Abstract
Hepatoma Derived Growth Factor (HDGF) is a nuclear protein with both mitogenic and angiogenic activity, it is highly expressed in the developing heart and vasculature. To date the mechanisms of HDGF’s function are unknown. Oligonucleotide microarray analysis was used to gain insights into HDGF function. Adenoviral expression of HDGF significantly (≥ 2 fold) downregulated a large group (66) of genes, and increased expression of a relatively small number of genes (9). Two groups of target genes which are involved in cardiovascular development and transcriptional regulation were validated by real time PCR, including the skeletal/cardiac muscle specific SET and MYND domain containing 1 (SMYD1) gene. This suggested that HDGF could function as a transcriptional repressor. In a one-hybrid system, GBD-HDGF significantly repressed reporter gene activity in a dose dependent manner. This demonstrated that HDGF has transcriptional repressive activity. Moreover, in G-7 myoblast cells, overexpression of a GFP-HDGF fusion specifically downregulated SMYD1 mRNA expression and the activity of the human SMYD1 promoter. HDGF repressed SMYD1 gene transcription through interaction with a transcriptional corepressor C-terminal binding protein (CtBP). Overexpressing of CtBP potentiated the trans-repressive activity of HDGF; on the other hand, knocking down CtBP attenuated the trans-repressive effect of HDGF. HDGF binds CtBP through a non-canonical binding motif (PKDLF) within the PWWP domain, as substitutional mutation of DL to AS abolished HDGF and CtBP interaction and diminished the trans-repressive effect of HDGF without affecting DNA binding. Finally, fluorescent microscopy studies showed that HDGF induced the nuclear accumulation of CtBP suggesting that HDGF forms a transcriptional complex with CtBP. Taken together, our data demonstrate that HDGF functions as a transcriptional repressor of the SMYD1 gene, through interaction with the transcriptional corepressor CtBP. Because of moderate conservation of the CtBP binding motif in HDGF family members, trans-repressive activity mediated by CtBP may be a common function among HDGF proteins.
Keywords: HDGF, SMYD1, CtBP, transcription, repressor
Introduction
HDGF is a developmentally regulated cardiovascular mitogen that is also re-expressed in vascular smooth muscle cells in vivo after vascular injury and in human atherosclerosis,1 suggesting an important role in cardiovascular development and human cardiovascular diseases. Originally HDGF was purified from the conditioned media of the human hepatoma cell line, HuH-7.2 HDGF appears to be a secreted protein in vitro, it is present in the conditioned media of HuH-7 cells, 2 mesenchymal kidney cells and COS-7 cells expressing recombinant HDGF.3 Although HDGF is a nuclear protein with nuclear targeting required for its mitogenic activity,4,5 the nuclear function of HDGF remains obscure.
From the primary structure, HDGF shows 32% homology with high mobility group-1 (HMG-1) but lacks an “HMG box”, which is responsible for DNA binding.2 Instead HDGF and its related proteins, HDGF related protein (HRP) 1, 2, 3, 4 and lens epithelium derived growth factor (LEDGF) contain a poorly characterized N-terminal PWWP domain. Although the function of the PWWP domain is unclear, from evolutional analysis, it was suggested that the PWWP domain may be involved in protein-protein and protein-DNA interactions.6,7 This notion was supported recently by NMR structural analysis of the HDGF and HRP3 PWWP domains.8,9 In addition, Qiu et al found that the PWWP domain of the DNA methyltransferase Dnmt3b, can bind to DNA.10-12 Our recent study has shown that HDGF binds a DNA element located in the promoter of SMYD1 gene specifically both in vitro and in vivo, and the PWWP domain was responsible for DNA binding.13
In the present study, we demonstrated that HDGF functions as a transcriptional repressor for the SMYD1 gene which is an essential gene for cardiogenesis.14 HDGF interacts with a transcriptional corepressor CtBP through a non-canonical binding motif (PKDLF). This motif is located within the PWWP domain and is variable in HDGF family proteins. CtBP mediates the transcriptional regulation by HDGF, since overexpressing of CtBP enhanced and knocking down of CtBP attenuated the trans-repressive effect of HDGF. A substitutional mutant of the binding motif abolished the transcriptional repression of HDGF. The fluorescent microscopy study showed that overexpression of HDGF induced the nuclear accumulation of CtBP, implying that HDGF and CtBP formed a transcriptional complex.
Results
Gene profiling in HDGF expressing SMC
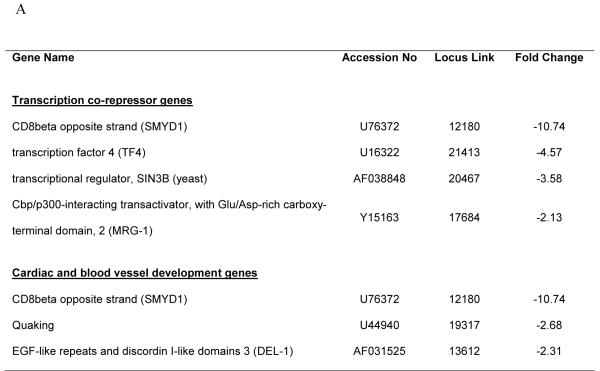
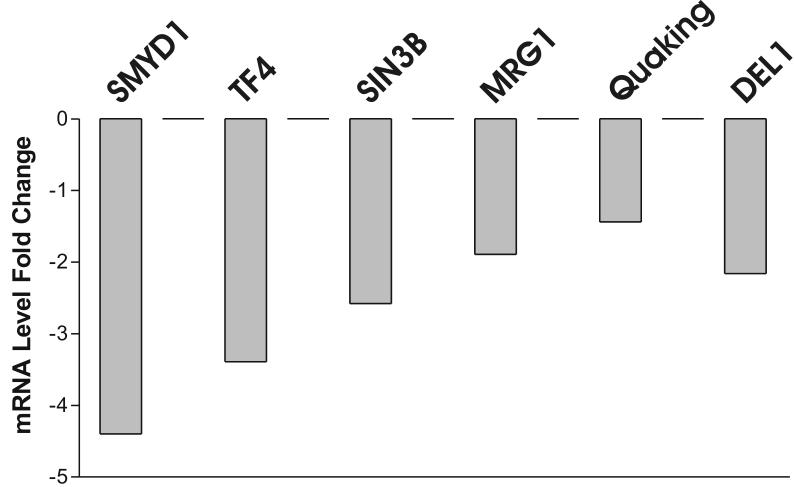
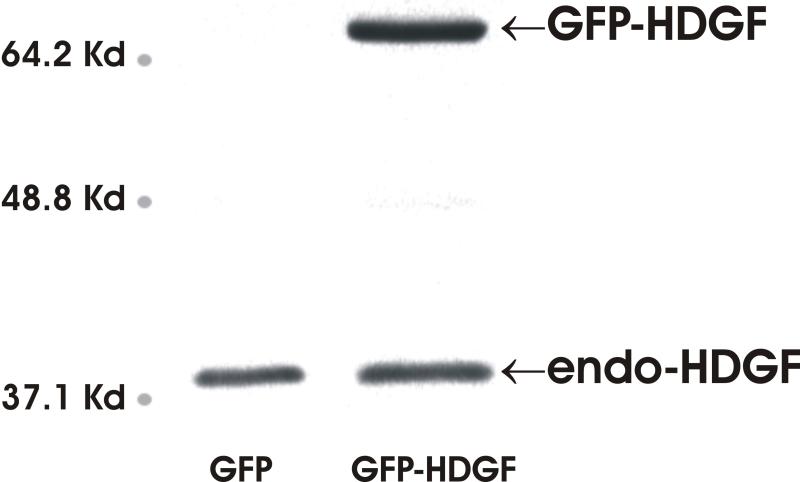
HDGF is mitogenic for fibroblasts, HuH-7 cells,2 aortic endothelial cells 3 and vascular smooth muscle cells.1 Recently HDGF was shown to be an angiogenic factor for pulmonary endothelial cells 15 and also involved in tumorgenesis,16,17 but the mechanisms remain largely unknown. We carried out oligonucleotide microarray analysis to study HDGF gene profiling in mouse primary aortic vascular smooth muscle cells infected with adenoviruses overexpressing a GFP-HDGF fusion or GFP as control for 18 hours. As a result, 75 genes demonstrated significant mRNA levels change (≥ 2 fold) (supplemental data). Surprisingly, 66 (88%) genes were decreased with only 9 genes upregulated by HDGF. Among them, many genes were potential target genes responsible for transcriptional regulation, cell cycle control, cell growth and differentiation etc (supplemental data). Analysis demonstrated two significant subgroups, a group of 3 genes (SMYD1, Quaking and DEL-1) necessary for normal cardiovascular development, and 4 genes (SMYD1, TF4, SIN3b and MRG-1) that are transcriptional co-repressors (Figure 1A). Down regulation of the listed 6 genes by HDGF overexpression was confirmed by Real Time PCR (Figure 1B) in vascular smooth muscle cells. A representative Western blot for HDGF is shown and demonstrated that GFP-HDGF was expressed 2-fold higher than endogenous HDGF (Figure 1C) in the microarray and validation experiments. Since HDGF is a nuclear protein, this result implies that HDGF could function predominately as a transcriptional repressor with specific activity for critical cardiovascular genes.
