Abstract
We report here the development and optimization of a simple 384-well colorimetric assay to measure H2O2 generated by the redox cycling of compounds incubated with reducing agents in high-throughput screening (HTS) assay buffers. The phenol red-horseradish peroxidase (HRP) assay readily detected H2O2 either added exogenously or generated by the redox cycling of compounds in dithiothreitol (DTT). The generation of H2O2 was dependent on the concentration of both the compound and DTT and was abolished by catalase. Although both DTT and tris(2-carboxyethyl)-phosphine sustain the redox cycling generation of H2O2 by a model quinolinedione, 6-chloro-7-(2-morpholin-4-yl-ethylamino)-quinoline-5,8-dione (NSC 663284; DA3003-1), other reducing agents such as β-mercaptoethanol, glutathione, and cysteine do not. The assay is compatible with HTS. Once terminated, the assay signal was stable for at least 5 h, allowing for a reasonable throughput. The assay tolerated up to 20% dimethyl sulfoxide, allowing a wide range of compound concentrations to be tested. The assay signal window was robust and reproducible with average Z-factors of ≥0.8, and the redox cycling generation of H2O2 by DA3003-1 in DTT exhibited an average 50% effective concentration of 0.830 μ 0.068 μM. Five of the mitogen-activated protein kinase phosphatase (MKP) 1 inhibitors identified in an HTS were shown to generate H2O2 in the presence of DTT, and their inhibition of MKP-1 activity was shown to be time dependent and was abolished or significantly reduced by either 100 U of catalase or by higher DTT levels. A cross-target query of the PubChem database with three structurally related pyrimidotriazinediones revealed active flags in 36–39% of the primary screening assays. Activity was confirmed against a number of targets containing active site cysteines, including protein tyrosine phosphatases, cathepsins, and caspases, as well as a number of cellular cytotoxicity assays. Rather than utilize resources to conduct a hit characterization effort involving several secondary assays, the phenol red-HRP assay provides a simple, rapid, sensitive, and inexpensive method to identify compounds that redox cycle in DTT or tris(2-carboxyethyl)phosphine to produce H2O2 that may indirectly modulate target activity and represent promiscuous false-positives from a primary screen.
Introduction
Compound interference is the generic terminology utilized by the HTS community to refer to the false-positives that are unavoidable in most, if not all, HTS assay formats.1–4 Compound interference encompasses many modes of action that include chemical and physical compound properties that may impact multiple assay formats and those that may be specific to a particular assay format. For example, compounds that are colored or are fluorescent have the potential to interfere with a wide variety of biochemical and cell-based assay formats.1,2,4,5 In general, direct compound interference is readily anticipated, and the resulting false-positives can be eliminated by either a simple counterscreen or by incorporating an internal reference into the primary screening assay.1,2,4 However, target protein activity may also be modulated indirectly by reactive products generated by the interaction of compounds with assay buffer component(s), and a more involved hit characterization process may be required to identify and eliminate these false-positives.3,6,7
For example, in HTS assays for protein targets with cysteine (Cys) residues that are crucial for either their catalytic or structural function, it is common practice to try to maintain the protein in its active form by including one of a variety of reducing agents in assay and/or enzyme storage buffers.8,9 Commonly utilized reducing agents include dithiothreitol (DTT), tris(2-carboxyethyl) phosphine (TCEP), β-mercaptoethanol (BME), reduced glutathione (GSH), or Cys.8,9 However, O2-containing aqueous solutions of DTT at neutral to acidic pH can generate H2O2 via a chain reaction.10 Furthermore, a number of compounds undergo redox cycling in DTT-containing buffers to generate significant amounts of H2O2.3,6,7 In an initial set of 20,000 compounds, it was found that 85% of the caspase-8 inhibitors were artifacts that inhibited the enzyme through the generation of H2O2 by redox cycling in the presence of DTT.3 The follow-up process to identify and eliminate such redox cycling compounds (RCCs) involved an extensive hit characterization effort, including: enzyme kinetics analysis to show that the inhibition was time dependent; exploring the concentration dependence of different reducing agents; testing whether the inclusion of catalase in the assay abolished inhibition; and examination of the time-dependent effects of the reducing agent on the ultraviolet/visible spectra of compounds to demonstrate that they were reduced.3,6,7
The Molecular Library Screening Center Network (MLSCN) is a component of the National Institutes of Health (NIH) Roadmap Initiative that has provided the broader scientific community access to HTS capabilities.8,11–14 The 10 screening centers of the pilot phase of the network screen multiple NIH-approved assays submitted by assay providers from the scientific community against a diverse compound chemical library that the NIH has assembled and distributes via a small molecule repository.5,8,11–14 The screening centers deliverables include: assays that have been developed and validated for HTS; data uploaded to the PubChem public database from screening the NIH compound library in these assays; and ultimately the generation of chemical probes to further the study of biology and disease through a follow-up process involving secondary assays, resynthesis of the hit(s), and chemical analog synthesis to improve physicochemical properties.5,8,11–14 Compounds that generate H2O2 by redox cycling in reducing agents may indirectly modulate (activate or inhibit) the activity of susceptible targets and would therefore be scored as false-positives.3,6,7 Furthermore, these compounds will likely exhibit nonselective or promiscuous bioactivity profiles against other targets with Cys residues susceptible to modulation by H2O2. We describe here the development and validation of a simple homogeneous 384-well assay to quantify H2O2 generated by the redox cycling of compounds and reducing agents that can be readily utilized to identify and eliminate these false-positives.
Materials and Methods
Reagents and supplies
H2O2 (30% wt/wt) (CAS number 7722-84-1), phenol red (catalog number P-2417), and horseradish peroxidase (HRP) (catalog number P2088-10KU) were from Sigma-Aldrich Co. (St. Louis, MO). Hanks' Balanced Salt Solution (HBSS) was from Hyclone (Logan, UT). Sodium hydroxide was from EMD (Gibbstown, NJ). Dimethyl sulfoxide (DMSO) (99.9% high-performance liquid chromatography grade under argon) was from Alfa Aesar (Ward Hill, MA). All other materials were reagent grade. The quinolinedione DA3003-1 [NSC 663284 or 6-chloro-7-(2-morpholin-4-yl-ethylamino)-quinoline-5,8-dione] was synthesized and obtained as described previously.7,15
384-well H2O2 assay
The 384-well-format H2O2 generation assay was adapted and developed from a tube-based assay to measure H2O2 production by chemically elicited peritoneal macrophages.16–18 The assay is based on the ability of HRP to catalyze the oxidation of phenol red by H2O2, producing a change in its absorbance at 610 nm after the assay reaction has been terminated and adjusted to alkaline pH by addition of 1 N NaOH.18 The miniaturized assay was performed in 384-well flat-bottomed clear polystyrene microtiter plates (120 μl well volume, catalog number 781101, Greiner Bio-One, Monroe, NC). Compounds and plate controls (100 μM H2O2 or 1% DMSO final concentration in the well) were diluted to the required concentrations in HBSS. DTT was diluted in HBSS from 200 mM stocks to the required concentrations (typically 0.5–1.0 mM final concentration in the well). The phenol red-HRP detection reagent contained 300 μg/ml phenol red and 180 μg/ml HRP in HBSS. The assay involved three liquid transfer steps into the 384-well plate performed on an Evolution-P3 liquid handling platform (Perkin Elmer, Waltham, MA) using a 384-well P30 dispensing head; this involved adding 20 μl each of compounds/controls, DTT, and the phenol red-HRP detection reagent to give a final assay volume of 60 μl. Compounds and DTT were incubated together at ambient temperature for a minimum of 15 min prior to addition of the phenol red-HRP detection reagent (100 μg/ml phenol red and 60 μg/ml HRP final concentration in the well). After an additional incubation period at ambient temperature, minimally 5 min, the assay was terminated by addition of 10 μl of 1 N NaOH, and the absorbance of the phenol red was measured at 610 nm in a SpectraMax M5 microtiter plate reader (Molecular Devices, Sunnyvale, CA).
