Abstract
We previously investigated the estrogen receptor α gene (ESR1) as a positional candidate for type 2 diabetes (T2DM), and found evidence for association between the intron 1-intron 2 region of this gene and type 2 diabetes and/or nephropathy in an African American (AA) population. Our objective was to comprehensively evaluate variants across the entire ESR1 gene for association in AA with T2DM and End Stage Renal Disease (T2DM-ESRD). One hundred fifty SNPs in ESR1, spanning 476 kb, were genotyped in 577 AA individuals with T2DM-ESRD and 596 AA controls. Genotypic association tests for dominant, additive, and recessive models, and haplotypic association, were calculated using a χ2 statistic and corresponding P value. Thirty-one SNPs showed nominal evidence for association (P< 0.05) with T2DM-ESRD in one or more genotypic model. After correcting for multiple tests, promoter SNP rs11964281 (nominal P=0.000291, adjusted P=0.0289), and intron 4 SNPs rs1569788 (nominal P=0.000754, adjusted P=0.0278) and rs9340969 (nominal P=0.00109, adjusted P=0.0467) remained significant at experimentwise error rate (EER) P<0.05 for the dominant class of tests. Twenty-three of the thirty-one associated SNPs cluster within the intron 4-intron 6 region. Gender stratification revealed nominal evidence for association with 35 SNPs in females (352 cases; 306 controls) and seven SNPs in males (225 cases; 290 controls). We have identified a novel region of the ESR1 gene that may contain important functional polymorphisms in relation to susceptibility to T2DM and/or diabetic nephropathy.
Keywords: association, African American, diabetes type 2, endocrinology, molecular genetics
Introduction
Diabetes is a growing epidemic whose risk factors include obesity, race, and family history (Arslanian 2002). An African American (AA) is twice as likely to develop diabetes as compared to a Caucasian peer (Brancati et al. 2000). Non-parametric analysis of a genome-wide scan for susceptibility of AA to T2DM provided evidence of linkage (LOD= 2.26) to 6q24-27, with the LOD-1 interval containing the gene encoding estrogen receptor α (ESR1) (Sale et al. 2004). ESR1 is a large, polymorphic gene that contains nearly 1,300 SNPs across its coding region spanning over 295 kb. The human ESR1 gene has been reported to exist in different isoforms, including deletion of one or more exons, duplication of one or more exons, alternative uses of 5′ untranslated exons, and through the use of ‘intronic’ exons (Hirata et al. 2003). An additional exon, “exon 9,” has been identified 4,374 bp downstream of the normal eight exon genomic region (Wang et al. 2005). The classical role for estrogen receptor α, the protein produced by ESR1, is to act as a transcription factor, although non-transcriptional effects have also been identified (Bjornstrom and Sjoberg 2002).
Previous studies suggest that sex hormones play a role in insulin resistance (Livingstone and Collison 2002). The only known human (male) with a null ESR1 mutation exhibits insulin resistance, impaired glucose tolerance, increased height, and was obese (Smith et al. 1994). Esr1 knock-out mice (male and female) also display moderate insulin resistance, impaired glucose tolerance, and obesity (Heine et al. 2000). Additionally, 17-beta estradiol (E2) appears to protect female rodents from hyperglycemia unless ovariectomized and reverses diabetes in male rodents (Louet et al. 2004). E2 replacement also displays renoprotective effects in streptozotocin (STZ) induced diabetic rats (Mankhey et al. 2005). Clinical studies in humans also provide evidence that E2 treatment improves glycemic control and insulin sensitivity in postmenopausal women with diabetes (Seed 2002) and reduces the incidence of diabetes when taken with progestin (Margolis et al. 2004).
ESR1 polymorphisms, primarily variants in the intron 1- intron 2 region, have shown positive association with numerous phenotypes including T2DM (Speer et al. 2001). However few studies have investigated T2DM and End Stage Renal Disease (ESRD) in AA. Our lab has previously shown significant association between ESR1 variants and T2DM-ESRD in AA (Gallagher et al. 2007). Seventeen ESR1 SNPs were genotyped across a 41 kb region spanning intron 1- intron 2. One SNP, rs1033182, and an independent 6-SNP haplotype of high linkage disequilibrium (LD) showed positive association with T2DM-ESRD; however these associations did not appear to be responsible for the linkage peak observed on 6q.
To comprehensively evaluate polymorphisms in the ESR1 gene for associations with T2DM-ESRD, we selected 150 SNPs spanning 476 kb (Build 35: 152,062,467-152,538,601), based on linkage disequilibrium (LD) data from the Pathobiological Determinants of Atherosclerosis in Youth (PDAY) study. These SNPs were genotyped in 577 AA individuals with T2DM-ESRD and 596 AA controls.