Figure 1.
Representative genes downregulated by HDGF. (A) Classified microarray results shown that a group of 4 genes serve as transcription co-repressor and a group of 3 involved in cardiac and blood vessel development. (B) Real Time PCR confirmed the microarray result. Mouse smooth muscle cells were infected with GFP-HDGF adenovirus and GFP virus as control for 18 hours, total RNA was extracted and reverse transcripted. Real Time PCR analysis was performed using gene specific primers for 7 genes listed in Table 1. Bar graph shows mRNA levels change. (C) Western blot using HDGF polyclonal antibody shows the expression of endogenous HDGF and GFP-HDGF fusion.
HDGF represses a one-hybrid reporter gene
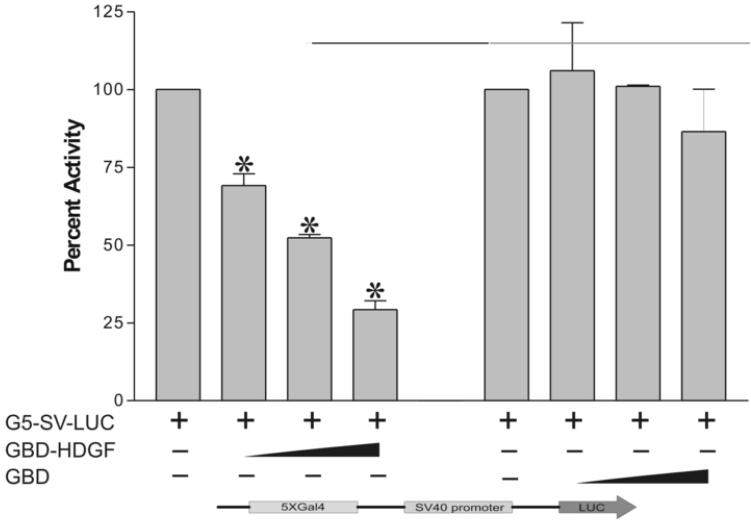
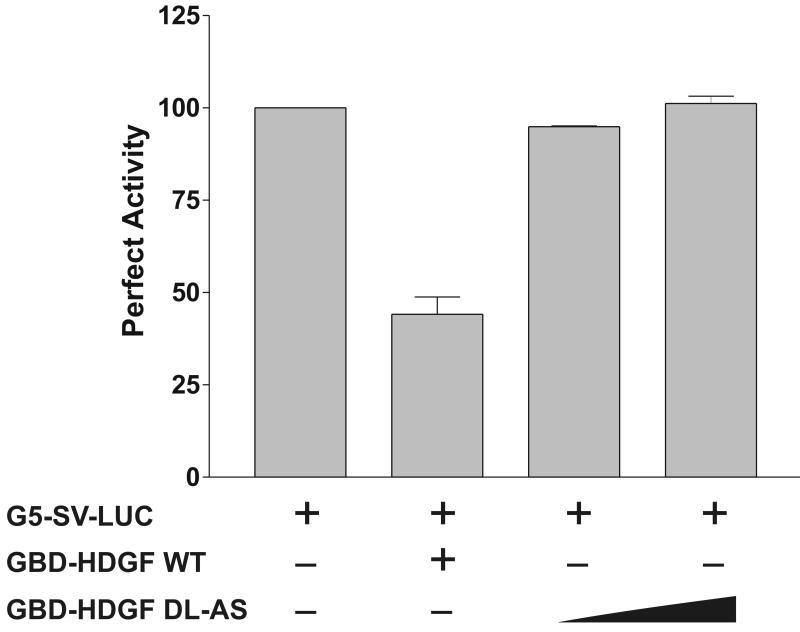
Based on our microarray results, we tested if HDGF functions as a transcriptional modulator. A one-hybrid reporter construct was created by inserting five tandem copies of the Gal4-DNA binding sites to the upstream of a heterologous SV40 promoter which controls a luciferase gene in the pGL3 control vector (G5-SV-LUC). The reporter gene was cotransfected into G-7 cells with the expression plasmid encoding the Gal4-DNA binding domain fused to HDGF (GBD-HDGF). Cotransfection of GBD-HDGF with the G5-SV-LUC reporter plasmid markedly repressed the reporter gene activity up to 76% in a dose-dependent manner (Figure 2), indicating that HDGF contains a transcriptional repressive domain.
Figure 2.
HDGF represses the expression of a one-hybrid reporter gene. The reporter gene G5-SV-LUC (Firefly luciferase 0.5 ug), pRL-CMV (Renilla luciferase 50 ng) and GBD-HDGF (0.5, 1, and 2 ug in left panel), or GBD (0.5, 1 and 2 ug in right panel) alone were cotransfected into G-7 cells. The dual-luciferase reporter assay system was used to measure the luciferase activities 24 hours after transfection. Percentage of firefly luciferase activity was calculated when luciferase activity with reporter gene alone was set at 100%. Firefly luciferase activity was normalized with constitutive renilla luciferase activity to correct transfection efficiency. Bars indicate mean ± SEM from 3 separate transfection assays with duplicate plates, *P < 0.05 by one-way ANOVA control versus GBD-HDGF.
This repression was specific to HDGF, because expression of the GBD alone did not affect reporter gene activity (Figure 2); whereas, HDGF without the Gal4 DNA binding domain, could not repress the luciferase reporter activity (data not shown). In addition, the positive control plasmid, Gal4-DBD fused to a VP16 activation domain (GBD-VP16), activated the reporter gene about 15-fold as compared with the reporter gene alone (data not shown), demonstrating that the G5-SV-LUC reporter was functional. All of these results indicate that HDGF can function as a transcriptional repressor.
HDGF represses SMYD1 gene expression
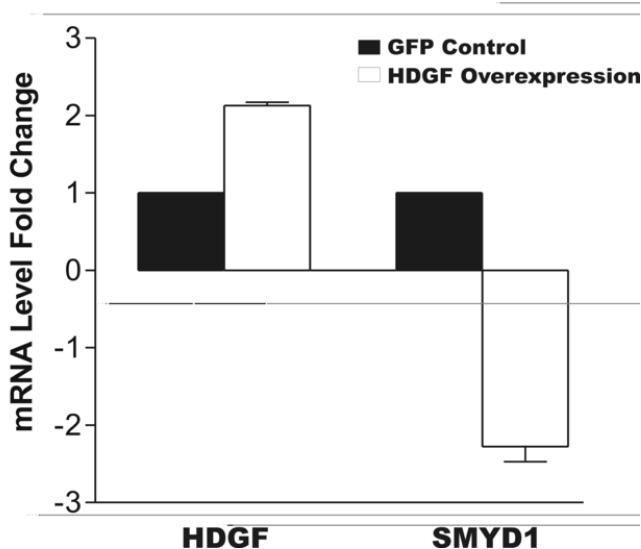
We chose the SMYD1 gene from our microarray studies to investigate the nuclear function of HDGF because of its significance to cardiovascular development. Since SMYD1 highly expressed in cardiac and skeletal muscle,11,18 we used G-7 myoblast cells (a skeletal muscle cell line) to validate the microarray result. With adenoviral expression of a GFP-HDGF fusion, real-time PCR demonstrated a 2 fold increase in HDGF mRNA that was associated with a 2.4 fold decrease in SMYD1 mRNA (Figure 3A). As a control, adenoviral expression of GFP did not significantly affect the expression of either HDGF or SMYD1.