Mitogen-activated protein kinase phosphatase (MKP) 1 assay
The development and implementation of a 384-well format MKP-1 assay in low-volume microtiter plates (catalog number 784076, Greiner Bio-One) with a final incubation volume of 15 μl have been previously described.8 In brief, the homogeneous fluorescence intensity assay involved three consecutive 5-μl additions performed on the Velocity11 (Menlo Park, CA) Vprep® outfitted with a 384-well transfer head: plate controls and compounds, MKP-1 enzyme (250 ng per well), and 3-O-methylfluorescein phosphate (OMFP) substrate (40 μM). Compounds were tested in an assay buffer comprising 30 mM Tris (pH 7.0), 75 mM NaCl, and 1.0 mM EDTA, with 1% each contributed by the diluted compounds and OMFP substrate to yield a final DMSO concentration of 2%. The phosphatase reactions were terminated after a 60-min incubation at ambient temperature by a 5-μl addition of 500 mM NaOH in deionized H2O performed on the Velocity11 Vprep outfitted with a 384-well transfer head, and the fluorescence intensity was measured on a SpectraMax M5 plate reader (excitation filter, 485 nm; emission filter, 525 nm; auto cutoff, 515 nm).
Data analysis assay development
We utilized GraphPad (San Diego, CA) Prism version 4.03 software to plot, fit data to curves, and analyze assay development data; perform linear regression analysis (y = mx + c); fit data to a fourth-order polynomial (y = A + B × x + C × x2 + D × x3 + E × x4) equation; fit data to the sigmoidal dose-response equation y = Bottom + (Top − Bottom)/(1 + 10((LogEC50 − x))); and fit data to the sigmoidal dose-response variable slope equation y = Bottom + (Top − Bottom)/(1 + 10((LogEC50 − x) × Hill slope)), where EC50 represents the 50% effective concentration. Unless otherwise noted the SD was indicated after mean values.
Results
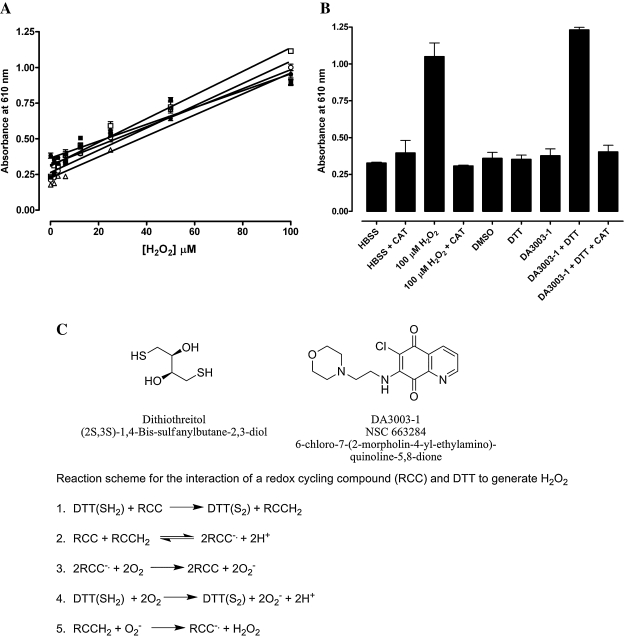
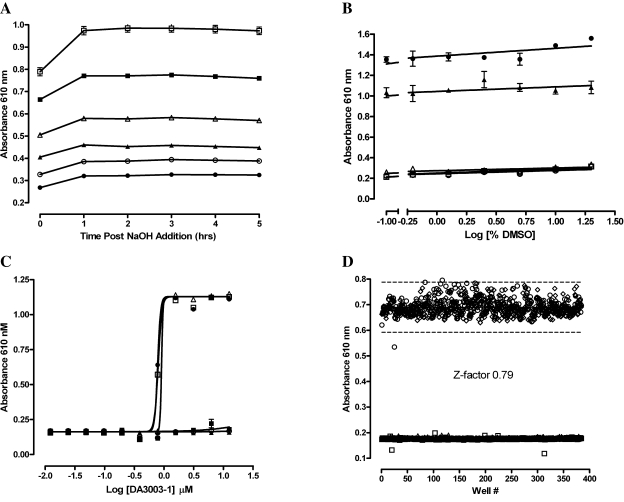
We first examined the performance of the miniaturized 384-well H2O2 detection assay with exogenously added H2O2 ranging from 1.5 to 100 μM (Fig. 1A). In six independent experiments, the HBSS assay buffer produced an average background absorbance of 0.27 μ 0.09, and addition of 100 μM H2O2 produced an average absorbance of 0.97 μ 0.08, providing an average signal to background ratio of 3.59. Linear regression analysis of the data produced an average r2 correlation coefficient of 0.946 μ 0.035 (Fig. 1A). These data indicated that the 384-well-format H2O2 assay was very reproducible and has a dynamic range suitable for detecting H2O2 in the 1–100 μM range. To establish that the assay could be used to detect the H2O2 produced by compounds capable of redox cycling in the presence of DTT, we utilized the quinolinedione DA3003-1, which has previously been shown to produce H2O2 via a redox cycling mechanism7,15 (Fig. 1B and C). When tested individually, neither 10 μM DA3003-1 nor 0.5 mM DTT produced an absorbance signal significantly different from the HBSS buffer control, and neither did the compound solvent DMSO (Fig. 1B). In combination, however, 10 μM DA3003-1 plus 0.5 mM DTT produced a higher absorbance signal than exogenously added H2O2 (100 μM), and both of these signals were completely abolished by the inclusion of 100 U/ml catalase in the assay (Fig. 1B). These data indicate that the H2O2 detection assay was capable of detecting the redox cycling of DA3003-1 in the presence of DTT, that both compound and reducing agent are required for the production of H2O2, that DA3003-1 and DTT can generate concentrations of H2O2 at or above 100 μM, and that the removal of H2O2 by catalase abolishes the change in absorbance of the phenol red-HRP detection reagent. A reaction scheme for the reaction of an RCC and DTT to generate H2O2 adapted from a previous publication6 is presented (Fig. 1C).
FIG. 1.
Phenol red-HRP detection of exogenously added H2O2 or the compound- and DTT-dependent redox cycling generation of H2O2. (A) Exogenous H2O2. In six independent experiments (each indicated by a unique symbol) conducted in triplicate wells per concentration, exogenously added H2O2 ranging from 1.5 to 100 μM (final concentration in well) was mixed with the phenol red-HRP detection reagent in HBSS assay buffer. After 45 min at ambient temperature the assay was terminated by addition of 10 μl of 1 N NaOH, and the absorbance of the phenol red was measured at 610 nm in the microtiter plate reader. The data presented are mean μ SEM values of triplicate wells (n = 3). A linear regression analysis of the data using GraphPad Prism software version 4.03 produced an average r2 correlation coefficient of 0.946 μ 0.035 (n = 6). (B) Compound- and DTT-dependent redox cycling generation of H2O2. In triplicate wells of a 384-well plate, 20 μl of the following components was mixed with 20 μl of HBSS assay buffer: HBSS buffer, 100 U of catalase (CAT), 100 μM H2O2, 100 μM H2O2 + 100 U of CAT, 1% DMSO, 0.5 mM DTT, 10 μM DA3003-1, 10 μM DA3003-1 + 0.5 mM DTT, and 10 μM DA3003-1 + 0.5 mM DTT + 100 U of CAT. The final concentrations of the components in the 60-μl assay volume are indicated. After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 45 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm in the microtiter plate reader. A bar graph of the mean μ SEM absorbance of triplicate wells (n = 3) is presented. (C) Chemical structures of DTT and DA3003-1 and a reaction scheme for the redox cycling generation of H2O2 (through the redox cycling interaction of an RCC and DTT to generate H2O2, adapted from a previous publication6). Addition of DTT(SH2) to RCCs results in the formation of the hydro-RCC (RCCH2) (step 1). When the RCCH2 and RCC are present together they undergo a comproportionation reaction to form a transient radical anion species (RCC−·) (step 2). The RCC−· radical anion reacts with oxygen (O2) to form superoxide (O2−) (step 3). DTT(SH2) also reacts with O2 in the buffer to generate superoxide (step 4). Superoxide can then oxidize the hydro-RCC resulting in the production of H2O2 (step 5).