Materials and Methods
Subjects
This study was conducted under Institutional Review Board approval from Wake Forest University School of Medicine, and adhered to the tenets of the Declaration of Helsinki. Identification, clinical characteristics, and recruitment of AA and European American (EA) patients and controls have been described previously (Yu et al. 1996). Briefly, 577 unrelated AA patients with T2DM born in North Carolina, South Carolina, Georgia, Tennessee or Virginia were recruited from dialysis facilities. Individuals with probable type 1 diabetes, identified as patients with a history of diabetic ketoacidosis, or who developed diabetes mellitus prior to age 25 and received continuous insulin therapy since diagnosis, were excluded. A diagnosis of T2DM was based upon participants reporting an initial diagnosis of diabetes mellitus after age 35 years, receiving oral hypoglycemic agents or dietary therapy without insulin for at least one year after initial diagnosis, and active treatment with diabetes medications. Cases had T2DM diagnosed at least 5 years prior to initiating renal replacement therapy, background or greater diabetic retinopathy and/or > 3+ proteinuria on urinalysis in the absence of other causes of nephropathy. Five hundred ninety-six unrelated AA controls born in North Carolina, South Carolina, Georgia, Tennessee or Virginia, who did not have a current diagnosis of T2DM or renal disease, were recruited. BMI data is not shown or used in analyses since this data was not available for the majority of controls, and in cases was largely based on self-report at the time of examination, reflecting weight on dialysis. Thirty-nine unrelated EA controls without known T2DM or renal disease were recruited using the same recruitment strategy and criteria as AA controls. DNA extraction was performed using the PureGene system (Gentra Systems, Minneapolis, MN). DNA was also obtained from 44 Yoruba Nigerians from the National Institute of General Medical Sciences (NIGMS) Human Variation Collection (Coriell Cell Repositories, Camden, NJ).
ESR1 SNP selection and genotyping
SNP selection was determined using data from the PDAY study (D Herrington and T Howard, personal communication). The PDAY study genotyped 160 SNPs across a 471 kb region spanning the ESR1 gene in a sample of 657 AA and 618 EA. This population was selected in preference to HapMap data due to the larger sample size and the population source (657 AA versus 30 Yoruba Nigerian trios). We used the aggressive tagging option (2- or 3-haplotype tagging of SNPs) of Tagger implemented in the program Haploview (de Bakker et al. 2005), to select SNPs which tag the 160 markers in the PDAY AA samples. We then forced the inclusion of the AA tag SNPs, and selected additional EA tag SNPs from the PDAY data, resulting in a total of 130 SNPs that tag the common 2- and 3- marker haplotypes in both AA and EA. We further enriched our SNP selection by including 20 high priority SNPs with an emphasis on potential functional variants, including one previously associated SNP, two promoter SNPs with a MAF > 0.05, 9 SNPs located in intron boundaries, 3 coding SNPs , one SNP located in an alternative promoter, 4 SNPs located in alternative exons of splice variants. The HapMap project (25) has genotyped 633 SNPs in the coding region of ESR1 (354 with a minor allele frequency > 0.05) in Yoruba Nigerian trios. Sixty nine of the 354 HapMap SNPs were genotyped in our data set, capturing 211 of the 354 alleles with a mean r2 of 0.973 using the pairwise tagging option of Haploview and 222 of 354 alleles with a mean r2 of 0.97 using the aggressive tagging option. In total, we genotyped one hundred fifty SNPs, spanning more than 476 kb, located in the coding and flanking regions of ESR1(extending 158,332 bp upstream and 22,081 bp downstream of the ESR1 coding region (NCBI Build 35)) in 577 AA individuals with T2DM-ESRD and 596 AA without a diagnosis for T2DM. One hundred twenty-three SNPs were genotyped using Illumina Inc.’s Custom Genotyping Service (San Diego, CA). Twenty seven additional SNPs were genotyped using iPlex methodology on a MassARRAY genotyping system (Sequenom Inc., San Diego, CA) (Buetow et al. 2001): rs9340799, rs2813545, rs3778609, rs6926750, rs3020343, rs2234693, rs9397456, rs9479134, rs9340774, rs9340775, rs9341068, rs9341069, rs2813563, rs2813562, rs3798577, rs9341070, rs9340973, rs2273206, rs2273207, rs9340804, rs3734807, rs9340902, rs9397459, rs6914438, rs9479119, rs11964281, rs17081692. The genotyping success rates for the 150 SNPs in the AA cases and controls range from 94.1-100%. Concordance rates for 46 replicate pairs were 100% for all SNPs, except rs9479134 where there were 2 discordant genotypes among 46 replicate pairs (95.7% concordance).
Genotyping for admixture analyses
Seventy biallelic Admixture Informative Markers (AIMs) were genotyped by Illumina Inc.’s Custom Genotyping Service (San Diego, CA) or using a MassARRAY genotyping system (Sequenom Inc., San Diego, CA) (Buetow et al. 2001) in 577 AA T2DM-ESRD cases and 596 AA controls, 44 Yoruba Nigerians and 39 EA controls (Supplementary Table 1). The genotyping success rates for the AIMs range from 94.9-100% in our AA cases, AA controls, Yoruba Nigerians, and EA controls. Primer sequences are available on request.
