictors. Our data support the consideration of genomic rearrangement screening in BRCA1 and BRCA2 for all non-Ashkenazi Jewish women with an estimated mutation prevalence over 10%.
The non-Ashkenazi Jewish group included sixteen (16/136, 12%) probands of African American ethnicity. Among the African American families with a negative sequencing result, we detected one family with a genomic rearrangement (deletion of exons 8–9) in BRCA1, representing 6% of all African American families and 16% of all mutations identified in this group (1/6). Our data underline the need for larger studies to explore the contribution of genomic rearrangements to the BRCA1 and BRCA2 mutation spectra among minority populations in the United States. The rate of genomic rearrangements identified in this series is consistent with previous series from Europe and Australia, but lower than a previous United States based series, in which 12% of probands without point mutations were found to have a rearrangement in BRCA1 and BRCA2 (48). The discrepancy likely is due to differences in the study eligibility criteria. The previous study included families with at least four cases of female breast cancer, ovarian cancer and/or male breast cancer, whereas the current study included patients with an estimated prevalence of over 10%, which encompasses families with as few as two case of female breast cancer. Nonetheless, the conclusion of both studies is the same – that non-Ashkenazi Jewish women without point mutations in BRCA1 and BRCA2 should be offered screening for genomic rearrangements in the clinical setting.
While genomic rearrangements significantly contribute to the total number of mutations in BRCA1 and BRCA2, we did not identify any large genomic rearrangements in the Ashkenazi Jewish probands negative for point mutations. Within the Ashkenazi Jewish group, 45 of 47 (96%) point mutations were founder mutations. Our data are consistent with previous studies, demonstrating that the founder mutations account for more than 90% of mutations in Ashkenazi Jewish women (50, 51). In total, reported in the literature and including our study, 99 Ashkenazi Jewish probands from high risk families have been negatively screened for genomic rearrangements (48). Thus, founder mutations continue to account for the vast majority of all mutations in this population. Even if 18% of all detectable non-founder mutations in our Ashkenazi Jewish cohort were genomic rearrangements, they nevertheless would represent very rare events, accounting for 0.72% of mutations based on the prevalence of non-founder mutations in the Ashkenazi Jewish population. These data suggest that routine screening for large genomic rearrangements does not appear warranted in Ashkenazi Jewish women. However, larger studies of this population need to be done.
Empiric predictive models, such as Myriad II, Couch and Manchester Scoring System are intended to estimate the possibility of a BRCA1 or BRCA2 mutation in a woman based on her family history and are used widely in clinical practice (5, 21, 52, 53). Currently these models do not account for genomic rearrangements and as such underestimate the number of women with identifiable mutations, particularly in BRCA1. In addition, understanding the proportion of mutation that are missed using current methods to identify BRCA1 or BRCA2 is useful when counseling patients about their chances of being undetected mutation carriers. Previous studies from the Breast Cancer Linkage Consortium have suggested that the percentage of linked families with a detectable point mutation may be as low as 65% (23). However, the study was limited as various mutation detection techniques were used, some of which only detect 60–65% of mutations as compared to sequencing as a gold standard (54). Nonetheless, these studies did not include genomic rearrangements and thought should be given to repeating them with improved mutation detection techniques and the larger spectrum of mutations now identified, so that women can be accurately counseled about their chances to carry an undetected mutation. We attempted to limit bias for and against mutations in our population by only including probands with a sample available in the laboratory and excluding any probands who themselves or any family member had prior genetic testing before being seen in CREP and having commercial testing. Due to our strict eligibility criteria, the number of probands available for study was restricted; as such, these findings will need to be replicated in a larger population. However, any such study will need to include systematic screening of a clearly defined population, so that the findings can be generally applied. While our study is based on a single high risk clinic population and thus limited, we have demonstrated that the number of observed mutations does not significantly different than those expected in the commercial testing population based on the Myriad II model. The Myriad II model has been demonstrated to perform similarly to BOADICEA and BRCAPRO; all of the models are limited in that they generally under predict mutations at the lower end of prediction probability and overestimate the upper ends (40, 55). In the Ashkenazi Jewish population, the models did under predict the number of mutations at the lower end the mutation prevalence spectrum. Most models are limited to first- and second- degree relatives; however, in clinical practice family history characteristics of more distant relatives are important in evaluating a family. Overall, 80% of potentially eligible probands provided a sample; 86% of the non-Ashkenazi Jewish probands. The sampled Ashkenazi Jewish probands were representative of the entire potentially eligible Ashkenazi Jewish group. The non-sampled non-Ashkenazi Jewish probands did not differ in their estimated mutation prevalence, but did have fewer point mutations than the sampled group. However, including the non-sampled probands in the overall total did not significantly affect the rate of point mutations. As such, our clinic and the data presented herein are representative of patients seen at high risk clinics across the United States.