Figure 3.
HDGF represses the expression of SMYD1 gene. A, G-7 cells were infected with 100 MOI adenovirus expressing GFP-HDGF fusion or GFP for 18 hours, total RNA were extracted and reverse transcribed for real-time PCR analysis. Gene specific primers of HDGF and SMYD1 were used to detect their mRNA levels. Specific primers amplifying the mouse TATA Box Binding Protein (TBP) were used to normalize gene expression levels. Bars indicate mean ± SEM from 2 separate experiments with triplicate assays. B, 1 ug pGL3-enhancer vector as CONT-LUC or SMYD1 promoter reporter SMYD1-LUC were cotransfected with 50 ng pRL-CMV with or without 3ug HDGF expression construct into G-7 cells. The dual-luciferase reporter assay system was used to measure the luciferase activities 24 hours after transfection. Percentage of firefly luciferase activity was calculated when SMYD1-LUC luciferase activity without HDGF was set at 100%. Firefly luciferase activity was normalized with constitutive renilla luciferase activity to correct transfection efficiency. Bars indicate mean ± SEM from 3 separate transfection assays with duplicate plates.
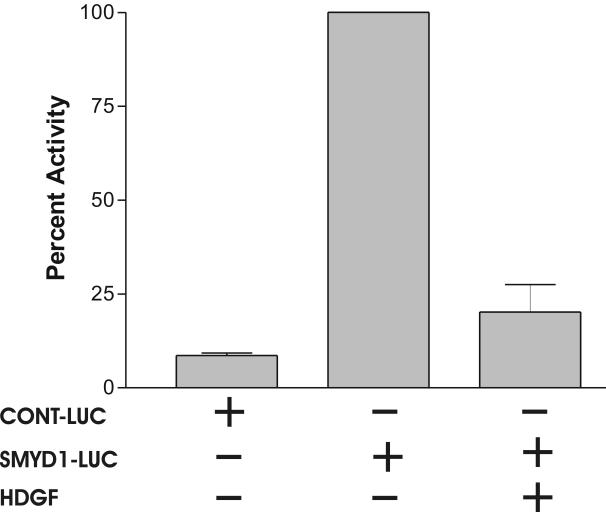
To further study the transcriptional regulation by HDGF, we PCR amplified a human SMYD1 gene promoter (-1084 to +37, +1 is the start codon) fragment, and inserted it upstream of the luciferase gene in the pGL3 enhancer vector to generate a SMYD1-LUC reporter. To test the SMYD1-LUC reporter for promoter activity, G-7 cells were transiently transfected with the empty pGL3 enhancer vector as a control. When expressed in G-7 cells, the SMYD1 promoter activated luciferase about 12 folds higher than the non-promoter control (Figure 3B). Importantly, the luciferase activity was decreased ∼80% when SMYD1-LUC was cotransfected with an HDGF expression vector in G-7 cells (Figure 3B). These results indicate that HDGF is a transcriptional repressor for the SMYD1 gene.
Association of CtBP with HDGF in vitro and in vivo
To study the mechanism of HDGF transcriptional repression, we identified that the HDGF N-terminal PWWP domain contained a non-canonical binding motif (aa 60PKDLF, conserved amino acids are underlined) for the transcriptional co-repressor CtBP. CtBP mediates repression by binding to transcription factors through a consensus PXDLS motif.19-22 HDGF protein family has 6 members, they are all nuclear proteins with different expression patterns, but the functions are largely unknown, except that the LEDGF was identified as a weak transcription coactivator. The PWWP domain in all 6 HDGF protein family members is highly conserved, however alignment analysis showed that the PKDLF motif is variable (table 2). HRP 2 and 3 have identical non-canonical CtBP binding motif as HDGF whereas LEDGF, HRP 1 and 4 are more variable.
Table 2.
Alignment of PWWP domains in 6 HDGF family members
 |
Note: PKDLF motifs were underlined
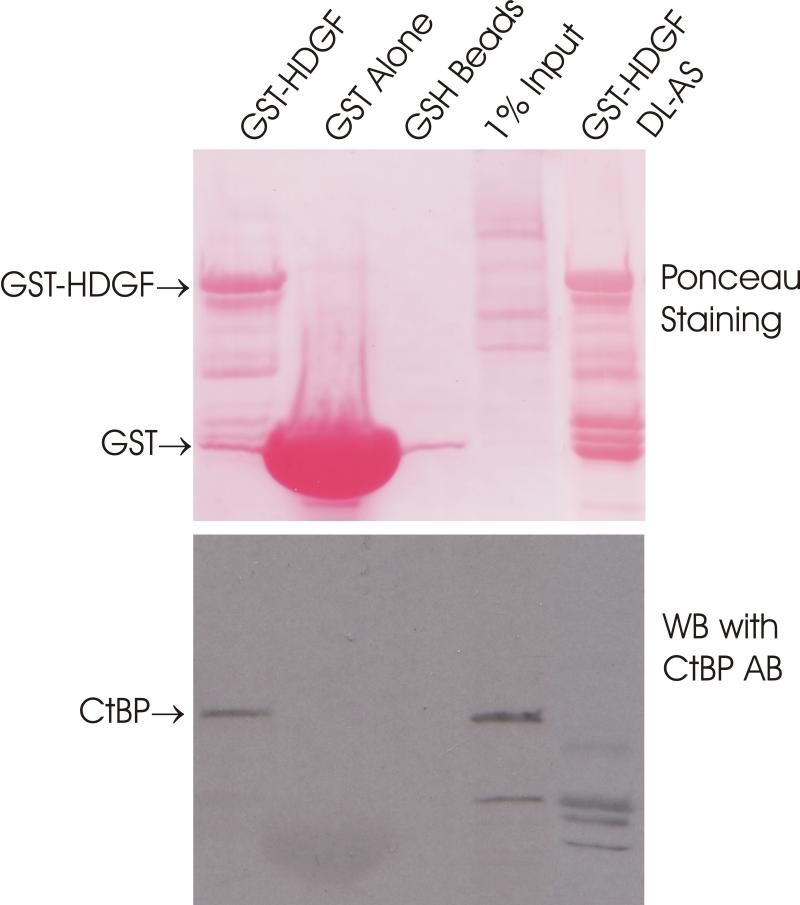
We used in vitro GST-HDGF pull down of MCF7 cell extracts to determine whether CtBP interacts with HDGF. As shown in Figure 4A, a GST-HDGF fusion protein binds CtBP whereas neither GST protein alone nor empty GSH beads could bind CtBP.
Figure 4.