Statistical analyses
Hardy Weinberg equilibrium (HWE) values were determined by calculating a χ2 statistic and corresponding P-value. Haplotype block structure was established using Haploview 3.2 (Barrett JC, et al. 2005), using the block definition from Gabriel et al. (Gabriel et al. 2002).
Unadjusted genotypic association for dominant, additive, and recessive models, and 2-and 3-SNP haplotypic association, were tested by calculating a χ2 statistic and corresponding P-value using the program SNPGWA (C Langefeld, personal communication). Due to a lack of validity of the large sample χ2 test statistic, only the dominant model was considered for SNPs with 10 or fewer individuals that were homozygous for the minor allele. To correct for multiple tests at the gene level we evaluated the association results using a permutation analysis (10,000 replicates) to simulate the null joint distribution of marker statistics, and applied MinP step down analysis on the ranked, adjusted P-values (Westfall and Young 1993). This procedure adjusts for multiple tests of SNPs within a gene while maintaining strong control over the Experiment-wise Error Rate (EER) at any desired level of significance.
Ancestral proportions were calculated using the program FRAPPE (Frequentist Estimation of individual ancestry proportion) (Tang et al. 2005) under a two population model. Estimates of “ancestral” allele frequencies were obtained from genotyped African and EA samples. Covariate adjustment of individual estimates of African ancestry for AA subjects was used in logistic regression tests of dominant, additive, and recessive models of association for all SNPs as implemented in the program SNPADMIX (C Langefeld, personal communication).
Results
Characteristics of the AA case and control populations are shown in Table 1. Controls were significantly younger than cases (P<0.0001), although they were significantly older than the mean age at T2DM diagnosis in cases (P<0.0001). Additionally, age data were unavailable for 148 controls recruited during the early phase of the study. There was a higher proportion of females (61%) in the cases than controls (51%), possibly due to a combination of higher prevalence of T2DM in women and participation bias.
Table 1.
Characteristics of African American study subjects
| Trait | T2DM-ESRD Cases (n= 577) |
Controls (n= 596) |
|---|---|---|
| % Female (n) | 61% (351) | 51% (305) |
| Age at exam (years + SD) (n*) | 62.2 + 10.3 (541) | 49.3 + 9.8 (448) |
| Age at T2DM diagnosis (years + SD) (n*) | 41.8 + 11.6 (544) | N/A |
| Age at ESRD diagnosis (years + SD) (n*) | 59.0 + 10.5 (560) | N/A |
n = number with data available. All cases were diagnosed with T2DM and ESRD.
Four SNPs were monomorphic in the AA cases and controls and were excluded from further analysis. Five SNPs deviated from expected Hardy Weinberg Equilibrium (HWE) proportions (P < 0.01) in the AA cases (rs9397459, rs9341070, rs9340969, rs1569788, and rs722208) and one SNP (rs9341070) deviated from HWE in the AA controls.
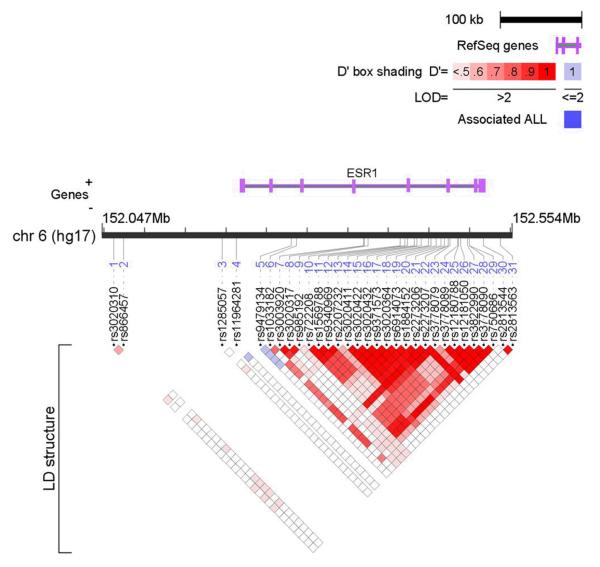
LD structure for the 146 ESR1 SNPs is shown in Supplementary Figure 1. Twenty-seven and 29 blocks of high LD were identified in the AA controls and cases, respectively, using the method of Gabriel et al. implemented in the program Haploview (Gabriel et al. 2002). Genotype frequencies and counts are shown in Supplementary Table 2, and single-SNP genotypic association results with corresponding odds ratios are presented in Supplementary Table 3. Thirty one SNPs showed nominal evidence of association (P<0.05) with T2DM-ESRD in one or more genotypic model (Table 2). Twenty-three of the thirty-one associated SNPs cluster together in the intron 4-intron 6 region (Figure 1). The LD structure for the 31 associated SNPs suggests high LD between most of the intron 4-intron 6 markers (Figure 2). The most significant genotypic associations were observed with an intron 4 SNP, rs1569788, (P=0.0003, dominant model; and P= 0.009; additive model) and a promoter SNP, rs11964281 (P= 0.0003, dominant model). Stratification based on gender reveals evidence for single SNP association with 35 SNPs in females (352 cases; 306 controls) and seven SNPs in males (225 cases; 290 controls), shown in Table 3.
