Abstract
Porphyromonas gingivalis peptidylarginine deiminase (PAD) catalyzes the deimination of peptidylarginine residues of various peptides to produce peptidylcitrulline and ammonia. P. gingivalis is associated with adult-onset periodontitis and cardiovascular disease, and its proliferation depends on secretion of PAD. We have expressed two recombinant forms of the P. gingivalis PAD in Escherichia coli, a truncated form with a 43-amino acid N-terminal deletion and the full-length form of PAD as predicted from the DNA sequence. Both forms contain a poly-His tag and Xpress epitope at the N-terminus to aid in detection and purification. The activities and stabilities of these two forms have been evaluated. PAD is cold sensitive; it aggregates within 30 min at 4 °C, and optimal storage conditions are at 25 °C in the presence of a reducing agent. PAD is not a metalloenzyme and does not need a co-factor for catalysis or stability. Multiple L-arginine analogs, various arginine-containing peptides, and free L-arginine were used to evaluate substrate specificity and determine kinetic parameters.
Keywords: Porphyromonas gingivalis, Peptidylarginine deiminase, PAD, Arginine deimination
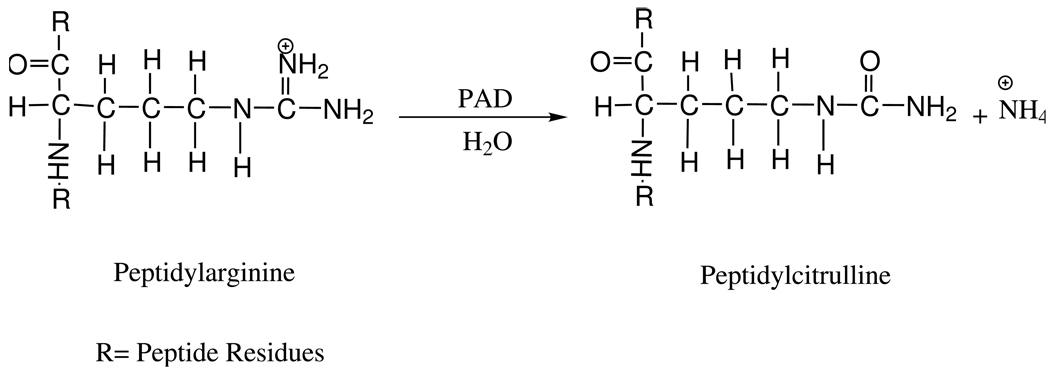
Porphyromonas gingivalis is a Gram negative, non-motile, anaerobic bacterium found in the oral cavity as normal microflora, and in the vasculature as a pathogen [1]. P. gingivalis has been implicated in major human diseases such as periodontitis and cardiovascular diseases [2–4]. The organism is associated with the initiation and progression of adult-onset periodontitis through secretion of a peptidylarginine deiminase (PAD1), E.C.3.5.3.15 [5–8], a virulence factor responsible for bacterial survival in the gum [9, 10]. PAD catalyzes the deimination of peptidylarginine residues of various polypeptides to produce peptidylcitrulline and ammonia (Scheme 1). The ammonia produced in this reaction serves to effectively control the local pH surrounding the pathogen, allowing secreted P. gingivalis R-gingipains, the major arginine-specific cysteine proteinases produced by the organism, to activate cardiac prekallikrein. Prekallikrein initiates the production of bradykinin; the presence of bradykinin increases vascular permeability allowing the bacteria to colonize the arteries, producing cardiovascular disease [11, 12].
Scheme 1.
PAD is encoded by a gene of 1671 base pairs, and the resultant protein is predicted to have a molecular mass of 61 kDa [13]. Following synthesis, the PAD from P. gingivalis is exported from the cell, and the only published report on this enzyme has dealt with the secreted form [14]. The PAD extracted from the bacterial growth medium was missing 43 amino acids from the N-terminus compared to the predicted protein sequence. Additionally, FMN was reported to be necessary for linearity in plots of product formation versus time and to improve enzyme stability [14]. Substrate specificity studies showed that the enzyme primarily catalyzes the deimination of peptidylarginine residues of various polypeptides, but is also able to catalyze the deimination of free L-arginine. The N-terminal 43 amino acids missing from the purified enzyme could have been removed during export from the cell, a process normally coupled to proteolysis, or perhaps by cleavage catalyzed by proteases also produced and secreted by the organism. Since cleavage of the N-terminus may be essential for catalytic activity, a comparison of both the truncated and full-length forms of PAD could be useful in understanding this phenomenon. As will be shown, however, the instability of the full-length form of PAD precludes detailed comparisons of the two enzyme forms.