Acknowledgments
We thank the families for participating in this study. We would like to acknowledge Dr. Barbara Weber who started the Cancer Risk Evaluation Program at the University of Pennsylvania.
Grant support: Breast Cancer Research Foundation (KLN), Associazione Italiana Ricerca sul Cancro (MDP), QVC Network and the Fashion Footwear Association of New York, Marjorie B. Cohen Foundation, National Cancer Institute Cancer Genetics Network [HHSN216200744000C] (SMD).
Footnotes
References
- 1.Ries LAG, Harkins D, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2003. 2006 [cited; Available from: http://seer.cancer.gov/csr/1975_2003/
- 2.Madigan MP, Ziegler RG, Benichou J, Byrne C, Hoover RN. Proportion of breast cancer cases in the United States explained by well-established risk factors. J Natl Cancer Inst. 1995;87:1681–5. doi: 10.1093/jnci/87.22.1681. [DOI] [PubMed] [Google Scholar]
- 3.Claus EB, Schildkraut JM, Thompson WD, Risch NJ. The genetic attributable risk of breast and ovarian cancer. Cancer. 1996;77:2318–24. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2318::AID-CNCR21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Struewing JP, Hartge P, Wacholder S, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–8. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 5.Couch FJ, DeShano ML, Blackwood MA, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997;336:1409–15. doi: 10.1056/NEJM199705153362002. [DOI] [PubMed] [Google Scholar]
- 6.Easton D. Breast cancer genes--what are the real risks? [news] Nat Genet. 1997;16:210–1. doi: 10.1038/ng0797-210. [DOI] [PubMed] [Google Scholar]
- 7.Antoniou A, Pharoah PD, Narod S, et al. Average Risks of Breast and Ovarian Cancer Associated with BRCA1 or BRCA2 Mutations Detected in Case Series Unselected for Family History: A Combined Analysis of 22 Studies. Am J Hum Genet. 2003;72:1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 9.Tavtigian SV, Simard J, Rommens J, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds [see comments] Nat Genet. 1996;12:333–7. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 10.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2 [see comments] [published erratum appears in Nature 1996 Feb 22;379(6567):749] Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 11.Nelson HD, Huffman LH, Fu R, Harris EL. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: systematic evidence review for the U.S. Preventive Services Task Force. Ann Intern Med. 2005;143:362–79. doi: 10.7326/0003-4819-143-5-200509060-00012. [DOI] [PubMed] [Google Scholar]
- 12.Pierce LJ, Levin AM, Rebbeck TR, et al. Ten-year multi-institutional results of breast-conserving surgery and radiotherapy in BRCA1/2-associated stage I/II breast cancer. J Clin Oncol. 2006;24:2437–43. doi: 10.1200/JCO.2005.02.7888. [DOI] [PubMed] [Google Scholar]
- 13.Rebbeck TR, Friebel T, Lynch HT, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group [see comment] J Clin Oncol. 2004;22:1055–62. doi: 10.1200/JCO.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 14.Rebbeck TR, Friebel T, Wagner T, et al. Effect of short-term hormone replacement therapy on breast cancer risk reduction after bilateral prophylactic oophorectomy in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2005;23:7804–10. doi: 10.1200/JCO.2004.00.8151. [DOI] [PubMed] [Google Scholar]
- 15.Domchek SM, Friebel TM, Neuhausen SL, et al. Mortality after bilateral salpingo-oophorectomy in BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Lancet Oncol. 2006;7:223–9. doi: 10.1016/S1470-2045(06)70585-X. [DOI] [PubMed] [Google Scholar]
- 16.Leach MO, Boggis CR, Dixon AK, et al. Screening with magnetic resonance imaging and mammography of a UK population at high familial risk of breast cancer: a prospective multicentre cohort study (MARIBS) Lancet. 2005;365:1769–78. doi: 10.1016/S0140-6736(05)66481-1. [DOI] [PubMed] [Google Scholar]
- 17.Kauff ND, Satagopan JM, Robson ME, et al. Risk-reducing salpingo-oophorectomy in women with a BRCA1 or BRCA2 mutation. N Engl J Med. 2002;346:1609–15. doi: 10.1056/NEJMoa020119. [DOI] [PubMed] [Google Scholar]
- 18.Couch FJ, DeShano ML, Blackwood MA, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer [see comments] N Engl J Med. 1997;336:1409–15. doi: 10.1056/NEJM199705153362002. [DOI] [PubMed] [Google Scholar]