Table 2.
Unadjusted and admixture adjusted genotypic P-values for the 31 significantly associated SNPs with T2DM-ESRD
| Marker | Physical Position* |
Location | Dominant P-value |
Dominant Admixture- Adjusted P-value |
Additive P-value |
Additive Admixture Adjusted P-value |
Recessive P-value |
Recessive Admixture Adjusted P-value |
|---|---|---|---|---|---|---|---|---|
| rs3020310 | 152062467 | 5′ UTR | 0.097 | 0.169 | 0.021 | 0.040 | 0.009 | 0.012 |
| rs866457 | 152074526 | 5′ UTR | 0.032 | 0.096 | 0.024 | 0.071 | 0.180 | 0.243 |
| rs1285057 | 152195905 | 5′ UTR | 0.496 | 0.687 | 0.176 | 0.264 | 0.035 | 0.038 |
| rs11964281 | 152213556 | Promoter | 0.0003 | 0.001 | -- | -- | -- | -- |
| rs9479134 | 152281109 | Intron 2 | 0.002 | 0.005 | -- | -- | -- | -- |
| rs1033182 | 152287147 | Intron 2 | 0.025 | 0.144 | -- | -- | -- | -- |
| rs3003920 | 152370431 | Intron 4 | 0.011 | 0.028 | 0.038 | 0.074 | 0.474 | 0.548 |
| rs3020317 | 152370854 | Intron 4 | 0.038 | 0.061 | 0.044 | 0.071 | 0.329 | 0.392 |
| rs985192 | 152375591 | Intron 4 | 0.061 | 0.102 | 0.025 | 0.043 | 0.069 | 0.086 |
| rs722208 | 152414998 | Intron 4 | 0.0008 | 0.003 | 0.018 | 0.063 | 0.898 | 0.760 |
| rs1569788 | 152420729 | Intron 4 | 0.0003 | 0.001 | 0.009 | 0.033 | 0.822 | 0.856 |
| rs9340969 | 152424243 | Intron 4 | 0.0005 | 0.002 | 0.013 | 0.045 | 0.885 | 0.794 |
| rs2207232 | 152432401 | Intron 5 | 0.002 | 0.002 | -- | -- | -- | -- |
| rs3020411 | 152435876 | Intron 5 | 0.002 | 0.008 | 0.003 | 0.016 | 0.172 | 0.425 |
| rs3020422 | 152440871 | Intron 5 | 0.002 | 0.007 | 0.003 | 0.018 | 0.211 | 0.492 |
| rs3020432 | 152450040 | Intron 5 | 0.002 | 0.007 | 0.003 | 0.018 | 0.211 | 0.492 |
| rs9371573 | 152452833 | Intron 5 | 0.002 | 0.007 | 0.003 | 0.016 | 0.180 | 0.450 |
| rs3020364 | 152459231 | Intron 5 | 0.0006 | 0.002 | 0.002 | 0.011 | 0.228 | 0.465 |
| rs6914073 | 152465351 | Intron 5 | 0.014 | 0.039 | 0.033 | 0.086 | 0.342 | 0.512 |
| rs1884152 | 152469293 | Intron 5 | 0.008 | 0.023 | 0.016 | 0.048 | 0.250 | 0.388 |
| rs2273206 | 152474425 | Intron 6 (Boundary) | 0.041 | 0.105 | 0.118 | 0.264 | 0.687 | 0.926 |
| rs2273207 | 152474439 | Intron 6 (Boundary) | 0.001 | 0.002 | -- | -- | -- | -- |
| rs3778079 | 152476342 | Intron 6 | 0.036 | 0.087 | 0.058 | 0.149 | 0.344 | 0.555 |
| rs3778089 | 152485874 | Intron 6 | 0.005 | 0.017 | 0.001 | 0.005 | 0.011 | 0.028 |
| rs12180788 | 152486892 | Intron 6 | 0.005 | 0.015 | 0.001 | 0.004 | 0.011 | 0.027 |
| rs12181050 | 152489247 | Intron 6 | 0.006 | 0.018 | 0.001 | 0.006 | 0.014 | 0.034 |
| rs3822990 | 152498078 | Intron 6 | 0.009 | 0.013 | 0.014 | 0.023 | 0.639 | 0.738 |
| rs3778090 | 152500141 | Intron 6 | 0.002 | 0.003 | -- | -- | -- | -- |
| rs750686 | 152500239 | Intron 6 | 0.025 | 0.062 | 0.095 | 0.229 | 0.817 | 0.860 |
| rs2813544 | 152517695 | 3′ UTR | 0.316 | 0.293 | 0.097 | 0.083 | 0.027 | 0.023 |
| rs2813563 | 152538601 | Exon 9 | 0.169 | 0.166 | 0.047 | 0.043 | 0.019 | 0.016 |
P-values <0.05 are shown in bold
NCBI Build 35 (May 2004).
Figure 1.