P. gingivalis PAD is believed to be evolutionarily unrelated to mammalian PAD despite the fact that both catalyze the same chemical reaction. However, both P. gingivalis PAD and mammalian peptidylarginine deiminase 4 (PAD4), belong to a novel superfamily, the guanidino-group modifying enzyme (GME) superfamily, along with arginine deiminase (ADI), glycine-arginine amidinotransferase (AT), Nω, Nω-dimethylarginine dimethylamino hydrolase (DDAH), agmatine deiminase (AIH), and arginine succinyltransferase (AstB) [15, 16]. Despite the lack of significant amino acid sequence similarity, it has been proposed that the core domain structure is similar for all members of the superfamily, and that an active site cysteine residue initiates nucleophilic attack on the guanidino carbon of the respective substrates [16].
The PAD from P. gingivalis is an attractive drug target, but attempts to design inhibitors have been hampered by a lack of knowledge regarding enzyme structure and catalytic mechanism. The objective of this work is to characterize the enzyme, with emphasis on the catalytic mechanism and cofactor requirements. The overexpression of recombinant PAD in Escherichia coli maximizes the amount of the enzyme that can be produced and reduces the steps in protein purification, giving a better yield of enzyme. Additionally, the availability of recombinant PAD provides the opportunity to employ mutational studies to identify residues important in catalysis.
Materials and methods
Materials
The genomic DNA from the strain BAA-308-5 (W83) of P. gingivalis was from the American Type Culture Collection. Ransom Hill Bioscience prepared the oligonucleotide primers. Ex Tag high fidelity DNA polymerase was from TAKARA, Inc. The pCRT7 TOPO TA expression kit, TOP10F’ competent cells, the E. coli BL21 Star (DE3)pLysS competent cells, and Anti-Xpress Antibody were from Invitrogen. Restriction enzyme NdeI was from New England Biolabs and the DNA purification kit used was from Qiagen. IPTG was from Research Products International Corp. Recombinant enterokinase and Ni+-NTA resin were from Novagen. The Superdex HiLoad-200 gel filtration FPLC column was from Pharmacia Biotech (GE). Dithiothreitol (DTT), flavin adenine mononucleotide (FMN), flavin adenine dinucleotide (FAD), nicotinamide adenine dinucleotide phosphate (NADPH), L-arginine, N-α-benzoyl arginine ethyl ester (BAEE), N-α-benzoyl arginine (NαBA), L-arginine, D-arginine, homoarginine, Nω-hydroxyl-L-arginine and agmatine were from Sigma. Bradykinin antagonist and bradykinin were from American Peptide Company, Inc. Nω, Nω-dimethyl-L-arginine was from Fisher Scientific. The tripeptides, P-R-F and P-F-R, were custom made by EZBiolab Inc., Westfield, IN. Citrulline and L-arginine hydroxamate were from ACROS organics. All other reagents were of the highest purity commercially available.
Cloning and expression of full-length and truncated forms of PAD
The PAD coding sequence was prepared from P. gingivalis genomic DNA by digesting 5 µg of chromosomal DNA with Sac I, followed by amplification by polymerase chain reaction (PCR). The sense and the antisense primers for the full-length form of PAD were 5’ATGAAAAAGCTTTTACAGGCT 3’ and 5’TTATTTGAGAATTTTCATTGTCTCACG 3’, respectively. The sense and the antisense primers for the truncated form were 5’GCATTCCAGAAACGAATCCCCCTGCA 3’ and 5’TTATTTTGAGAATTTTCATTGTCTCACG 3’, respectively. PCR conditions were as described in the manufacturer’s protocol for Ex Tag polymerase with 200 ng of chromosomal DNA template and 125 ng of each primer. Ex Tag polymerase adds a single deoxyadenosine to the 3’ends of PCR products. PCR was performed with the following temperature program: 94°C for 5 min followed by 94 °C for 30 sec per cycle, 48 °C for 30 cycles then 36 °C for 10 cycles (30 sec per cycle), 72 °C for 2 min per cycle, followed by a 5 min hold at 72 °C. The PCR products were subsequently cloned into the linearized vector pCRT7/NT-TOPO, which has two single overhanging 3’ deoxythymidine residues, and adds a poly-His and an Xpress epitope tag to the N-terminus of inserted proteins. The resulting plasmids were transformed into TOP10F’ competent cells.