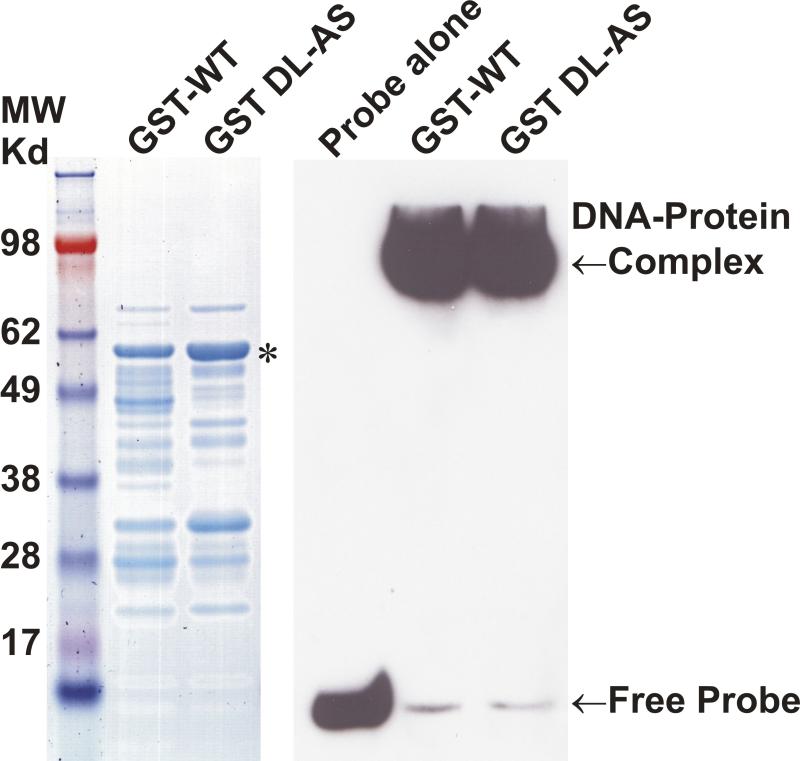
HDGF interacted with CtBP. A, GST-HDGF fusion proteins (lane 1, wild type, lane 5, DL-AS mutant) were conjugated to glutathione-agarose beads and incubated with whole cell lysate of MCF-7 cells, as described under “Experimental Procedures.” After washing and recovery of the beads, associated proteins were resolved by SDS-PAGE and analyzed by western blot with anti-CtBP antibody. Top panel shows Ponceau staining of the nitrocellulose membrane. B, Endogenous HDGF or CtBP were immunoprecipitated from MCF-7 cell lysates with either monoclonal anti-HDGF antibody, or monoclonal anti-CtBP antibody, the immunoprecipitated protein complexes were detected by immunoblotting with Rabbit polyclonal anti-HDGF or anti-CtBP antibodies. C, Flag-CtBP was co-expressed with either HA-HDGF wild type or DL-AS mutant form in MCF-7 cells, co-immunoprecipitation were done by either IP with HA antibody, western blotting with Flag antibody, or IP with Flag antibody, western blotting with HA antibody. Western blotting with HA and Flag antibodies for whole cell lysate were also shown. D, 62DL-AS mutant HDGF binds to DNA. 0.25 pmol 5′ end biotin labeled probes for SMYD1 promoter (nucleotides -688 to -609) were incubated with 1 ug GST-HDGF fusion protein (wild type or 62DL-AS mutant) in the reaction buffer on ice for 30 min. The complex were resolved in 5% PAGE and transferred onto a nylon membrane. The biotin end-labeled DNA probe is detected using a streptavidin-horseradish peroxidase conjugate and chemiluminescent substrate. The left panel shows the coomassie staining of the fusion proteins, star indicates the full length of HDGF fusion proteins. E, 62DL-AS mutant does not have the trans-repressive activity. Wild type (2ug) or 62DL-AS mutant forms of GBD-HDGF expression constructs (1 and 2 ug) were cotransfected with G5-SV-LUC (0.5 ug) and pRL-CMV (50 ng), dual-luciferase assay was performed as described in Figure 2.
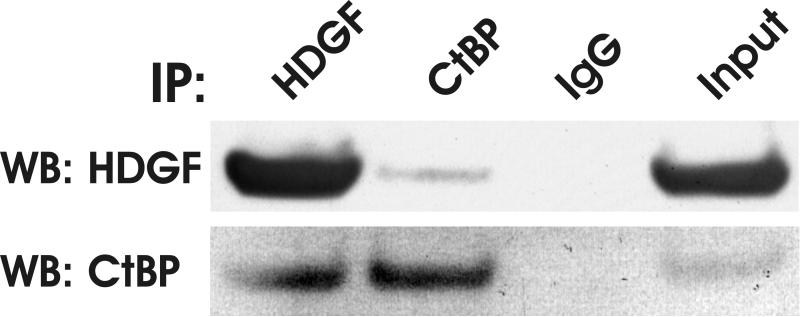
The possible interaction of CtBP with HDGF in vivo was tested further by co-immunoprecipitation (co-IP). As shown in Figure 4B, endogenous HDGF can be detected in an immunocomplex which was IPed with a CtBP monoclonal antibody. Similarly, endogenous CtBP can be detected in an immunocomplex which was IPed with a HDGF monoclonal antibody. Co-immunoprecipitation with overexpressed HA-HDGF and Flag-CtBP showed the same result (Figure 4C). Taken together, these experiments demonstrated that HDGF interacted with CtBP.
To determine whether HDGF binds to CtBP through the 60PKDLF motif, we mutated amino acids 62DL to AS to disrupt the potential CtBP binding motif in HDGF. As a result, the 62DL-AS mutation totally abolished HDGF binding with CtBP using the GST pull down assay (Figure 4A, lane 5). The diminished interaction was also proved by co-immunoprecipitation assay of tagged HDGF and CtBP (Figure 4C, lane 3).
In our previous study, we found that HDGF binds to a DNA element which is located in the SMYD1 gene promoter via the HDGF N-terminal PWWP domain.13 Since DNA binding is essential for transcriptional regulation, we tested whether the DL-AS mutant altered the DNA binding activity of HDGF. As shown in Figure 4D, in an EMSA assay, the 62DL-AS mutant form of a GST-HDGF fusion protein bound the SMYD1 promoter DNA binding sequence as tightly as wild type HDGF.
To test the functional importance of the HDGF 62DL-AS mutant, we subcloned the HDGF 62DL-AS substitutional mutant into the pM vector to express a GBD fusion for transcription assays. As shown in Figure 4E, compared with wild type HDGF, the 62DL-AS mutant totally lost the transcriptional repressive activity. This data demonstrated that CtBP plays an important role in the transcriptional regulation by HDGF.
CtBP mediates the trans-repressive activity of HDGF
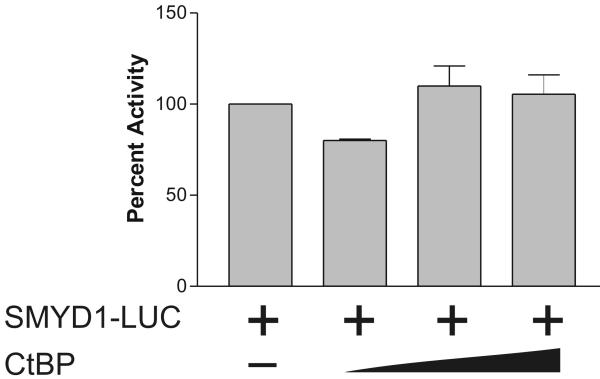
CtBP functions as a transcriptional corepressor, 21 to test the functional effect of the interaction of CtBP with HDGF in G-7 cells, co-transfection of Flag-CtBP was used with the GBD-HDGF one hybrid reporter gene system. As shown in Figure 5A, GBD-HDGF alone represses the luciferase reporter, whereas, when CtBP was co-expressed, the repressive effect of HDGF was potentiated in a dose dependent manner. As a control, without GBD-HDGF, CtBP alone had no effect on either one-hybrid reporter gene activity (Figure 5A, right panel) or SMYD1 promoter activity (Figure 5B). These experiments indicate that the HDGF-CtBP complex is functional.
Figure 5.