ESR1 unadjusted genotypic association results for dominant, additive, and recessive models for 146 SNPs. Marker position (bp) is plotted along the x-axis while the -log[P-value] is plotted along the y-axis. The black dotted line represents the -log [P-value] threshold of 1.3 (corresponding to a P-value of 0.05).
Figure 2.
LD structure of the 31 associated SNPs of the ESR1 gene in the AA Controls (n = 596) using the program LocusView (T. Petryshen, A. Kirby, M. Ainscow, unpublished software). D’ values are displayed in the squares: Empty squares represent a pairwise D’ = 1; blue empty squares represent D’ values =1 but LOD <2; red squares represent high pairwise LD, coloring down to white squares of low pairwise LD.
Table 3.
Gender stratified unadjusted and admixture adjusted genotypic P-values for the significantly associated SNPs with T2DM-ESRD
| a.Females | ||||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Physical Position* |
Location | Dominant P-value |
Dominant Admixture- Adjusted P-value |
Additive P-value |
Additive Admixture- Adjusted P-value |
Recessive P-value |
Recessive Admixture- Adjusted P-value |
| rs866457 | 152074526 | 5′ UTR | 0.018 | 0.096 | 0.006 | 0.041 | 0.036 | 0.087 |
| rs1999805 | 152160477 | 5′ UTR | 0.999 | 0.814 | 0.214 | 0.285 | 0.030 | 0.033 |
| rs1415193 | 152184751 | 5′ UTR | 0.047 | 0.268 | -- | -- | -- | -- |
| rs2504065 | 152187280 | 5′ UTR | 0.705 | 0.713 | 0.184 | 0.141 | 0.045 | 0.022 |
| rs1285057 | 152195905 | 5′ UTR | 0.959 | 0.513 | 0.482 | 0.821 | 0.043 | 0.040 |
| rs11964281 | 152213556 | Promoter | 0.002 | 0.004 | -- | -- | -- | -- |
| rs2077647 | 152221190 | Exon 1 | 0.044 | 0.076 | 0.072 | 0.112 | 0.384 | 0.433 |
| rs11155818 | 152276243 | Intron 2 | 0.038 | 0.073 | -- | -- | -- | -- |
| rs1033182 | 152287147 | Intron 2 | 0.027 | 0.196 | -- | -- | -- | -- |
| rs3020317 | 152370854 | Intron 4 | 0.036 | 0.055 | 0.013 | 0.022 | 0.045 | 0.064 |
| rs985192 | 152375591 | Intron 4 | 0.044 | 0.065 | 0.009 | 0.014 | 0.015 | 0.022 |
| rs3020403 | 152387828 | Intron 4 | 0.031 | 0.167 | -- | -- | -- | -- |
| rs726283 | 152395122 | Intron 4 | 0.004 | 0.033 | -- | -- | -- | -- |
| rs7754762 | 152403650 | Intron 4 | 0.111 | 0.203 | 0.026 | 0.073 | 0.032 | 0.082 |
| rs722208 | 152414998 | Intron 4 | 0.0006 | 0.002 | 0.009 | 0.030 | 0.651 | 0.925 |
| rs1569788 | 152420729 | Intron 4 | 0.0007 | 0.003 | 0.008 | 0.026 | 0.587 | 0.813 |
| rs9340969 | 152424243 | Intron 4 | 0.0007 | 0.003 | 0.006 | 0.022 | 0.511 | 0.724 |
| rs9340973 | 152424810 | Intron 4 (Boundary) | 0.043 | 0.069 | -- | -- | -- | -- |
| rs7755185 | 152431728 | Intron 5 | 0.135 | 0.169 | 0.024 | 0.040 | 0.028 | 0.046 |
| rs2207232 | 152432401 | Intron 5 | 0.008 | 0.008 | -- | -- | -- | -- |
| rs3020411 | 152435876 | Intron 5 | 0.009 | 0.033 | 0.003 | 0.014 | 0.028 | 0.074 |
| rs3020422 | 152440871 | Intron 5 | 0.007 | 0.026 | 0.002 | 0.012 | 0.028 | 0.074 |
| rs3020432 | 152450040 | Intron 5 | 0.007 | 0.026 | 0.002 | 0.012 | 0.028 | 0.074 |
| rs9371573 | 152452833 | Intron 5 | 0.007 | 0.026 | 0.002 | 0.011 | 0.020 | 0.063 |
| rs3020364 | 152459231 | Intron 5 | 0.002 | 0.007 | 0.002 | 0.010 | 0.082 | 0.184 |
| rs6914073 | 152465351 | Intron 5 | 0.040 | 0.075 | 0.174 | 0.332 | 0.987 | 0.717 |
| rs1884152 | 152469293 | Intron 5 | 0.022 | 0.047 | 0.100 | 0.217 | 0.831 | 0.873 |
| rs2273207 | 152474439 | Intron 6 (Boundary) | 0.003 | 0.005 | -- | -- | -- | -- |
| rs3778089 | 152485874 | Intron 6 | 0.011 | 0.036 | 0.003 | 0.014 | 0.021 | 0.058 |
| rs12180788 | 152486892 | Intron 6 | 0.012 | 0.039 | 0.003 | 0.014 | 0.020 | 0.056 |
| rs12181050 | 152489247 | Intron 6 | 0.012 | 0.039 | 0.004 | 0.018 | 0.029 | 0.076 |
| rs3822990 | 152498078 | Intron 6 | 0.012 | 0.022 | -- | -- | -- | -- |
| rs3778090 | 152500141 | Intron 6 | 0.001 | 0.002 | -- | -- | -- | -- |
| rs750686 | 152500239 | Intron 6 | 0.017 | 0.053 | 0.022 | 0.064 | 0.219 | 0.321 |
| rs1543403 | 152520817 | 3′ UTR | 0.174 | 0.199 | 0.677 | 0.658 | 0.035 | 0.038 |
| b.Males | ||||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Physical Position* |
Location | Dominant P-value |
Dominant Admixture- Adjusted P-value |
Additive P-value |
Additive Admixture- Adjusted P-value |
Recessive P-value |
Recessive Admixture- Adjusted P-value |
| rs9479134 | 152281109 | Intron 2 | 0.006 | 0.021 | -- | -- | -- | -- |
| rs1709183 | 152286109 | Intron 2 | 0.071 | 0.072 | 0.035 | 0.036 | 0.108 | 0.113 |
| rs2175898 | 152289065 | Intron 2 | 0.029 | 0.028 | -- | -- | -- | -- |