- 19.Thompson D, Easton D. Breast Cancer Linkage C. Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet. 2001;68:410–9. doi: 10.1086/318181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson D, Easton D. Breast Cancer Linkage C. Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomarkers Prev. 2002;11:329–36. [PubMed] [Google Scholar]
- 21.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–90. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 22.Nanda R, Schumm LP, Cummings S, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–33. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 23.Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–89. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrij-Bosch A, Peelen T, van Vliet M, et al. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients [published erratum appears in Nat Genet 1997 Dec;17(4):503] Nat Genet. 1997;17:341–5. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- 25.Puget N, Torchard D, Serova-Sinilnikova OM, et al. A 1-kb Alu-mediated germ-line deletion removing BRCA1 exon 17. Cancer Res. 1997;57:828–31. [PubMed] [Google Scholar]
- 26.Puget N, Sinilnikova OM, Stoppa-Lyonnet D, et al. An Alu-mediated 6-kb duplication in the BRCA1 gene: a new founder mutation? Am J Hum Genet. 1999;64:300–2. doi: 10.1086/302211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unger MA, Nathanson KL, Calzone K, et al. Screening for genomic rearrangements in families with breast and ovarian cancer identifies BRCA1 mutations previously missed by conformation-sensitive gel electrophoresis or sequencing. Am J Hum Genet. 2000;67:841–50. doi: 10.1086/303076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agata S, Viel A, Della Puppa L, et al. Prevalence of BRCA1 genomic rearrangements in a large cohort of Italian breast and breast/ovarian cancer families without detectable BRCA1 and BRCA2 point mutations. Genes Chromosomes Cancer. 2006;45:791–7. doi: 10.1002/gcc.20342. [DOI] [PubMed] [Google Scholar]
- 29.Preisler-Adams S, Schonbuchner I, Fiebig B, Welling B, Dworniczak B, Weber BH. Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genet Cytogenet. 2006;168:44–9. doi: 10.1016/j.cancergencyto.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Gad S, Caux-Moncoutier V, Pages-Berhouet S, et al. Significant contribution of large BRCA1 gene rearrangements in 120 French breast and ovarian cancer families. Oncogene. 2002;21:6841–7. doi: 10.1038/sj.onc.1205685. [DOI] [PubMed] [Google Scholar]
- 31.de la Hoya M, Gutierrez-Enriquez S, Velasco E, et al. Genomic rearrangements at the BRCA1 locus in Spanish families with breast/ovarian cancer. Clin Chem. 2006;52:1480–5. doi: 10.1373/clinchem.2006.070110. [DOI] [PubMed] [Google Scholar]
- 32.Thomassen M, Gerdes AM, Cruger D, Jensen PK, Kruse TA. Low frequency of large genomic rearrangements of BRCA1 and BRCA2 in western Denmark. Cancer Genet Cytogenet. 2006;168:168–71. doi: 10.1016/j.cancergencyto.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Woodward AM, Davis TA, Silva AG, Kirk JA, Leary JA. Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. J Med Genet. 2005;42:e31. doi: 10.1136/jmg.2004.027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann C, John AL, Klaes R, et al. Large BRCA1 gene deletions are found in 3% of German high-risk breast cancer families. Hum Mutat. 2004;24:534. doi: 10.1002/humu.9291. [DOI] [PubMed] [Google Scholar]
- 35.Anonymous. The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. The BRCA1 Exon 13 Duplication Screening Group. Am J Hum Genet. 2000;67:207–12. [PMC free article] [PubMed] [Google Scholar]
- 36.Gutierrez-Enriquez S, de La Hoya M, Martinez-Bouzas C, et al. Screening for large rearrangements of the BRCA2 gene in Spanish families with breast/ovarian cancer. Breast Cancer Res Treat. 2006 doi: 10.1007/s10549-006-9376-8. [DOI] [PubMed] [Google Scholar]