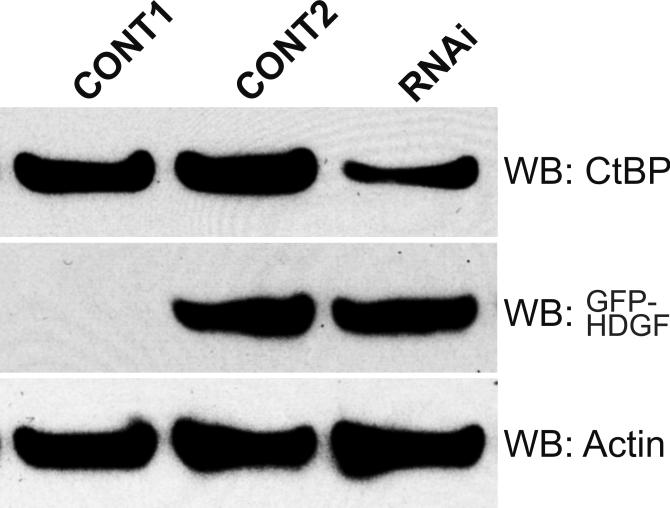
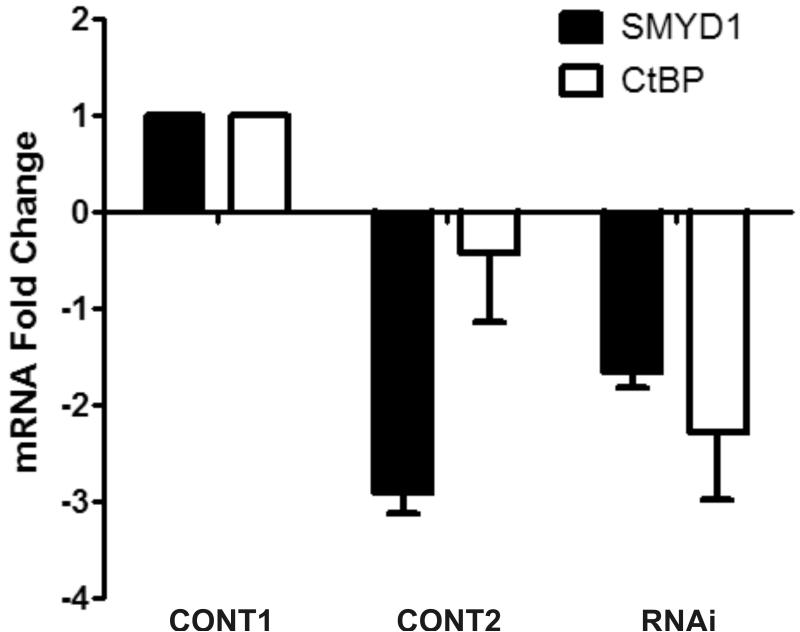
CtBP mediated the trans-repressive activity of HDGF. A, Reporter gene construct G5-SV-LUC (0.5 ug) and pRL-CMV (50 ng) were cotransfected with (left panel) or without GBD-HDGF (right panel) and increasing amounts of CtBP expression construct (0.5, 1, 2 ug) as indicated in the figure in G7 cells, the dual-luciferase assay was performed as described in Fig 2. *P < 0.05 by one-way ANOVA control versus CtBP (Left panel). B, CtBP alone does not repress SMYD1 gene promoter activity. SMYD1 promoter reporter SMYD1-LUC (1ug) was cotransfected with 50 ng pRL-CMV and increasing amount of CtBP expression construct (1, 2, 3 ug) into G-7 cells. The dual-luciferase reporter assay was performed as described in Fig 3. C, HDGF expression constructs (2ug) were cotransfected with 0, 3 or 4 ug Flag-CtBP in G7 cells for 48 hours, real-time PCR were performed as described in Fig 3 to measure the mRNA levels change of SMYD1 gene. D, 1 ug GFP-HDGF was cotransfected with 1 ug of either conrol or specific miR targeting CtBP expression vectors into MCF-7 cells which cultured in 6 wells plate using lipofectamine 2000 reagent. 72 hours after transfection, the protein lysate were subjected to SDS-PAGE and further western blotting using specific antibodies against CtBP, GFP or Actin as control. Control 1 used empty GFP vector instead of GFP-HDGF. E, Total RNA extracted from MCF-7 cells described in 4D was subjected to real-time PCR. The mRNA levels of both SMYD1 and CtBP in control 1 (GFP vector plus control miR vector) were set to 1.
CtBP also enhanced the repressive effect of HDGF on SMYD1 gene transcription directly. As shown in Figure 5C, overexpressing of HDGF alone downregulated SMYD1 gene transcription, whereas when CtBP was co-expressed, the mRNA level of SMYD1 gene was decreased further as compared with overexpressing of HDGF alone.
To test whether CtBP is essential for the trans-repressive activity of HDGF, we used RNAi strategy to knock down the endogenous CtBP protein level about 70% in MCF-7 cells (Figure 5D). When overexpressing HDGF in MCF-7 cells in the presence of control RNAi reagent, the SMYD1 RNA decreased 3 fold. However, in the presence of RNAi specific for CtBP, the trans-repressive effect of HDGF was dramatically attenuated (Figure 5E) to only 1.8 fold. This experiment strongly indicated that CtBP mediated the transcriptional repression of HDGF.
HDGF induced CtBP accumulation in the nucleus
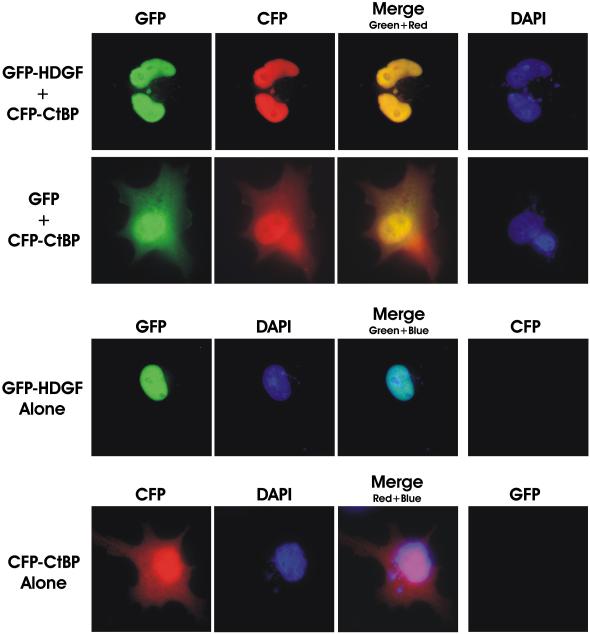
CtBP was first identified as a cellular phosphoprotein that binds to the C-terminal of the human adenovirus E1A proteins,19,23 subsequent studies found that CtBP functions as a transcriptional corepressor and localizes to both the nucleus and cytoplasm.24-26 CtBP can translocate to the nucleus by binding to PXDLS motif containing partner proteins, such as a transcription factor BKLF.27 We therefore examined whether the nuclear protein HDGF can direct CtBP to the nuclear compartment. As shown in the representative fluorescent micrographs in Figure 6, expression of a CFP-CtBP fusion was diffusely distributed to both nuclear and cytoplasmic compartments. However, in the presence of coexpressed GFP-HDGF, almost all of the CFP-CtBP translocated to nucleus. As a control, coexpressing GFP alone did not change the diffuse distribution pattern of CtBP (Fig. 6). Taken all together HDGF alters the distributive pattern of CtBP in the cells strongly suggesting that HDGF and CtBP formed a transcriptional complex in the nuclei.
Figure 6.
HDGF induced the nuclear accumulation of CtBP. MCF-7 cells grown on cover slips were transfected with 0.5 ug of CFP-CtBP and GFP-HDGF (top panel) or GFP (2nd panel) for 48 hours, cells were fixed in 1% paraformaldehyde and the cover slips were mounted in DAPI containing medium, DAPI, CFP and GFP images are shown, with the merged images of CFP and GFP fluorescence (overlay). MCF-7 cells transfected with GFP-HDGF alone (3rd panel) or CFP-CtBP alone (bottom panel) was used as control. The merged image of GFP-HDGF (green) and DAPI (blue) in 3rd panel is showing the nuclear localization of GFP-HDGF. The image of the bottom panel is showing the diffused distributive pattern of CFP-CtBP in both nucleus and cytoplasm.