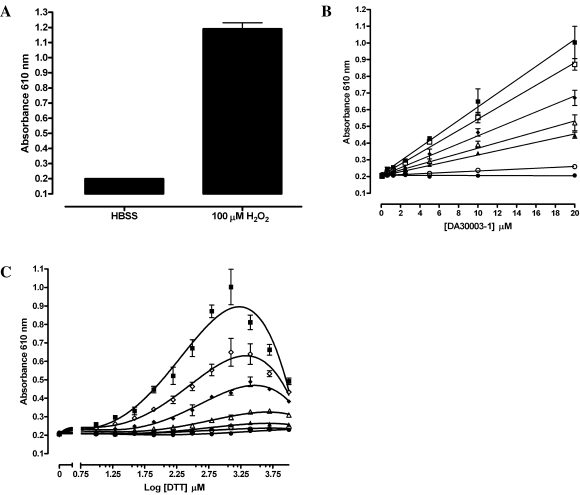
To determine the optimal concentrations of DTT and DA3003-1 required for redox cycling H2O2 generation, we conducted concentration-response assays with the indicated amounts of DA3003-1 and DTT incubated together (Fig. 2). Neither DA3003-1 nor DTT individually produced an absorbance at 610 nm significantly different from either the HBSS or DMSO background signals at any of the concentrations tested. In the presence of DA3003-1, however, DTT in the 0.2–1.0 mM concentration range was able to support the generation of readily detectable levels of H2O2 (Fig. 2B). At DTT concentrations in the 0.2–1.0 mM range, the generation of H2O2 increases linearly with increasing compound concentration, and more H2O2 was produced with higher DTT concentrations (Fig. 2B). At 5.0 and 10.0 mM DTT, however, there was a significant decrease in the absorbance signal that was only partially overcome at the higher compound concentrations of 10 and 20 μM (Fig 2C). With almost all of the DA3003-1 concentrations tested, the DTT concentration response for H2O2 production was biphasic (Fig. 2C). From 0.2 to 1 mM DTT, the generation of H2O2 at each compound concentration increases with higher DTT concentrations, but at DTT concentrations of 5 and 10 mM, the assay signal decreased with higher DTT concentrations (Fig. 2C). These data confirmed that both DA3003-1 and DTT were required for the redox cycling generation of H2O2 and that the amount produced varied in a concentration-dependent manner (Fig. 2B and C).
FIG. 2.
Concentration responses for DA3003-1- and DTT-dependent redox cycling generation of H2O2. (A) Exogenous H2O2. In triplicate wells 20 μl of HBSS or 100 μM H2O2 (final concentration in well) was mixed together with 20 μl of HBSS assay buffer. After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 45 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm in the microtiter plate reader. A bar graph of the mean μ SEM absorbance of triplicate wells (n = 3) is presented. (B) DA3003-1 concentration–response. In triplicate wells 20 μl of DA3003-1 at the indicated concentrations, ranging between 0.625 and 20 μM, was mixed together with 20 μl of the following DTT concentrations in HBSS: 0 μM (•), 9.8 μM (○), 78 μM (▴), 156 μM (▵), 312 μM (♦), 625 μM (□), and 1,250 μM (▪). The final concentrations of the components in the 60-μl assay volume are indicated. After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 45 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm in the M5 microtiter plate reader. The data are presented as mean μ SEM absorbance of triplicate wells (n = 3) together with a linear regression analysis of the data using GraphPad Prism software version 4.03. The r2 correlation coefficients of the linear analysis ranged between 0.92 and 0.98. (C) DTT concentration–response. In triplicate wells 20 μl of DTT at the indicated concentrations, ranging between 9.8 and 10,000 μM, was mixed together with 20 μl of the following DA3003-1 concentrations in HBSS: 0 μM (•), 0.625 μM (○), 1.25 μM (▴), 2.5 μM (▵), 5.0 μM (♦), 10.0 μM (□), and 20.0 μM (▪). The final concentrations of the components in the 60-μl assay volume are indicated. After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 45 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm in the microtiter plate reader. The data are presented as the mean μ SEM absorbance of triplicate wells (n = 3) fit to a fourth-order polynomial (y = A + B × x + C × x2 + D × x3 + E × x4) equation using GraphPad Prism software version 4.03. The data were fit to curves to illustrate the biphasic concentration response of DTT, and except with no DA3003-1 and at 0.625 μM DA3003-1, the r2 correlation coefficient of the fourth-order polynomial equation fit of the data exceeded 0.9.
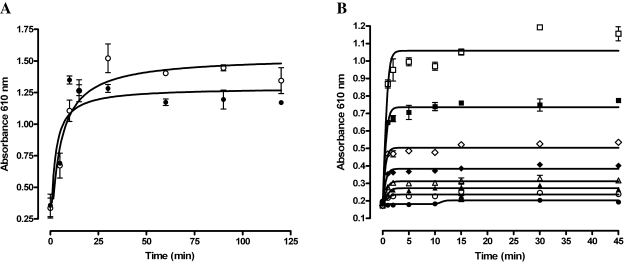
To measure the time course for DA3003-1- and DTT-dependent H2O2 generation, 10 μM DA3003-1 and 0.8 mM DTT were incubated together for the indicated times prior to addition of phenol red-HRP detection reagent (Fig. 3A). Plates were then incubated for an additional either 5 or 45 min before addition of 1 N NaOH and measurement of the absorbance at 610 nm (Fig. 3A). During the first 15–20 min, the amount of H2O2 detected increases with the preincubation time of compound and DTT prior to addition of the phenol red-HRP reagent, and further preincubation produced no significant increase in signal (Fig. 3A). It did not appear to make a significant difference whether the detection reagent was incubated for 5 or 45 min after the incubation of DA3003-1 and DTT together before addition of the 1 N NaOH and measurement of the absorbance at 610 nm (Fig. 3A). To confirm this observation and determine the time course of phenol red-HRP detection, the indicated concentrations of exogenous H2O2 were added to wells, and the detection was terminated with 1 N NaOH at the indicated times after addition of the phenol red-HRP detection reagent before the absorbance was measured at 610 nm (Fig. 3B). The phenol red-HRP detection of the exogenously added H2O2 attained the maximum signal within 2 min, and prolonged incubation with the detection reagent prior to addition of the NaOH provided no additional increase in signal (Fig. 3B). Based on these observations we would recommend an assay procedure where the test compound and reducing agent are incubated together at ambient temperature for a minimum of 15 min prior to addition of the phenol red-HRP detection reagent and that the assay can be terminated with the NaOH stop reagent after an additional 5-min incubation.
FIG. 3.