| rs3020368 | 152463303 | Intron 5 | 0.018 | 0.017 | -- | -- | -- | -- |
| rs2982896 | 152491606 | Intron 6 | 0.031 | 0.032 | -- | -- | -- | -- |
| rs2813545 | 152524431 | 3′ UTR | 0.078 | 0.074 | 0.025 | 0.025 | 0.040 | 0.043 |
| rs2813562 | 152538434 | Exon 9 | 0.171 | 0.167 | 0.034 | 0.034 | 0.017 | 0.019 |
P-values <0.05 are shown in bold
NCBI Build 35 (May 2004).
P-values <0.05 are shown in bold
NCBI Build 35 (May 2004).
Admixture proportions and individual admixture estimates for the AA cases and controls were calculated based on the genotyping results of 70 admixture informative markers (AIMs). The mean proportion of African ancestry was estimated to be 0.817 (SD 0.133) and 0.791 (SD 0.131) in the AA cases and controls, respectively. Admixture adjustments were performed for all tests of association, including the gender stratified results. In the combined sample set, 25 of the 31 SNPs remained significant in one or more genotypic model of association after adjusting for admixture. Additionally, 26 of 35 and 7 of 7 SNPs remained significantly associated in one or more genotypic model of association after admixture adjustments in the female and male sample sets respectively.
Discussion
We have investigated 150 SNPs in the ESR1 gene for association with T2DM-ESRD in an African American population. Multiple SNPs were associated across the intron 4-intron 6 region, as well as promoter and intron 2. Additionally, sex-specific associations were detected, primarily in female T2DM-ESRD patients.
The data revealed positive association with an intron 2 SNP (rs9479134, P=0.002, dominant model) and a clustering of associated SNPs in intron 4 (six associated SNPs), intron 5 (eight associated SNPs) and intron 6 (nine associated SNPs). The dominant genotypic model supported evidence for association with T2DM-ESRD (P= 0.025) for the previously associated intron 2 SNP, rs1033182 (Gallagher et al. 2007), however upon adjusting for admixture, significance was no longer present (P=0.144). Although the initial findings were not successfully replicated after admixture adjustments, the lack of significance may be attributable to the smaller sample size used for this study (577 cases and 596 controls versus 851 cases and 635 controls). Thirteen SNPs (rs6902771, rs4870056, rs9322331, rs2234693, rs9340799, rs7774230, rs12664989, rs1514348, rs11155818, rs1709183, rs1033182, rs2175898, rs11155819) were genotyped in both studies with 541 AA cases and 426 AA controls common to both investigations. Markers rs1033182 and rs9479134 are separated by only 6 kb, therefore the significant association with rs9479134, not genotyped in the initial study, (P= 0.002, dominant model) suggest that additional variants near rs1033182 may play a role in T2DM-ESRD susceptibility. Twenty-four of the 31 nominally associated SNPs, including two of the most significantly associated SNPs, were located in the intron 4-intron 6 region. Introns 4, 5, and 6 are very large, with lengths greater than 67 kb, 49 kb, and 33 kb respectively. Previous studies show that exons 4-8 code for the ligand binding domain of the estrogen receptor α protein (Hirata et al. 2003). Stratification based on gender revealed evidence for association at the P < 0.05 significance level with 35 SNPs in females and seven SNPs in males. The most significant SNP, rs1569788, had an unadjusted P-value for the dominant genotypic model of 0.0003, 0.0007, and 0.13 in the combined, female, and male sample sets respectively, with corresponding odds ratios of 0.64 (CI: 0.51-0.82), 0.57 (CI: 0.42-0.79), and 0.76 (0.53-1.09), suggesting that the A allele for this SNP has a protective effect. Although the effect in males was in the same direction, the lack of significance may be due to limited power. Results from Breslow-Day tests of homogeneity of odds ratios suggest that only four SNPs: rs726283 (P=0.0051), rs3020403 (P=0.0067), rs6912184 (P=0.0331) and rs3020404 (P=0.0412), had significantly different odds ratios between the two sexes; none of the four were associated in the combined data set. Although estrogens are present in both males and females, they are usually present at significantly higher concentrations in pre-menopausal females as compared to similar aged males. Given the functional effects of estrogens on female biology and the higher proportion of female participants in this study, it is not surprising that a greater number of significant associations with T2DM-ESRD were observed with female patients.