Several plasmids were isolated from TOP10F’cells using the Qiagen midi prep kit and analyzed by restriction mapping with NdeI, which has two restriction sites in the plasmid, one located in the PAD DNA sequence and the other in the pCRT7/NT-TOPO vector sequence. The predicted sizes of the DNA fragments were used to determine the correct orientation of the coding sequence. Plasmids carrying truncated and full-length forms of PAD were transformed into BL21 Star (DE3)pLysS competent cells; the resultant cells were designated SR001 and SR002, respectively.
SR001 and SR002 cells were grown at 37 °C with shaking at 250 rpm in LB media supplemented with 34 µg/mL chloramphenicol and 100 µg/mL ampicillin. Protein expression was induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) followed by incubation for 6 hr at 37 °C, with continued shaking at 250 rpm. The cells were harvested by centrifugation at 4200 rpm (H-6000 6 liter rotor, Sorvall RC-3B) at 4 °C for 25 min and frozen at −80 °C until needed for protein purification.
Enzyme purification
Using the Novagen protocol, cell pellets (42 gm) were thawed, then resuspended with 6 mL of lysis buffer (50 mM Tris/HCl pH 7.8, 400 mM NaCl, 100 mM KCl, 10% glycerol, 0.5% Triton X-100 and 10 mM imidazole) /liter of culture. Final concentrations of 1 mM PMSF, 20 µg/mL of DNase I and 30 µg/mL of RNase A were added to the cell suspension. The cell suspension was then incubated at room temperature for 30 min. After incubation, the cell suspension was sonicated twice for 20 sec at 70% duty cycle (Sonifier 350 from Branson Sonic Power) and PMSF was then added to a final concentration of 0.5 mM. The cell debris was centrifuged at 13000 rpm for 30 min at 20 °C (SS-34 rotor, Sorvall RC-5B) and the supernatant was recovered. The 80 mL of supernatant was mixed with 15 mL of Ni+-NTA resin that had been equilibrated with binding buffer (400 mM NaCl, 50 mM Tris/HCl pH 7.8, and 10 mM imidazole) and gently shaken for 1 hr at room temperature to ensure that all of the PAD fusion protein was bound to the resin. The mixture was then packed into a column, and the excess liquid was eluted from the column by gravity flow. The resin was washed once with two column volumes of binding buffer and four times with two column volumes of washing buffer consisting of 400 mM NaCl, 50 mM Tris/HCl, 25 mM imidazole, pH 7.8. PAD was eluted using 2 column volumes of elution buffer consisting of 400 mM NaCl, 50 mM Tris/HCl, and 250 mM imidazole at pH 7.8. The Ni+-NTA purified enzyme was applied to an Amicon Ultra-15 YM-50 centrifugal filter apparatus, which has a 50,000 molecular weight cutoff, and spun at 4500 rpm (SS-34, RC-5) for 16 min at 20 °for each wash with gel filtration buffer consisting of 50 mM potassium phosphate, 0.5 mM β-mercaptoethanol, and 150 mM NaCl at pH 7.8. A total of six washes were performed, resulting in a 1:1,000,000 dilution of small molecules present in solution. The 10-fold concentrated protein sample was then applied to a Superdex-200 gel filtration column that had been equilibrated with 3 column volumes of gel filtration buffer. The column was developed with the same buffer, and the fractions containing protein, as determined by A280, were tested for enzyme activity. The fractions that tested positive for PAD activity were concentrated 10-fold with the Amicon Ultra-15 YM-50. The purity of the protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining.
The enzyme was stored at 25 °C with 0.5 mM β-mercaptoethanol or 1 mM DTT and 0.1% sodium azide in 50 mM potassium phosphate buffer, pH 7.8. The poly-His and Xpress epitope tags were not cleaved from the enzyme. Only the samples of PAD that were stable (no visible precipitation), with at least 85% purity and with 100% activity compared with the Vmax obtained with 150 µM NαBA were used to perform experiments.