Time course of DA3003-1 and DTT redox cycling H2O2 generation and of phenol red-HRP detection. (A) Time course of DA3003-1 and DTT redox cycling H2O2 generation. In triplicate wells, 20 μl of DA3003-1 (10 μM final concentration) in HBSS was mixed together with 20 μl of DTT (0.8 mM final concentrtion) in HBSS assay buffer. The DA3003-1 and DTT were incubated together at ambient temperature for the indicated times, ranging from 0 to 120 min, prior to addition of phenol red-HRP detection reagent. Plates were then either incubated for an additional 5 min (○) or 45 min (•) before addition of 10 μl of 1 N NaOH to terminate the assay and measurement of the absorbance at 610 nm in the microtiter plate reader. The data are presented as mean μ SEM absorbance of triplicate wells (n = 3). (B) Time course of phenol red-HRP detection of H2O2. In triplicate wells, 20 μl of the indicated concentrations of exogenous H2O2 was added to wells containing 20 μl of HBSS. After addition of the phenol red-HRP detection reagent (20 μl), the reaction was terminated with 10 μl of 1 N NaOH at the indicated times, and the absorbance at 610 nm was measured in the microtiter plate reader. Final H2O2 concentrations in HBSS were: 0 μM (•), 1.562 μM (○), 3.125 μM (▴), 6.25 μM (▵), 12.5 μM (♦), 25 μM (⋄), 50 μM (▪), and 100 μM (□). The data are presented as mean μ SEM absorbance of triplicate wells (n = 3). The time course data in (A) and (B) were fit to curves using the sigmoidal dose-response equation y = Bottom + (Top − Bottom)/(1 + 10(LogEC50 − x)) using GraphPad Prism software version 4.03.
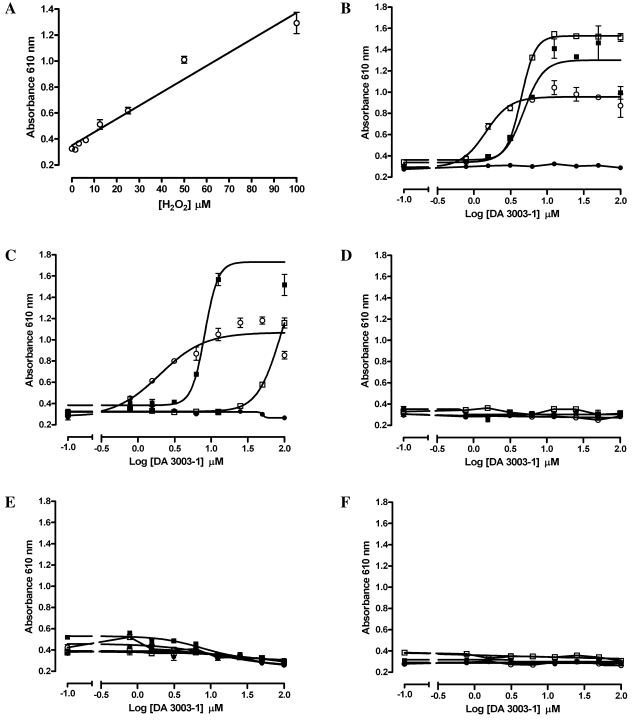
We next wanted to investigate the ability of other reducing agents to support redox cycling and H2O2 generation by DA3003-1 (Fig. 4). For comparison, the absorbance signal produced by exogenously added H2O2 at a concentration ranging from 1.5 to 100 μM is presented (Fig. 4A). The reducing agents tested included DTT (Fig. 4B), TCEP (Fig. 4C), GSH (Fig. 4D), Cys (Fig. 4E), and BME (Fig. 4F). The indicated concentrations of DA3003-1 were incubated together with the various reducing agents at 0, 0.2, 0.4, or 0.8 mM for 15 min prior to addition of the phenol red-HRP detection reagent, and the assay was then stopped with 1 N NaOH after another 5 min (Fig. 4B–F). Only DTT and TCEP were able to support the redox cycling production of H2O2 in the presence of DA3003-1 (Fig. 4B and C).
FIG. 4.
Other reducing reagents. (A) Exogenous H2O2. In triplicate wells per concentration, exogenously added H2O2 ranging from 1.5 to 100 μM (final concentration) was mixed with 20 μl of the phenol red-HRP detection reagent in HBSS assay buffer. After 5 min at ambient temperature the assay was terminated by addition of 10 μl of 1 N NaOH, and the absorbance of the phenol red was measured at 610 nm with a microtiter plate reader. The data are presented as mean μ SEM absorbance of triplicate wells (n = 3). A linear regression analysis of the data using the GraphPad Prism software version 4.03 produced an r2 correlation coefficient of 0.963. (B–F) Different reducing reagents: (B) DTT, (C) TCEP, (D) GSH, (E) Cys, and (F) BME. In triplicate wells 20 μl of DA3003-1 at the indicated concentrations, ranging between 0.78 and 100 μM, were mixed together with 20 μl of the following concentrations of the different reducing reagents in HBSS: 0 μM (•), 200 μM (○), 400 μM (□), and 800 μM (▪). The final concentrations of the components in the 60-μl assay volume are indicated. After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 5 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm with a microtiter plate reader. The data are presented as mean μ SEM absorbance of triplicate wells (n = 3). The data for DTT (B) and TCEP (C) were fit to curves using the sigmoidal dose-response variable slope equation y = Bottom + (Top − Bottom)/(1 + 10((LogEC50 − x) × Hill slope)) using GraphPad Prism software version 4.03.
We conducted a series of experiments to see if the phenol red-HRP H2O2 detection assay would be compatible with HTS. To examine how long the assay signal was stable after addition of the 1 N NaOH stop reagent, we measured the absorbance at 610 nm at 1-h intervals for up to 5 h (Fig. 5A). Relative to the readings immediately post-termination, there was a slight but reproducible increase in the absorbance at 610 nm when the plate was reread 1 h after termination of the reaction, and thereafter the signal was stable for 5 h (Fig. 5A) and even as long as 24 h (data not shown). These data indicate that throughput and capacity would not be compromised by signal stability. The detection of both exogenously added H2O2 and H2O2 generated via the redox cycling of DA3003-1 in the presence of 0.8 mM DTT was tolerant to DMSO concentrations ranging from 0.3% up to 20% (Fig. 5B), indicating that a wide range of compound/DMSO concentrations could be screened in the assay. The DA3003-1 concentration response for the redox cycling-dependent generation of H2O2 in the presence of 0.8 mM DTT was very reproducible and produced an average EC50 of 0.830 μ 0.068 μM from three independent experiments conducted in triplicate (Fig. 5C). To further verify signal reproducibility and the magnitude of the assay signal window, we utilized a rigorous procedure8,9,13 to measure the Z-factor statistical parameter19 in three independent experiments using two full 384-well plates of maximum (100 μM H2O2) and minimum (HBSS) signals for each experiment (Fig. 5D). The Z-factors generated in these experiments were 0.84, 0.79, and 0.79, indicating that the assay signal window had the required robustness and reproducibility for HTS.
FIG. 5.