Although the dominant, additive, and recessive genotypic models of association were tested for each SNP, only the dominant model P-values were applicable to SNPs with 10 or fewer individuals that were homozygous for the minor allele. Of the thirty-one nominally associated SNPs (Table 2), six SNPs (rs11964281, rs9479134, rs1033182, rs2207232, rs2273207, and rs3778090) had 10 or fewer individuals that were homozygous for the minor allele (Supplementary Table 2) where only the dominant model was considered. After correcting for multiple tests, promoter SNP rs11964281 (nominal P=0.000291, adjusted P=0.0289), and intron 4 SNPs rs1569788 (nominal P=0.000754, adjusted P=0.0278) and rs9340969 (nominal P=0.00109, adjusted P=0.0467) remained significant at EER P<0.05 for the dominant class of tests. Under a composite null hypothesis of no association at a SNP under any model, implemented using a minimum P statistic (min { Padd, Pdom, Prec } ), only rs11964281 maintained significance at EER P<0.05, with a composite null gene-level adjusted P value of 0.0377. The clustering of associated SNPs in the intron 4 region along with the highly significant P-values for rs1569788 and rs9340969, provides encouraging data that variants in this region may be functionally relevant. Additionally, an in silico search for putative transcription factor binding sites for the promoter SNP rs11964281 suggests that the presence of the T allele creates a binding site for the transcription factor HSF2, which is not observed when the C allele is present (TFSearch: www.cbrc.jp/research/db/TFSEARCH.html) .
In an attempt to eliminate spurious associations due to admixture, covariate adjustments using estimated ancestral proportions were performed for significantly associated SNPs (Tables 2 and 3). Twenty-five, twenty-six, and seven SNPs remained nominally associated in one or more genotypic model of association after admixture adjustments were performed in the combined, female, and male sample sets respectively. All seven markers remained significant in at least one model of association in the males; however, markers rs1033182, rs866457, rs3020317, rs3778079, rs2273206, and rs750686 were no longer significantly associated with T2DM-ESRD in the combined set, while markers rs1415193, rs2077647, rs11155818, rs1033182, rs3020403, rs7754762, rs9340973, rs6914073, and rs750686 were no longer significant in the female sample set, suggesting a modest but important influence of admixture on overall results.
One limitation of this study was that our ascertainment scheme did not permit us to distinguish whether associations were with T2DM and/or nephropathy. Associations with both phenotypes are plausible, and require investigation in independent populations. Additionally, the majority of controls were not tested for diabetes, thus the control sample probably contains a small proportion of undiagnosed T2DM cases. While this has not impacted our ability to detect positive associations with ESR1, this is likely to have reduced our power to detect more subtle influences of additional variants, and led to underestimation of odds ratios. Although genome wide association studies (GWAS) have not supported evidence for association with ESR1 variants and T2DM at the genome wide significance level, two GWAS have identified nominal association with variants within ESR1 and T2DM. In the 2007 report by Saxena et. al, four ESR1 SNPs (rs6930355, rs3798571, rs9479191, and rs3778089) were reported to have P values of association with T2DM ranging from P=0.0157 to 0.0177, in a Scandinavian population (Saxena et. al 2007). One of those SNPs, rs3778089 (P=0.0177) was also associated in our study (P=0.005, dominant model). In a second GWAS, the Wellcome Trust Case Control Consortium also identified seven ESR1 SNPs with nominal association with T2DM with P values ranging from 0.049 to 0.00049 (Wellcome Trust Case Control Consortium, 2007). One of the seven associated SNPs (rs9397459, P=0.049) was also included in our study, however it was not significantly associated in our population (P=0.415).
In a comprehensive evaluation of the entire ESR1 gene, including the promoter and a distal alternative exon region, we have detected significant associations with T2DM-ESRD in AA. To date, this study represents the largest genetic evaluation of ESR1 variants in relation to T2DM and/or ESRD susceptibility. By analyzing variants throughout the gene, we were able to identify novel regions of association spanning intron 4 - intron 6. The clustering of multiple significantly associated SNPs in high LD in the same region of the gene supports the validity of these findings. These results indicate that this region warrants further examination to investigate the biological relevance of variation in these intronic regions.