- 37.Casilli F, Tournier I, Sinilnikova OM, et al. The contribution of germline rearrangements to the spectrum of BRCA2 mutations. J Med Genet. 2006;43:e49. doi: 10.1136/jmg.2005.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agata S, Dalla Palma M, Callegaro M, et al. Large genomic deletions inactivate the BRCA2 gene in breast cancer families. J Med Genet. 2005;42:e64. doi: 10.1136/jmg.2005.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahti-Domenici J, Rapakko K, Paakkonen K, et al. Exclusion of large deletions and other rearrangements in BRCA1 and BRCA2 in Finnish breast and ovarian cancer families. Cancer Genet Cytogenet. 2001;129:120–3. doi: 10.1016/s0165-4608(01)00437-x. [DOI] [PubMed] [Google Scholar]
- 40.Barcenas CH, Hosain GM, Arun B, et al. Assessing BRCA carrier probabilities in extended families. J Clin Oncol. 2006;24:354–60. doi: 10.1200/JCO.2005.02.2368. [DOI] [PubMed] [Google Scholar]
- 41.Hendrickson BC, Judkins T, Ward BD, et al. Prevalence of five previously reported and recurrent BRCA1 genetic rearrangement mutations in 20,000 patients from hereditary breast/ovarian cancer families. Genes Chromosomes Cancer. 2005;43:309–13. doi: 10.1002/gcc.20189. [DOI] [PubMed] [Google Scholar]
- 42.Rohlfs EM, Puget N, Graham ML, et al. An Alu-mediated 7.1 kb deletion of BRCA1 exons 8 and 9 in breast and ovarian cancer families that results in alternative splicing of exon 10. Genes Chromosomes Cancer. 2000;28:300–7. doi: 10.1002/1098-2264(200007)28:3<300::aid-gcc8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 43.Ward BD, Hendrickson BC, Judkins T, et al. A multi-exonic BRCA1 deletion identified in multiple families through single nucleotide polymorphism haplotype pair analysis and gene amplification with widely dispersed primer sets. J Mol Diagn. 2005;7:139–42. doi: 10.1016/S1525-1578(10)60020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrois M, Bieche I, Mazoyer S, Champeme MH, Bressac-de Paillerets B, Lidereau R. Real-time PCR-based gene dosage assay for detecting BRCA1 rearrangements in breast-ovarian cancer families. Clin Genet. 2004;65:131–6. doi: 10.1111/j.0009-9163.2004.00200.x. [DOI] [PubMed] [Google Scholar]
- 45.Borg A, Dorum A, Heimdal K, Maehle L, Hovig E, Moller P. BRCA1 1675delA and 1135insA account for one third of Norwegian familial breast-ovarian cancer and are associated with later disease onset than less frequent mutations. Dis Markers. 1999;15:79–84. doi: 10.1155/1999/278269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudkin TM, Hamel N, Galvez M, et al. The frequent BRCA1 mutation 1135insA has multiple origins: a haplotype study in different populations. BMC Med Genet. 2006;7:15. doi: 10.1186/1471-2350-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallon-Christersson J, Cayanan C, Haraldsson K, et al. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Molec Genet. 2001;10:353–60. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh T, Casadei S, Coats KH, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–88. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 49.Mazoyer S. Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum Mutat. 2005;25:415–22. doi: 10.1002/humu.20169. [DOI] [PubMed] [Google Scholar]
- 50.Kauff ND, Perez-Segura P, Robson ME, et al. Incidence of non-founder BRCA1 and BRCA2 mutations in high risk Ashkenazi breast and ovarian cancer families. J Med Genet. 2002;39:611–4. doi: 10.1136/jmg.39.8.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phelan CM, Kwan E, Jack E, et al. A low frequency of non-founder BRCA1 mutations in Ashkenazi Jewish breast-ovarian cancer families. Hum Mutat. 2002;20:352–7. doi: 10.1002/humu.10123. [DOI] [PubMed] [Google Scholar]
- 52.Evans DG, Lalloo F, Wallace A, Rahman N. Update on the Manchester Scoring System for BRCA1 and BRCA2 testing. J Med Genet. 2005;42:e39. doi: 10.1136/jmg.2005.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antoniou AC, Pharoah PP, Smith P, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–90. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eng C, Brody LC, Wagner TM, et al. Interpreting epidemiological research: blinded comparison of methods used to estimate the prevalence of inherited mutations in BRCA1. J Med Genet. 2001;38:824–33. doi: 10.1136/jmg.38.12.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capalbo C, Ricevuto E, Vestri A, et al. BRCA1 and BRCA2 genetic testing in Italian breast and/or ovarian cancer families: mutation spectrum and prevalence and analysis of mutation prediction models. Ann Oncol. 2006;17(Suppl 7):vii34–vii40. doi: 10.1093/annonc/mdl947. [DOI] [PubMed] [Google Scholar]