HTS compatibility: signal stability, DMSO tolerance, EC50 curves, and assay signal window Z-factor determination. (A) Phenol red-HRP H2O2 signal stability. In six wells, 20 μl of the indicated concentrations of exogenous H2O2 were added to wells containing 20 μl of HBSS. Five minutes after addition of 20 μl of the phenol red-HRP detection reagent the assay was terminated with 10 μl of 1 N NaOH, and the absorbance at 610 nm was measured with a microtiter plate reader. Final H2O2 concentrations in HBSS were: 0 μM (•), 6.25 μM (○), 12.5 μM (▴), 25 μM (▵), 50 μM (▪), and 100 μM (□). The data are presented as mean μ SEM absorbance of six wells (n = 6). The plate was read immediately after termination and reread every hour thereafter for up to 5 h. (B) DMSO tolerance. In triplicate wells of a 384-well plate, 20 μl of the following components were mixed together with 20 μl of HBSS assay buffer containing the indicated concentrations of DMSO ranging from 0% to 20%: HBSS buffer (▵), 100 μM H2O2 (▴), 0.8 mM DTT (□), 10 μM DA3003-1 (○), and 10 μM DA3003-1 + 0.8 mM DTT (•). The final concentrations of the components in the 60-μl assay volume are indicated. After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 5 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm with a microtiter plate reader. A line graph of the mean μ SEM absorbance of triplicate wells (n = 3) versus the log10 of the percentage DMSO is presented. (C) DA3003-1 EC50 curves with or without 0.8 mM DTT. In triplicate wells, 20 μl of the indicated concentrations of DA3003-1, ranging from 0.12 to 12.5 μM, were added to wells containing 20 μl of HBSS with or without 0.8 mM DTT (final concentration in assay well). After a 15–20-min incubation at ambient temperature, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for an additional 5 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm with a microtiter plate reader. The data are presented as mean μ SEM absorbance of triplicate wells (n = 3). Three independent experiments were conducted, and the data for H2O2 generation by DA3003-1 with or without 0.8 mM DTT were fit to curves using the sigmoidal dose-response variable slope equation y = Bottom + (Top − Bottom)/(1 + 10((LogEC50 − x) × Hill slope)) using GraphPad Prism software version 4.03: Test 1 no DTT (▴), Test 1 + 0.8 mM DTT (▵), Test 2 no DTT (▪), Test 2 + 0.8 mM DTT (□), Test 3 no DTT (•), and Test 3 + 0.8 mM DTT (○). (D) Assay signal window Z-factor determination. To rigorously determine the assay signal window and variability of the phenol red-HRP H2O2 detection HTS assay, we assayed two full 384-well plates each of maximum (100 μM H2O2 + 1% DMSO) and minimum (HBSS + 1% DMSO) signal controls in independent experiments conducted on three consecutive days. The data from the four plates run on day 2 are presented in a scatter plot of absorbance at 610 nm versus well number: maximum plate 1 (•), maximum plate 2(○), minimum plate 1 (□), minimum plate 2 (▪); solid lines, mean absorbance 610 nm of maximum or minimum plates 1 and 2 (n = 768); dashed lines, mean μ 3 × SEM (n = 768) absorbance 610 nm of maximum or minimum plates 1 and 2. The phenol red-HRP H2O2 detection HTS assay Z-factor19 was calculated as described previously.8,9,13 The Z-factors generated in the independent experiments were 0.84, 0.79, and 0.79, indicating that the assay signal window has the required robustness and reproducibility for HTS.
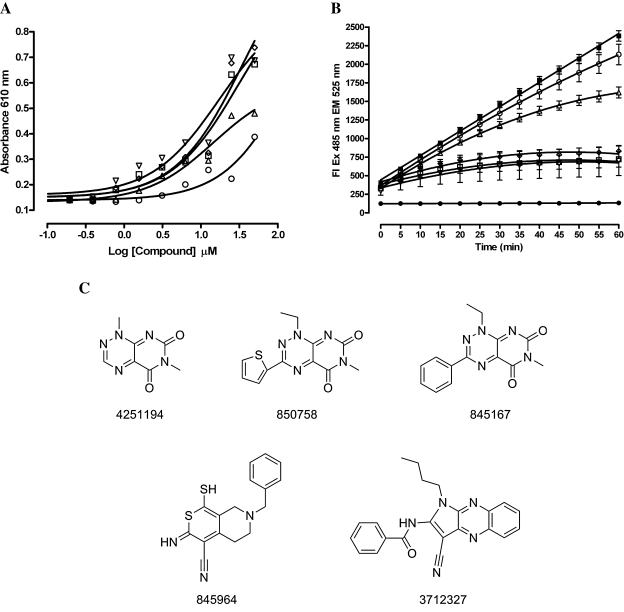
A HTS of 65,000 compounds from the NIH's compound library in a MKP-1 assay identified 46 concentration-dependent inhibitors of MKP-1 with 50% inhibitory concentration (IC50) values of <50 μM.8 The HTS and IC50 data have been uploaded to the PubChem database and are publicly available via the PubChem assay identity number (AID) 374 and 551, respectively. As protein tyrosine phosphatase (PTPs) are susceptible to reversible oxidation and inactivation by H2O26,7,20–27 and the compounds were screened in an MKP-1 assay buffer containing 1 mM DTT,8 we examined the ability of the MKP-1 confirmed actives to generate H2O2 in the presence of 1 mM DTT (AID 676). Five of the MKP-1 active compounds incubated in 1 mM DTT generated readily detectable levels of H2O2 in 10-point concentration-response assays (Fig. 6A). Consistent with the hypothesis that the inhibition of MKP-1 by these compounds was mediated by the redox cycling generation of H2O2, the degree of inhibition of MKP-1 activity was shown to be time dependent and increased over the course of the assay incubation period (Fig. 6B). The three most potent redox cycling compounds were structurally related pyrimidotriazinediones (Fig. 6C): 4251194, 1,6-dimethylpyrimido[5,6-e][1,2,4]triazine-5,7-dione; 850758, 1-ethyl-6-methyl-3-thiophen-2-ylpyrimido[5,6-e][1,2,4]triazine-5,7-dione; and 845167, 1-ethyl-6-methyl-3-phenylpyrimido[5,6-e][1,2,4]triazine-5,7-dione. The other redox cycling compounds were 845964, 3-amino-7-(phenylmethyl)-1-sulfanylidene-6,8-dihydro-5H-thiopyrano[5,4-c]pyridine-4-carbonitrile, and 3712327, N-(1-butyl-3-cyanopyrrolo[5,4-b]quinoxalin-2-yl)benzamide (Fig. 6C). For these compounds, addition of 100 U of catalase to the MKP-1 assay or raising the DTT concentration in the MKP-1 assay to 25 mM either abolished the inhibition of MKP-1 or shifted the potencies of the MKP-1 IC50 values higher (Table 1).
FIG. 6.
Redox cycling generation of H2O2 by selected MKP-1 inhibitors, time-dependent inhibition of MKP-1, and chemical structures. (A) Redox cycling generation of H2O2 by selected MKP-1 inhibitors and DTT. In singlet wells 20 μl of the MKP-1 inhibitors at the indicated concentrations, ranging between 0.2 and 50 μM (final concentration), was mixed together with 20 μl of 0.5 mM DTT (final concentration) in HBSS: 3712327 (○), 845964 (▵), 845167 (□), 850758 (⋄), and 4251194 (▿). After 15–20 min, 20 μl of phenol red-HRP detection reagent was added, and the assay was incubated for 5 min at ambient temperature before termination by addition of 10 μl of 1 N NaOH and measurement of the absorbance at 610 nm with a microtiter plate reader. The concentration–response data (n = 1) were fit to curves using the sigmoidal dose–response equation y = Bottom + (Top − Bottom)/(1 + 10(LogEC50 − x)) using GraphPad Prism software version 4.03. (B) Time-dependent inhibition of MKP-1 activity. Recombinant MKP-1 enzyme (250 ng per well) was incubated with 40 μM OMFP and the indicated compounds at 10 μM in a final volume of 15 μl in an assay buffer comprising 30 mM Tris (pH 7.0), 75 mM NaCl, and 1.0 mM EDTA, for up to 60 min at ambient temperature in triplicate wells of a 384-well low-volume microtiter plate, and the fluorescent intensity at 485 nm excitation/525 nm emission was acquired on the M5 plate reader over time: maximum plate control (▪), minimum plate control (NaOH-inhibited enzyme) (•), 3712327 (○), 845964 (▵), 845167 (□), 850758 (⋄), and 4251194 (▿). The mean μ SEM fluorescent intensity of triplicate wells (n = 3) for each time point and compound are plotted together with the resulting linear or nonlinear regression line or curve fits produced using GraphPad Prism software version 4.03. The maximum (▪) and minimum (•) plate controls exhibited r2 correlation coefficients of 0.9985 and 0.9131 for the linear analysis, respectively. The data for 3712327 (•), 845964 (▵), 845167 (□), 850758 (⋄), and 4251194 (▿) were all fit to curves using the sigmoidal dose–response equation y = Bottom + (Top − Bottom)/(1 + 10(LogEC50 − x)) using GraphPad Prism software version 4.03. (C) Chemical structures. The chemical structures of the five compounds capable of redox cycling in DTT: 4251194, 1,6-dimethylpyrimido[5,6-e][1,2,4]triazine-5,7-dione; 850758, 1-ethyl-6-methyl-3-thiophen-2-ylpyrimido[5,6-e][1,2,4]triazine-5,7-dione; 845167, 1-ethyl-6-methyl-3-phenylpyrimido[5,6-e][1,2,4]triazine-5,7-dione; 845964, 3-amino-7-(phenylmethyl)-1-sulfanylidene-6,8-dihydro-5H-thiopyrano[5,4-c]pyridine-4-carbonitrile; and 3712327, N-(1-butyl-3-cyanopyrrolo[5,4-b]quinoxalin-2-yl)benzamide.