Supplementary Material
Acknowledgments
We thank the patients and staff of the Southeastern Kidney Council/ESRD Network 6 and individuals recruited as controls for their participation. Thanks also to recruiters Joyce Byers and Mitzie Spainhour, technician Candace Gordon, programmer Matt Stiegert, and Mark Hansen and colleagues at Illumina Inc. This work was supported by grants DK066358, DK072550, the Wake Forest University General Clinical Research Center M01 RR07122, and a Career Development Award from the American Diabetes Association (MMS).
References
- Arslanian SA. Metabolic differences between Caucasian and African-American children and the relationship to type 2 diabetes mellitus. J Pediatr Endocrinol Metab. 2002;15(Suppl 1):509–17. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Signal transducers and activators of transcription as downstream targets of nongenomic estrogen receptor actions. Mol Endocrinol. 2002;16:2202–14. doi: 10.1210/me.2002-0072. [DOI] [PubMed] [Google Scholar]
- Brancati FL, Kao WH, Folsom AR, Watson RL, Szklo M. Incident type 2 diabetes mellitus in African American and white adults: the Atherosclerosis Risk in Communities Study. Jama. 2000;283:2253–9. doi: 10.1001/jama.283.17.2253. [DOI] [PubMed] [Google Scholar]
- Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98:581–4. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gallagher CJ, Keene KL, Mychaleckyj JC, Langefeld CD, Hirschhorn JN, Henderson BE, Gordon CJ, Freedman BI, Rich SS, Bowden DW, Sale MM. Investigation of the estrogen receptor-alpha gene with type 2 diabetes and/or nephropathy in African-American and European-American populations. Diabetes. 2007;56:675–84. doi: 10.2337/db06-0303. [DOI] [PubMed] [Google Scholar]
- Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, Cooke PS. Increased adipose tissue in male and female estrogen receptor-alpha knockout mice. Proc Natl Acad Sci U S A. 2000;97:12729–34. doi: 10.1073/pnas.97.23.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata S, Shoda T, Kato J, Hoshi K. Isoform/variant mRNAs for sex steroid hormone receptors in humans. Trends Endocrinol Metab. 2003;14:124–9. doi: 10.1016/s1043-2760(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Livingstone C, Collison M. Sex steroids and insulin resistance. Clin Sci (Lond) 2002;102:151–66. doi: 10.1042/cs1020151. [DOI] [PubMed] [Google Scholar]
- Louet JF, LeMay C, Mauvais-Jarvis F. Antidiabetic actions of estrogen: insight from human and genetic mouse models. Curr Atheroscler Rep. 2004;6:180–5. doi: 10.1007/s11883-004-0030-9. [DOI] [PubMed] [Google Scholar]
- Mankhey RW, Bhatti F, Maric C. 17beta-Estradiol replacement improves renal function and pathology associated with diabetic nephropathy. Am J Physiol Renal Physiol. 2005;288:F399–405. doi: 10.1152/ajprenal.00195.2004. [DOI] [PubMed] [Google Scholar]
- Margolis KL, Bonds DE, Rodabough RJ, Tinker L, Phillips LS, Allen C, Bassford T, Burke G, Torrens J, Howard BV. Effect of oestrogen plus progestin on the incidence of diabetes in postmenopausal women: results from the Women’s Health Initiative Hormone Trial. Diabetologia. 2004;47:1175–87. doi: 10.1007/s00125-004-1448-x. [DOI] [PubMed] [Google Scholar]
- Sale MM, Freedman BI, Langefeld CD, Williams AH, Hicks PJ, Colicigno CJ, Beck SR, Brown WM, Rich SS, Bowden DW. A genome-wide scan for type 2 diabetes in African-American families reveals evidence for a locus on chromosome 6q. Diabetes. 2004;53:830–7. doi: 10.2337/diabetes.53.3.830. [DOI] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bostrom K Bengtsson, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parihk H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Seed M. The choice of hormone replacement therapy or statin therapy in the treatment of hyperlipidemic postmenopausal women. Atheroscler. 2002;(Suppl 3):53–63. doi: 10.1016/s1567-5688(01)00009-5. [DOI] [PubMed] [Google Scholar]
- Smith EP, Boyd J, Frank GR, Takahashi H, Cohen RM, Specker B, Williams TC, Lubahn DB, Korach KS. Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N Engl J Med. 1994;331:1056–61. doi: 10.1056/NEJM199410203311604. [DOI] [PubMed] [Google Scholar]
- Speer G, Cseh K, Winkler G, Vargha P, Braun E, Takacs I, Lakatos P. Vitamin D and estrogen receptor gene polymorphisms in type 2 diabetes mellitus and in android type obesity. Eur J Endocrinol. 2001;144:385–9. doi: 10.1530/eje.0.1440385. [DOI] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochem Biophys Res Commun. 2005;336:1023–1027. doi: 10.1016/j.bbrc.2005.08.226. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing: Examples and methods for p-value adjustment. John Wiley & Sons Inc.; New York: 1993. [Google Scholar]
- Yu H, Bowden DW, Spray BJ, Rich SS, Freedman BI. Linkage analysis between loci in the renin-angiotensin axis and end-stage renal disease in African Americans. J Am Soc Nephrol. 1996;7:2559–64. doi: 10.1681/ASN.V7122559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.