Discussion
Growth factors play a fundamental role in embryogenesis and regeneration. HDGF 2 represents the original member of a new family of growth factors which now includes HRP-1,28 HRP-2,29 HRP-3,30 p52/75/LEDGF 31 and HRP-4.32 They have a highly conserved N-terminal PWWP domain, termed homologous with the amino terminus of HDGF (hath) region, but diverse C-termini. They are all nuclear proteins and have mitogenic activity; with the addition that LEDGF was identified as a weak transcriptional coactivator.33 The function of HDGF and the other family members are largely unknown. Although the expression pattern of HDGF has suggested possible roles in development,34 vascular injury 1 and cancer,16,17,35-37 the mechanism of function of HDGF in these processes is unknown. In this study, we discovered that HDGF transcriptionally represses SMYD1 gene expression by forming a transcriptional complex with the corepressor CtBP.
HDGF has a PWWP domain at its N-terminal.2 The PWWP domain was characterized by a region of sequence similarity of about 90 amino acids containing a highly conserved PWWP motif (90 residues containing the Pro-Trp-Trp-Pro core). It has been found in more than 60 eukaryotic proteins.38 PWWP was thought homologous with chromatin-binding (chromo) and malignant brain tumor (MBP) domains because they originate from a common ancestor.39 Interestingly, Chromo, MBP and most PWWP domain containing proteins are involved in chromatin function.39,40 For example, BS69,41 ICO4,42 Dnmt3a and Dnmt3b43 were all found to function as transcriptional repressors. The PWWP domain may play a key role in directing DNA binding. Recently, in Dnmt3b the PWWP domain was identified as a novel chromatin/chromosome targeting module 12 and it can bind to DNA directly.10 In our recent study, we found that HDGF binds to SMYD1 gene promoter specifically,13 combined with the finding in the present study that HDGF functions as a transcriptional repressor, our data is clearly in accord with previous PWWP studies.
Transcriptional repression is an important mechanism in the regulation of gene expression. Gene specific repressors mediate their effect by binding to a target gene promoter, then recruiting corepressors. Corepressor CtBP was first identified as a cytoplasmic protein which bound to the C-terminal adenovirus E1A protein.23 CtBP has been shown to play a significant role in the regulation of cellular genes involved in growth and differentiation.21 Studies in Drosophila show that CtBP has an essential role in the tissue specific expression of genes in the embryo.44 The mechanism for transcriptional repression by CtBP is still unclear, however the repression effect is mediated in both histone deacetylase-dependent and independent manner.21 CtBP interacts with many transcriptional factors through an optimal PXDLS consensus motif.19-22 In our study, we found that HDGF contains a non-canonical PXDLS (PKDLF) motif in the N-terminal PWWP domain and binds CtBP. Mutation of the critical CtBP binding residues DL to AS abolish CtBP and HDGF interaction and diminish HDGF transcriptional repression activity. This identifies HDGF as a CtBP binding protein via a functional PKDLF CtBP binding motif. The PKDLF motif is present but non-canonical in all 6 known HDGF proteins. Besides HDGF, only HRP 2 and 3 have identical motifs. This indicated that the 6 HDGF members may have different mechanisms of functions, which is consistent with the diverse expression pattern of HDGF family members. Whether HRP 2 and 3 interact with and function through CtBP still needs to be determined. However the discovery of HDGF and CtBP binding and a non-canonical CtBP binding domain in all HDGF proteins opens up new insights into the function of the whole protein family.
HDGF is a developmentally regulated, nuclear targeted mitogen, and nuclear translocation is required for its mitogenic activity.2,4,5 In the present study, we have determined that HDGF is a transcriptional repressor. Most mitogens, exemplified by PDGF, increase the expression of downstream genes and other growth factors such as FGF to further amplify the proliferative effect. However, HDGF appears to repress unique subsets of genes that regulate terminal cardiovascular differentiation such as SMYD1. This suggests that HDGF’s mitogenic effect may be based on preventing some aspects of terminal differentiation, including repression of pathways that result in a differentiated cell phenotype that is no longer proliferating or migrating. In support HDGF is broadly expressed in the media of the developing aorta, but not in the adult aorta.1 Similarly, HDGF expression is not detectable in the carotid artery of the adult rat but is re-expressed after vascular injury.1 In the developing gut, we have shown that expression of a GFP-HDGF fusion protein resulted in the decreased expression of a panel of terminal differentiation markers.45 Suggesting that in the gut, expression of HDGF blocks terminal differentiation of the gut epithelia. Taken together, these data suggest that HDGF could serve as a conserved developmental pathway, which can be re-activated with injury to affect the developmental state of cardiovascular and possibly cancer cells.
In summary, we have discovered that HDGF’s nuclear mechanism of action is as a transcriptional repressor, it specifically represses SMYD1 gene expression, and likely other critical cardiovascular genes, by recruiting corepressor CtBP through a PKDLF motif in the PWWP domain. Collectively our data provide the first evidence for the nuclear function of HDGF.
Materials and Methods
Plasmids Construction
Flag-hCtBP-pcDNA3 is a gift from Dr. R. Goodman (Oregon Health Sciences University). CFP-CtBP is a gift from Dr. David Wotton (University of Virginia). HDGF expression constructs (pK7-GFP-HDGF and pKH3-HDGF) were described before.4 One hybrid reporter G5-SV-LUC was constructed by inserting 5 tandem copies of Gal4 DNA binding sites (from pBind vector, Promega) upstream of SV-40 promoter in pGL3-control vector (Promega). Human HDGF was PCR amplified from IMAGE clone 5587366 (ATCC) and subcloned in frame with Gal4 DNA binding domain in pM vector (Clontech). The primers were derived from the same IMAGE clone sequence (Forward 5′-ATGGGATCCGTTCGCGTCCAACCGGCAG-3′, Reverse 5′-TATTCTAGACTACAGGCTCTCATAGATCTCTG-3′), 5′ ends were introduced into appropriate restriction enzyme sites. The promoter sequence of the human SMYD1 gene was derived from The UCSC Genome Browser (http://genome.ucsc.edu/), primers (Forward 5′-AGCATACCAGCATCACCAGC-3′, Reverse 5′-TGAAGACCTCCACGTTCTCC-3′) were designed to amplify the fragment from -1084 to +37 (+1 is the start codon). The PCR product was directly cloned into pCRScript (Stratagene), then subcloned into pGL3 enhancer (Promega) to generate SMYD1-LUC reporter. A HDGF 62DL-AS mutations in pGEX and pM vectors were generated by PCR method (Quick Change site-directed mutagenesis, Stratagene). All new constructs were confirmed by sequencing.
Microarray analysis
Conditions for infection of mouse primary aortic vascular smooth muscle cells with Ad-GFP-HDGF and Ad-GFP were described previously.14 After 18 hours infection, total RNA was extracted using RNeasy mini Kit (QIAGEN). cRNA was prepared from T7-transcribed cDNA made from 10 ug of the total RNA and hybridized for expression analysis to the Affymetrix U74Av2 Gene-Chip. Microarray analyses were done at Biomolecular Research Facility, University of Virginia. The detail protocols were listed online (http://www.healthsystem.virginia.edu/internet/biomolec/genechipprotocols.cfm). Program dChip was used for data analysis. Experiments were repeated twice.