Table 1.
MKP-1 Inhibition Activity Profile: MKP-1 IC50, Effects of Catalase and Higher DTT, and EC50 for Redox Cycling H2O2 Generation
| |
MKP-1 + |
|
|
|||||
|---|---|---|---|---|---|---|---|---|
| |
1 mM DTT |
1 mM DTT + catalase |
25 mM DTT |
H2O2 generation |
||||
| PubChem SID | % inhibition at 50 μM | IC50 (μM) | % inhibition at 50 μM | IC50 (μM) | % inhibition at 50 μM | IC50 (μM) | % activity at 50 μM | EC50 (μM) |
| 4251194 | 94.40 | 0.184 | −27.25 | >50 | 97.95 | 5.684 | 85.817 | 10.714 |
| 850758 | 94.42 | 0.158 | −92.22 | >50 | 98.18 | 4.427 | 93.886 | 13.738 |
| 845167 | 95.59 | 0.197 | −66.27 | >50 | 96.16 | 4.901 | 82.819 | 13.77 |
| 845964 | 95.19 | 1.128 | 47.72 | >50 | 35.71 | >50 | 52.776 | 34.554 |
| 3712327 | 79.92 | 23.536 | 69.91 | 32.374 | −5.25 | >50 | 36.786 | >50 |
A cross-target query of PubChem revealed that the three most potent redox active compounds, the pyrimidotriazinediones, produced active flags in 36–39% of the assays against which they were screened, while the two less potent redox active compounds, 845964 and 3712327, produced active flags in 17–22% of their screens (Table 2). Furthermore, these redox active compounds have exhibited confirmed activity against a number of targets with active site Cys residues, including PTPs, cathepsins, and caspases, as well as in a number of cellular cytotoxicity assays (Table 2).
Table 2.
Cross-Target Query of PubChem Database for Redox Cycling Compounds
| PubChem SID | Number of assays | Number of active flags | Selected confirmed targets |
|---|---|---|---|
| 4251194 | 164 | 64 (39%) | MKP-1; MKP-3; Cdc25B; cathepsins S, B, and L; caspases 1 and 7; HSP90; several cytotoxicity models and other targets |
| 850758 | 196 | 71 (36%) | MKP-1; MKP-3; Cdc25B; cathepsins S, B, and L; HSP90; several cytotoxicity models and other targets |
| 845167 | 221 | 85 (38.5%) | MKP-1; MKP-3; Cdc25B; cathepsins S, B, and L; caspases 1 and 7; HSP90; several cytotoxicity models and other targets |
| 845964 | 172 | 38 (22.1%) | MKP-1; MKP-3; Cdc25B; cathepsins S, B; caspase 7; HSD17B4; HADH2 |
| 3712327 | 202 | 36 (17.8%) | MKP-1; MKP-3; Cdc25B; HSD174B4; HSP90; several cytotoxicity models and other targets |
HADH2, hydroxyacyl-coenzyme A dehydrogenase type II; HSD174B4, 17β-hydroxysteroid dehydrogenase 4; HSP, heat shock protein.
Discussion
We report here the development and optimization of a simple 384-well colorimetric assay to measure H2O2 generated by the redox cycling of compounds incubated with reducing agents in HTS assay buffers. The assay was based on the ability of HRP to catalyze the oxidation of phenol red by H2O2, thereby changing its absorbance at alkaline pH.16–18 The phenol red-HRP assay readily detected exogenously added H2O2 in the 1.5–100 μM concentration range (Figs. 1A, 2A, and 4A) and H2O2 generated when the quinolinedione DA3003-1 was incubated with DTT (Figs. 1B, 2B and C, 4B, and 5C). The detection of H2O2 either exogenously added or endogenously generated by DA3003-1 and DTT was abolished by addition of catalase (Fig. 1B). The redox cycling generation of H2O2 required the presence of both DA3003-1 and DTT, and the amount of H2O2 produced varied in a concentration-dependent manner for both components (Figs. 1B, 2B and C, and 4B). The amount of H2O2 produced by the redox cycling of DA3003-1 in the presence of DTT reached a steady-state level after 15–20 min of incubation, and thereafter addition of the phenol red-HRP detection reagent achieved a maximum signal within 2 min (Fig. 3). Although both DTT and TCEP supported the redox cycling generation of significant levels of H2O2 by DA3003-1, incubation with other reducing agents such as BME, GSH, and Cys failed to produce levels of H2O2 significantly over background (Fig. 4). The phenol red-HRP H2O2 detection assay is compatible with HTS: the assay signal was stable post-termination of the assay, the assay tolerated up to 20% DMSO, the assay signal window was robust and reproducible (Z-factors ≥0.8), and the average EC50 of 0.830 μ 0.068 μM for the redox cycling generation of H2O2 by DA3003-1 in DTT was reproducible (Fig. 5). Five confirmed actives from an MKP-1 HTS generated readily detectable levels of H2O2 in the presence of DTT (Fig. 6A), and the inhibition of MKP-1 activity was shown to be time dependent (Fig. 6B). Three of the five redox cycling compounds were structurally related pyrimidotriazinediones (Fig. 6C). The inhibition of MKP-1 by the redox cycling compounds was abolished or significantly reduced either by addition of catalase or by raising the DTT concentration in the MKP-1 assay to 25 mM (Table 1). A cross-target query of PubChem database for the bioactivity profiles of these five redox cycling compounds yielded active flags in 17–39% of the assays that they were screened against, with confirmed activity against a number of targets with active site Cys residues, including PTPs, cathepsins, and caspases, as well as a number of cellular cytotoxicity assays (Table 2). Many of the selected dose-response confirmation assays listed in Table 2 were secondary assays conducted to evaluate the target family specificity of putative confirmed actives, e.g., PTPs and Cys proteases. This simple colorimetric assay therefore provides a rapid, sensitive, and cheap method to identify compounds that interact with reducing agents like DTT or TCEP to produce H2O2 that may indirectly modulate target activity and represent false-positives in a primary screen.
With the commonly utilized reducing agent DTT, disulfide formation was strongly facilitated, and sulfur-centered radicals derived from DTT were chemically generated in aqueous solution in the neutral to acidic pH range.10 In the presence of O2 these sulfur-centered radicals derived from DTT were transformed by a chain reaction into dihydroxydithiane and H2O2.10 A number of compounds have been shown to interact with DTT in solution to establish a reactive oxygen intermediate (ROI) generating cycle that produces H2O2 (Fig. 1C).3,6,7 The inhibition of PTPα by ortho-quinones was shown to be mediated by the generation of H2O2 through a cyclic redox mechanism that required both the compound and DTT.6 ortho-Quinone-based inhibition of PTPα was time dependent and was sensitive to both superoxide dismutase and catalase.6 ortho-Quinones produced H2O2 in the presence of strong reducing agents including DTT and TCEP, but not when weaker reducing agents like GSH or Cys were present.6 It was further proposed that the cellular activity of the ortho-quinones might be due their interaction with cellular thiol donors, such as GSH, glutaredoxin, and thioredoxin, or other cell-associated enzyme systems that might be able to reduce quinones and catalyze the generation of H2O2.6 The CD45 receptor PTP inhibitor, 9,10-phenanthrenedione, was also shown to generate significant amounts of H2O2 in the presence of DTT.6 The in vitro inhibition of Cdc25B by cell-active quinolinediones was shown to be irreversible, time dependent, and sensitive to pH, catalase, and reducing agents.7,15 In the absence of exogenous reducing agent, the quinones did not inhibit Cdc25B in vitro, and addition of catalase abolished Cdc25B inhibition in the presence of reducing agents, implicating redox cycling and the generation of H2O2 as the mode of action.7,15 Interestingly, addition of these quinone compounds to cells was shown to directly trigger the production of intracellular ROI. An HTS of >700,000 compounds to identify inhibitors of caspase-8 produced an unusually high hit rate (∼1%), and for an initial set of 20,000 compounds, it was found that 85% of the inhibitors were artifacts that inhibited the enzyme through the generation of H2O2 by redox cycling in the presence of DTT.3 Quinone and quinonoid compound inhibition of caspase-8 was shown to be time dependent, abolished by catalase, and required strong reducing agents such as DTT or TCEP.3 These quinone and quinonoid compounds failed to inhibit caspase-8 in the presence of either GSH or Cys, but were rapidly reduced in the presence of DTT as indicated by changes in their ultraviolet/visible spectra.3 It was concluded the quinone and quinonoid compounds were reduced by DTT and established a ROI generating cycle that produced H2O2 that in turn inhibited the caspase-8 by oxidizing and inactivating the active site Cys of the enzyme.3 In each case, the identification and elimination of redox cycling compounds from the hit lists for these targets required a hit characterization process that involved several secondary follow-up assays.3,6,7
We utilized the phenol red-HRP H2O2 detection assay to identify five redox cycling compounds in the 46 confirmed actives from the MKP-1 HTS (Fig. 6).8 Three of the five redox cycling compounds were structurally related pyrimidotriazinediones (Fig. 6C). The inhibition of MKP-1 activity for the five compounds was time dependent, abolished by catalase, or significantly reduced at higher DTT concentrations (Fig. 6 and Table 1). The inhibition of Cdc25B by redox-active quinolinediones was also reversed by higher DTT concentrations, and the initial oxidation of the PTP active site Cys by H2O2 produced a sulfenic acid form that was reversible in the presence of excess DTT.6,7 The optimal DTT concentration for H2O2 generation varied with the concentration of DA3003-1, but in general the amount of H2O2 produced at 10 mM DTT was reproducibly less than with lower DTT concentrations (Fig. 2C). Although we currently cannot explain the apparent suppression of H2O2 generation by the redox cycling of DA3003-1 in buffers containing 10 mM DTT (Fig. 2C), the lower production of H2O2 combined with the ability of excess DTT to reverse the initial oxidation of active site Cys residues6 may both be contributing to the reduced MKP-1 inhibition observed in assays with 25 mM DTT (Table 1). Overall these data would support the selection of weaker reducing agents such as GSH or Cys rather than DTT for the HTS assay buffers of protein targets with active site Cys residues susceptible to oxidation by H2O2. If, however, the use of DTT is unavoidable, our data would suggest that it should be utilized at concentrations ≥10 mM to try to minimize the number of false-positives due to the generation of H2O2 by redox cycling compounds.
H2O2 is a relatively mild oxidant that can oxidize the Cys residues of proteins to Cys sulfenic acid or disulfide that can be reduced back to Cys by cellular thiol donors such as GSH, glutaredoxin, and thioredoxin.20,21,25,28–30 The pKa of sulfhydryl groups of most Cys residues in proteins is ∼8.5, and they do not react at physiologically significant rates with H2O2 unless the reaction is catalyzed.20,21,23,25–30 The Cys thiolate anion is much more readily oxidized by H2O2, and for some signaling proteins, most notably the PTPs, the Cys has been clearly shown to be preferentially in a thiolate form.20,21,23,25–30 The 107 PTPs found in the human genome are defined by the active site sequence C(X)5R(S/T), with X being any amino acid, and they are critical regulators of mammalian cell proliferation, differentiation, and apoptosis.24 The active site Cys of PTPs is required for catalytic activity and performs a nucleophilic attack on the phosphotyrosine residues of the substrate to form a covalent thiol-phosphate intermediate followed by hydrolysis and the release of the phosphate.20,21,23,25–27 Because of the unique environment and the invariant arginine, PTP active site Cys residues have an unusually low pKa of 4.7–5.4 and exist as thiolate anions at neutral pH, which enhances catalytic function as a nucleophile but increases susceptiblity to inactivation by H2O2.20,21,23,25–27 The PTP Cys thiolate reacts with H2O2 to yield the sulfenic acid of Cys, which can be reduced back to the active thiolate species by cellular thiol donors such as GSH.24 Oxidation to the sulfenic form renders the PTP enzyme inactive against phosphorylated substrates. Sulfenic acids are highly reactive and readily undergo further oxidation to sulfinic and sulfonic forms, which are irreversibly oxidized and inactive.20,21,23,25–27 The specific and reversible oxidation and inactivation of PTPs by H2O2 have now been demonstrated for numerous PTPs involved in the regulation of key signaling pathways controlling normal physiological processes and disease progression: PTP1B, PTPα, LAR, VHR, PTEN, SHP-2, Cdc25B, and MKP-3.6,7,20,21,23,25–27 Other proteins that may have critical Cys residues in the thiolate form that are susceptible to reaction with H2O2 include activator protein 1 and nuclear factor κB transcription factors, caspases, and some protein kinase C isoforms.21,25,28–30 The protein serine-threonine phosphatases are metallo-enzymes regulated by specific regulatory subunits, but some family members (PP2B, PP1, and PP2A) may be inactivated by H2O2, which has been attributed to the formation of a disulfide bond between redox-sensitive Cys residues of the proteins.25 A number of protein serine-threonine kinases and protein tyrosine kinases have been shown to be activated by H2O2: epidermal growth factor receptor, mitogen-activated protein kinase, c-Ret, Abl, Src, and LCK.21,25,28–30 H2O2 is now considered to be a ubiquitous intracellular messenger at subtoxic concentrations that can modulate (activate/inhibit) the activity of a variety of proteins, including protein phosphatases, protein kinases, Cys proteases, transcription factors, phospholipases, ion channels, and G proteins.20,21,23–30 As many of the assays screened by the MLSCN (and uploaded to PubChem) involve target classes susceptible to modulation by H2O2, it is perhaps not surprising that the five compounds identified in the phenol red-HRP H2O2 detection assay exhibit promiscuous bioactivity profiles (Table 2).
We describe here the development, optimization, and validation of a simple, rapid, and inexpensive assay to identify compounds capable of the redox cycling generation of H2O2 in the presence of DTT. This single assay can be used to identify and eliminate redox cycling compounds from primary screening hit lists that previously required a hit characterization process involving several secondary follow-up assays. The Pittsburgh Molecular Library Screening Center has utilized this assay to identify redox cycling compounds among the confirmed hits from several of our screening campaigns (Pubchem AIDs 672, 682, 683, 876, and 936) and to profile >197,000 compounds from the NIH's small molecule library (PubChem AIDs 878 and 1234, authors' manuscript in preparation). Given that H2O2 is considered to be a ubiquitous intracellular messenger at subtoxic concentrations that can modulate (activate/inhibit) the activity of a variety of proteins, the annotation of the NIH small molecule library should prove useful for future mining of the data in PubChem. In addition, it will be interesting to see how many redox cycling compounds are in the NIH's compound library and whether they will also exhibit promiscuous biological activity against other targets screened by the MLSCN. The availability of additional compound classes capable of redox cycling should provide probes to investigate whether compound interactions with cellular thiol donors and the generation of H2O2 may be the basis of activity in cellular assays, as has been proposed for ortho-quinones and quinolinediones.6,7
Acknowledgments
The work reported here was supported by NIH MLSCN grant U54MH074411.