Real Time PCR
G-7 cells were infected with adenovirus to express GFP or GFP-HDGF fusions for 18 hours.14 Total RNA was extracted from infected G-7 cells using Tri Reagent (MRC Inc). 1ug total RNA was reverse transcribed using iScript cDNA synthesis kit (Bio-Rad). Real-time quantitative PCR was performed using an iCycler iQ Real-Time PCR Detection System (Bio-Rad). Triplicates of each sample were subjected to a standard PCR protocol (95°C 10 sec, 55°C 15 sec, 72°C 20 sec X 40 cycles, followed by melting curve analysis to ensure the purity of the products) using iQ SYBR Green Supermix (Bio-Rad), primers were derived from the specific cDNA sequence in GenBank database, primer sequences are: Mouse SMYD1 Forward 5′-TCGCTCCGAGGGTTTGTATC-3′, Reverse 5′-TGTAGCCATCAACCATCCTC-3′. Mouse HDGF Forward 5′-TACAAGTGCGGAGACCTGGTGTTT-3′, Reverse 5′-AGGCCTTGACTGTAGGGTTGTTCT-3′. Human SMYD1 Forward 5′-TATAGCCGTCCACCATCCTC-3′, Reverse 5′-TGTAGCCATCAACCATCCTC-3′. Human CtBP Forward 5′-ACGACTTCACCGTCAAGCAGATGA-3′, Reverse 5′-TAAAGCTGAAGGGTTCCGACTCGT-3′. Specific primers amplifying the mouse TATA Box Binding Protein (TBP) or human RNA polymerase II (RPII) were used to normalize gene expression levels.46 Relative mRNA level changes were calculated by the 2-ΔΔCt method.47
Cell Culture, Transient Transfection and Reporter Gene Assay
G-7 myoblast cells were purchased from ATCC and maintained in Dulbecco’s Modified Eagle’s medium (DMEM) with 10% fetal calf serum and 10% horse serum (ATCC). Transient transfection assays were performed using FuGENE6 (Roche) according to the manufacturer’s instructions. G-7 cells in 6 well plates were transfected with 0.5-1 ug of reporter construct and various amounts of HDGF or CtBP in mammalian expression vectors and 50 ng of pRL-CMV (Promega) as internal control, the total DNA concentration was held constant by adding empty vector. 24 hours after transfection, Luciferase assay were performed using a Dual-Luciferase Reporter Assay Systems (Promega). Luciferase activity was measured using 20 ul cell lysate in a Monolight 2010 luminometer (Analytical Luminescence Laboratories) as described in the manufacturer’s manual. Firefly luciferase activity was normalized for Renilla luciferase activity. All transfection data are represented as the mean ± SEM of at least 3 independent experiments with each performed in duplicate.
GST Interaction Assays and Western Blot
GST-HDGF fusion proteins were expressed in the BL21 (DE3) strain of Escherichia coli using the pGEX-4T2 vector (Amersham Pharmacia Biotech). 1 mg of MCF7 whole cell lysate was incubated with GST-HDGF protein bound to glutathione-agarose beads (Sigma) in GST binding buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40) and protease inhibitors (Complete; Roche) overnight at 4°C. Beads were collected by centrifugation and washed six times in GST binding buffer. Bound proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane for immunodetection with a rabbit anti-CtBP antibody 48 (Santa Cruz) and visualized with ECL reagent (Amersham Biosciences).
co-immunoprecipitation Assays
MCF-7 cells were lysed in complete Lysis-M lysis buffer (Roche). Endogenous proteins were immunoprecipitated from MCF-7 cell lysates using monoclonal anti-HDGF or CtBP (Santa Cruz) antibodies coupled to protein A/G beads (Santa Cruz) and washed three times with cold PBS. Precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with polyclonal anti-HDGF or CtBP antibodies (rabbit, Santa Cruz). For IP with tagged proteins, HA-HDGF (3 ug, wild type or DL-AS mutant) and Flag-CtBP (3 ug) were cotransfected into MCF-7 cells using Fugene HD reagent for 48 hours, cell lysate was prepared as above. Co-immunoprecipitation was done by IP with goat HA agarose beads (Santa Cruz), blotting with rabbit anti-Flag antibody (Genetex) or IP with mouse Flag agarose beads (Sigma), blotting with rabbit anti-HA antibody (Santa Cruz).
Gene Knocking down
Modified miRNA specifically target human CtBP was purchased from invitrogen (Hmi403944). The double strand oligo was cloned into Block-iT Pol II miR RNAi expression Vector (invitrogen). The control and CtBP miR expression vectors were cotransfected with pK7-GFP-HDGF expression vector into MCF-7 cells using lipofectamine 2000 transfection reagent as suggested by the manufacture. 72 hours after transfection, the total RNA was extracted using RNeasy mini kit (Qiagen), and subjected to reverse transcribe and real-time PCR as described above.
Electrophoretic Mobility Shift (EMSA)
EMSA was carried out as described before.13 GST-HDGF wild type and 62DL-AS mutant fusion proteins were expressed in the Rosetta strain of Escherichia coli (Novagen) using the pGEX-4T2 vector (Amersham Pharmacia Biotech), EMSA was performed according to manufacturer’s instruction (Pierce, LightShift Chemiluminescent EMSA Kit). Briefly, 1 ug purified recombinant HDGF protein was incubated with 0.25 pmol 5′ end biotin labeled double-stranded oligonucleotide in the reaction buffer (10 mM Tris pH 7.5, 50 mM KCl, 1 mM DTT, 5% glycerol, 0.05% NP-40, 0.1 ug sheared salmon sperm DNA) for 30 min on ice. The reaction mixture was loaded onto a 5% non-denaturing polyacrylamide gel. The gel was run in 0.1XTBE buffer at 200 V for 2 hours. DNA-protein complex were transferred to a nylon membrane in 0.5XTBE buffer and UV crosslinked. The biotin end-labeled DNA probe was detected using a streptavidin-horseradish peroxidase conjugate and the chemiluminescent substrate. The 80 bp double stranded oligo probe is the same as we described before13 which is located at -688 to -609 of SMYD1 promoter (+1 is the start codon).
Fluorescence Cell Image
MCF-7 cells were split onto cover slips in 6-well plates and transfected with 0.5 ug of GFP or CFP tagged proteins using Fugene6 (Roche). After 48 hours transfection, cells were fixed in 1% paraformaldehyde/PBS for 10 minutes at room temperature. The slides then were mounted in SlowFade Gold antifade mounting reagent containing DAPI (Molecular Probe). Cells were imaged using Olympus BX61 motorized upright microscope with GFP, CFP and DAPI filter sets, images were visualized and captured with a Roper Photometrics CoolSnap HQ CCD camera controlled by IPLab4.0 software. Images were processed using IPLab and Adobe Photoshop CS.
Supplementary Material
Table 1.