Abbreviations
- 3712327
N-(1-butyl-3-cyanopyrrolo[5,4-b]quinoxalin-2-yl)benzamide
- 4251194
1,6-dimethylpyrimido[5,6-e][1,2,4]triazine-5,7-dione
- 845167
1-ethyl-6-methyl-3-phenylpyrimido[5,6-e][1,2,4]triazine-5,7-dione
- 845964
3-amino-7-(phenylmethyl)-1-sulfanylidene-6,8-dihydro-5H-thiopyrano[5,4-c]pyridine-4-carbonitrile
- 850758
1-ethyl-6-methyl-3-thiophen-2-ylpyrimido[5,6-e][1,2,4]triazine-5,7-dione
- AID
assay identity number
- BME
β-mercaptoethanol
- Cys
cysteine
- DA3003-1
NSC 663284 or 6-chloro-7-(2-morpholin-4-yl-ethylamino)-quinoline-5,8-dione
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EC50
50% effective concentration
- GSH
reduced glutathione
- HBSS
Hanks' Balanced Salt Solution
- HRP
horseradish peroxidase
- IC50
50% inhibitory concentration
- MKP
mitogen-activated protein kinase phosphatase
- MLSCN
Molecular Library Screening Center Network
- NIH
National Institutes of Health
- OMFP
O-methylfluorescein phosphate
- PTP
protein tyrosine phosphatase
- RCC
redox cycling compound
- ROI
reactive oxygen intermediate
- TCEP
tris(2-carboxyethyl)phosphine.
References
- 1.Carpenter JW. Laethem C. Hubbard FR. Eckols TK. Baez M. McClure D, et al. Configuring radioligand receptor binding assays for HTS using scintillation proximity assay technology. Methods Mol Biol. 2002;190:31–49. doi: 10.1385/1-59259-180-9:031. [DOI] [PubMed] [Google Scholar]
- 2.Johnston PA. Johnston PA. Cellular platforms for HTS: three case studies. Drug Discov Today. 2002;7:353–363. doi: 10.1016/s1359-6446(01)02140-7. [DOI] [PubMed] [Google Scholar]
- 3.Smith GK. Barrett DG. Blackburn K. Cory M. Dallas WS. Davis R, et al. Expression, preparation, and high-throughput screening of caspase-8: discovery of redox-based and steroid diacid inhibition. Arch Biochem Biophys. 2002;399:195–205. doi: 10.1006/abbi.2002.2757. [DOI] [PubMed] [Google Scholar]
- 4.Trask OJ., Jr Baker A. Williams RG. Nickischer D. Kandasamy R. Laethem C, et al. Assay development and case history of a 32K-biased library high-content MK2-EGFP translocation screen to identify p38 mitogen-activated protein kinase inhibitors on the ArrayScan 3.1 imaging platform. Methods Enzymol. 2006;414:419–439. doi: 10.1016/S0076-6879(06)14023-9. [DOI] [PubMed] [Google Scholar]
- 5.Simeonov A. Jadhav A. Thomas CJ. Wang Y. Huang R. Southall NT, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 6.Bova MP. Mattson MN. Vasile S. Tam D. Holsinger L. Bremer M, et al. The oxidative mechanism of action of ortho-quinone inhibitors of protein-tyrosine phosphatase alpha is mediated by hydrogen peroxide. Arch Biochem Biophys. 2004;429:30–41. doi: 10.1016/j.abb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Brisson M. Nguyen T. Wipf P. Joo B. Day BW. Skoko JS, et al. Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol. 2005;68:1810–1820. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- 8.Johnston PA. Foster CA. Shun TY. Skoko JJ. Shinde S. Wipf P, et al. Development and implementation of a 384-well homogeneous fluorescence intensity high-throughput screening assay to identify mitogen-activated protein kinase phosphatase-1 dual-specificity protein phosphatase inhibitors. Assay Drug Dev Technol. 2007;5:319–332. doi: 10.1089/adt.2007.066. [DOI] [PubMed] [Google Scholar]
- 9.Tierno MB. Johnston PA. Foster C. Skoko JJ. Shinde SN. Shun TY, et al. Development and optimization of high-throughput in vitro protein phosphatase screening assays. Nat Protoc. 2007;2:1134–1144. doi: 10.1038/nprot.2007.155. [DOI] [PubMed] [Google Scholar]
- 10.Lal M. Rao R. Fang X. Schuchmann HP. Sonntag CV. Radical-induced oxidation of dithiothreitol in acidic oxygenated solution: a chain reaction. J Am Chem Soc. 1997;119:5735–5739. [Google Scholar]
- 11.Austin CP. Brady LS. Insel TR. Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 12.Inglese J. Auld DS. Jadhav A. Johnson RL. Simeonov A. Yasgar A, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston PA. Johnston PA. Shun TY. Shinde S. Lazo JS. Huryn DM, et al. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 14.Lazo JS. Roadmap or roadkill: a pharmacologist's analysis of the NIH Molecular Libraries Initiative. Mol Interv. 2006;6:240–243. doi: 10.1124/mi.6.5.1. [DOI] [PubMed] [Google Scholar]
- 15.Guo J. Parise RA. Joseph E. Lan J. Pan SS. Joo B, et al. Pharmacology and antitumor activity of a quinolinedione Cdc25 phosphatase inhibitor DA3003-1 (NsC 663284) Anticancer Res. 2007;27:3067–3073. [PubMed] [Google Scholar]
- 16.Johnston PA. Adams DO. Hamilton TA. Regulation of the Fc-receptor-mediated respiratory burst: treatment of primed murine peritoneal macrophages with lipopolysac-charide selectively inhibits H2O2 secretion stimulated by immune complexes. J Immunol. 1985;135:513–518. [PubMed] [Google Scholar]
- 17.Johnston PA. Adams DO. Hamilton TA. Regulation of respiratory burst in murine peritoneal macrophages: differential sensitivity to phorbol diesters by macrophages in different states of functional activation. Cell Immunol. 1986;100:400–410. doi: 10.1016/0008-8749(86)90039-0. [DOI] [PubMed] [Google Scholar]
- 18.Pick E. Keisari Y. A simple colorimetric method for the measurement of hydrogen peroxide produced by cells in culture. J Immunol Methods. 1980;38:161–170. doi: 10.1016/0022-1759(80)90340-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.Denu JM. Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 21.Forman HJ FJ. Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287:C246–C256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 22.Kwon J. Lee SR. Yang KS. Ahn Y. Kim YJ. Stadtman ER, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci U S A. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SR. Yang KS. Kwon J. Lee C. Jeong W. Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 24.Ostman A. Hellberg C. Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 25.Rhee SG. Bae YS. Lee SR. Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000(53):PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 26.Salmeen A. Andersen JN. Myers MP. Meng TC. Hinks JA. Tonks NK, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 27.Seth D. Rudolph J. Redox regulation of MAP kinase phosphatase 3. Biochemistry. 2006;45:8476–8487. doi: 10.1021/bi060157p. [DOI] [PubMed] [Google Scholar]
- 28.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 29.Rhee SG. Chang TS. Bae YS. Lee SR. Kang SW. Cellular regulation by hydrogen peroxide. J Am Soc Nephrol. 2003;14(8 Suppl 3):S211–S215. doi: 10.1097/01.asn.0000077404.45564.7e. [DOI] [PubMed] [Google Scholar]
- 30.Rhee SG. Kang SW. Jeong W. Chang TS. Yang KS. Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]