Primers used for Real Time PCR
| Gene | Accession No | Forward Primer | Reverse Primer |
|---|---|---|---|
| SMYD1 | U76372 | 5′-TCGCTCCGAGGGTTTGTATC-3′ | 5′-TGTAGCCATCAACCATCCTC-3′ |
| TF4 | U16322 | 5′-TACTGTAGCCTGCATCCACACGAA-3′ | 5′-GGTGCTGTAATGGTTTGTGCCACT-3′ |
| SIN3B | AF038848 | 5′-ACCAGAAGGAACAGCTGCACACTA-3′ | 5′-GCCGAGAGCGTTTCTTGTTGTGTT-3′ |
| MRG-1 | Y15163 | 5′-GGTGTTCCGAGCAGAAATCGCAAA-3′ | 5′-GTTGCAATCTCGGAAGTGCTGGTT-3′ |
| Quaking | U44940 | 5′-AAGTTACTGGTACCTGCGGCTGAA-3′ | 5′-GGGCTGGTGATTTAATGTTGGCGT-3′ |
| DEL-1 | AF031525 | 5′-AAGGTGACATTTGCAACCCGAACC-3′ | 5′-TGCAACCTCCACAACACTAGAGCA-3′ |
Acknowledgments
This research was supported by The National Heart, Lung and Blood Institute Grant R01HL-69938 (Dr. A. Everett) and Flight Attendant Medical Research Institute (FAMRI, Dr. J. Yang).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Everett AD, Lobe DR, Matsumura ME, Nakamura H, McNamara CA. Hepatoma-derived growth factor stimulates smooth muscle cell growth and is expressed in vascular development. J. Clin. Invest. 2000;105:567–575. doi: 10.1172/JCI7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura H, Izumoto Y, Kambe H, Kuroda T, Mori T, Kawamura K, Yamamoto H, Kishimoto T. Molecular cloning of complementary DNA for a novel human hepatoma-derived growth factor. Its homology with high mobility group-1 protein. J. Biol. Chem. 1994;269:25143–25149. [PubMed] [Google Scholar]
- 3.Oliver JA, Al-Awqati Q. An endothelial growth factor involved in rat renal development. J. Clin. Invest. 1998;102:1208–1219. doi: 10.1172/JCI785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everett AD, Stoops T, McNamara CA. Nuclear targeting is required for hepatoma-derived growth factor-stimulated mitogenesis in vascular smooth muscle cells. J. Biol. Chem. 2001;276:37564–37568. doi: 10.1074/jbc.M105109200. [DOI] [PubMed] [Google Scholar]
- 5.Kishima Y, Yamamoto H, Izumoto Y, Yoshida K, Enomoto H, Yamamoto M, Kuroda T, Ito H, Yoshizaki K, Nakamura H. Hepatoma-derived growth factor stimulates cell growth after translocation to the nucleus by nuclear localization signals. J. Biol. Chem. 2002;277:10315–10322. doi: 10.1074/jbc.M111122200. [DOI] [PubMed] [Google Scholar]
- 6.Stec I, Nagl SB, van Ommen GB, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 7.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 8.Nameki N, Tochio N, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Fujikura Y, Saito M, Ikari M, Watanabe M, Terada T, Shirouzu M, Yoshida M, Hirota H, Tanaka A, Hayashizaki Y, Guntert P, Kigawa T, Yokoyama S. Solution structure of the PWWP domain of the hepatoma-derived growth factor family. Protein Sci. 2005;14:756–764. doi: 10.1110/ps.04975305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukasik SM, Cierpicki T, Borloz M, Grembecka J, Everett AD, Bushweller JH. High resolution structure of the HDGF PWWP domain: a potential DNA binding domain. Protein Sci. 2006;15:314–23. doi: 10.1110/ps.051751706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 12.Ge Y, Pu M, Gowher H, Wu H, Ding J, Jeltsch A, Xu G. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279:25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Everett AD. Hepatoma-derived growth factor binds DNA through the N-terminal PWWP domain. BMC Mol. Biol. 2007;31:101. doi: 10.1186/1471-2199-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett AD, Narron JV, Stoops T, Nakamura H, Tucker A. Hepatoma-derived growth factor is a pulmonary endothelial cell-expressed angiogenic factor. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;286:L1194–1201. doi: 10.1152/ajplung.00427.2003. [DOI] [PubMed] [Google Scholar]
- 16.Okuda Y, Nakamura H, Yoshida K, Enomoto H, Uyama H, Hirotani T, Funamoto M, Ito H, Everett AD, Hada T, Kawase I. Hepatoma-derived growth factor induces tumorigenesis in vivo through both direct angiogenic activity and induction of vascular endothelial growth factor. Cancer Sci. 2003;94:1034–1041. doi: 10.1111/j.1349-7006.2003.tb01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Ren H, Yuan P, Lang W, Zhang L, Mao L. Down-regulation of hepatoma-derived growth factor inhibits anchorage-independent growth and invasion of non-small cell lung cancer cells. Cancer Res. 2006;66:18–23. doi: 10.1158/0008-5472.CAN-04-3905. [DOI] [PubMed] [Google Scholar]
- 18.Hwang I, Gottlieb DP. The Bop gene adjacent to the mouse CD8b gene encodes distinct Zinc-finger proteins expressed in CTLs and in muscle. J. Immunology. 1997;58:1165–1174. [PubMed] [Google Scholar]
- 19.Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 21.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 22.Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Criqui-Filipe P, Ducret C, Maira SM, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weigert R, Silletta MG, Spano S, Turacchio G, Cericola C, Colanzi A, Senatore S, Mancini R, Polishchuk EV, Salmona M, Facchiano F, Burger KN, Mironov A, Luini A, Corda D. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- 26.Turner J, Crossley M. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays. 2001;23:683–690. doi: 10.1002/bies.1097. [DOI] [PubMed] [Google Scholar]
- 27.Verger A, Quinlan KG, Crofts LA, Spanò S, Corda D, Kable EP, Braet F, Crossley M. Mechanisms directing the nuclear localization of the CtBP family proteins. Mol. Cell. Biol. 2006;26:4882–94. doi: 10.1128/MCB.02402-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda T, Tanaka H, Nakamura H, Nishimune Y, Kishimoto T. Hepatoma-derived growth factor-related protein (HRP)-1 gene in spermatogenesis in mice. Biochem. Biophys. Res. Commun. 1999;262:433–437. doi: 10.1006/bbrc.1999.1115. [DOI] [PubMed] [Google Scholar]
- 29.Izumoto Y, Kuroda T, Harada H, Kishimoto T, Nakamura H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- 30.Ikegame K, Yamamoto M, Kishima Y, Enomoto H, Yoshida K, Suemura M, Kishimoto T, Nakamura H. A new member of a hepatoma-derived growth factor gene family can translocate to the nucleus. Biochem. Biophys. Res. Commun. 1999;266:81–87. doi: 10.1006/bbrc.1999.1733. [DOI] [PubMed] [Google Scholar]
- 31.Singh DP, Kimura A, Chylack LT, Jr, Shinohara T. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene. 2000;242:265–273. doi: 10.1016/s0378-1119(99)00506-5. [DOI] [PubMed] [Google Scholar]
- 32.Dietz F, Franken S, Yoshida K, Nakamura H, Kappler J, Gieselmann V. The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem. J. 2002;366:491–500. doi: 10.1042/BJ20011811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinohara T, Singh DP, Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 34.Everett AD. Identification, cloning, and developmental expression of hepatoma-derived growth factor in the developing rat heart. Dev. Dyn. 2001;222:450–458. doi: 10.1002/dvdy.1204. [DOI] [PubMed] [Google Scholar]
- 35.Uyama H, Tomita Y, Nakamura H, Nakamori S, Zhang B, Hoshida Y, Enomoto H, Okuda Y, Sakon M, Aozasa K, Kawase I, Hayashi N, Monden M. Hepatoma-derived growth factor is a novel prognostic factor for patients with pancreatic cancer. Clin. Cancer Res. 2006;12:6043–6048. doi: 10.1158/1078-0432.CCR-06-1064. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto S, Tomita Y, Hoshida Y, Takiguchi S, Fujiwara Y, Yasuda T, Doki Y, Yoshida K, Aozasa K, Nakamura H, Monden M. Expression of hepatoma-derived growth factor is correlated with lymph node metastasis and prognosis of gastric carcinoma. Clin. Cancer Res. 2006;12:117–122. doi: 10.1158/1078-0432.CCR-05-1347. [DOI] [PubMed] [Google Scholar]
- 37.Ren H, Tang X, Lee JJ, Feng L, Everett AD, Hong WK, Khuri FR, Mao L. Expression of hepatoma-derived growth factor is a strong prognostic predictor for patients with early-stage non-small-cell lung cancer. J. Clin. Oncol. 2004;22:3230–3237. doi: 10.1200/JCO.2004.02.080. [DOI] [PubMed] [Google Scholar]
- 38.Stec I, Nagl SB, van Ommen GJB, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 39.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 40.Slater LM, Allen MD. Bycroft M. Structural variation in PWWP domains. J. Mol. Biol. 2003;330:571–576. doi: 10.1016/s0022-2836(03)00470-4. [DOI] [PubMed] [Google Scholar]
- 41.Masselink H, Bernards R. The adenovirus E1A binding protein BS69 is a corepressors of transcription through recruitment of N-CoR. Oncogene. 2000;19:1538–1546. doi: 10.1038/sj.onc.1203421. [DOI] [PubMed] [Google Scholar]
- 42.Vary JC, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Bio. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b Are Transcriptional Repressors That Exhibit Unique Localization Properties to Heterochromatin. J. Biol. Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 44.Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lepourcelet M, Tou L, Cai L, Sawada J, Lazar AJ, Glickman JN, Williamson JA, Everett AD, Redston M, Fox EA, Nakatani Y, Shivdasani RA. Insights into developmental mechanisms and cancers in the mammalian intestine derived from serial analysis of gene expression and study of the hepatoma-derived growth factor (HDGF) Development. 2005;132:415–427. doi: 10.1242/dev.01579. [DOI] [PubMed] [Google Scholar]
- 46.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitschea A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Deltour S, Pinte S, Guerardel C, Wasylyk B, Leprince D. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol. Cell. Biol. 2002;22:4890–4901. doi: 10.1128/MCB.22.13.4890-4901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.