Molecular mass determination by gel filtration
Gel filtration chromatography was used to estimate the molecular mass of purified PAD, using the Superdex-200 column and an Amersham Pharmacia Biotech FPLC system. The column was equilibrated with the buffer system described above, and standardized using Blue Dextran (2,000 kDa), E. coli transcription factor Rho (282 kDa), β-amylase (200 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome C (12.4 kDa).
SDS-PAGE and Western blotting analysis
SDS-PAGE was performed using 10% pre-cast gels (Cambridge). Two sets of protein size markers were used: the full range molecular weight markers from Amersham (recombinant proteins from 10 to 260 kDa) and the low range molecular weight markers (phosphorylase b, 97 kDa; bovine serum albumin, 66 kDa; ovalbumin, 45 kDa; trypsin inhibitor, 20 kDa; lysozyme, 10 kDa).
Western blots were prepared using an antibody against the Xpress epitope that is fused to PAD. The samples were first subjected to SDS-PAGE, and then the proteins were transferred electrophoretically to a polyvinylidene difluoride (PVDF) membrane (Millipore) at 100 V for 1 hr. The Western blotting was carried out following the anti-Xpress antibody protocol from Invitrogen. Recombinant PAD was detected by using ECL Plus (Amersham), and following manufacturer’s protocol. Chemiluminescent signals were visualized by exposing the membrane to X-ray film (blue basic autorad film, double emulsion; ISCBioExpress) for different periods of time (1 sec to 5 min).
Poly-His tag cleavage
Following the Novagen protocol for removal of poly-His and Xpress epitope tags using enterokinase, 2 µL of 10 µg/µL PAD were mixed with 5 µL of 10X rEK cleavage buffer and 1 unit of recombinant enterokinase to a final volume of 50 µL. The reaction mixture was incubated at room temperature for 2 hr. The removal of the poly-His and Xpress tags was verified by Western blot analysis for the Xpress epitope. The intact and cleaved proteins were compared in catalytic activity and overall stability.
Enzyme activity assays
All activity measurements were performed at least in triplicate; the results shown in each figure are the mean of the multiple experiments. Enzymatic activity was determined by measuring product formation using a colorimetric assay that detects formation of the ureido group of citrulline [17], adapted for use in microcentrifuge tubes. The reaction was carried out in 50 mM CHES/HCl, pH 9.5 containing 10 mM DTT, 50 µg of protein and the appropriate concentration of substrate in a total volume of 200 µL. The period of incubation was 10 min at 37 °C. A 400 µL portion of freshly prepared color developing reagent containing 1 volume of Solution A (80 mM diacetyl monoxime and 2 mM thiosemicarbazide) and 3 volumes Solution B (3 M phosphoric acid, 6 M sulfuric acid, 2 mM ammonium iron (III) disulfate) was used to quench the reaction. The samples were incubated at 95 °C for 15 min, and cooled for 3 min on the benchtop. The absorbance of the samples was then measured at 540 nm.
The enzymatic activity, V, in the incubation volume of 200 µL was calculated using equation 1, where Acit is the measured absorption, A0 is the absorption of blank, B is the slope of the calibration curve and T is the time of enzymatic reaction [17]. Calibration curves were generated using L-citrulline and N-α-benzoyl citrulline methyl ester as standards.
| (1) |
Ammonia production was determined using a modified version of the spectrophotometric assay described by Özer (1985) that couples the production of NH4+ by PAD to the reaction catalyzed by glutamate dehydrogenase [18]. The reaction was monitored by following the decrease in absorbance at 340 nm as NADPH is converted to NADP+, with the oxidation of one mole of NADPH directly proportional to the production of one mole of NH4+. The assay contained 45 mM CHES/HCl pH 9.5, 5 mM DTT, 10 mM α-ketoglutarate, 0.15 mM NADPH (ε = 6220 M−1cm−1 at 340 nm), 1 mM 5'-GDP (an activator of glutamate dehydrogenase), 14 units/mL glutamate dehydrogenase, 100 µg of PAD and the appropriate concentration of substrate in a total volume of 1 mL. The reaction was initiated with addition of PAD; the period of incubation was 5 or 10 min at 37 °C, depending on the experiment. The rate of product formation was calculated by equation 2
| (2) |
All activity measurements were performed at least in triplicate; the results shown in each figure are the mean of the multiple experiments.
Thin-layer chromatography
An enzyme reaction was carried out with 1, 10 and 20 mM L-arginine, 150 µg of PAD, 10 mM DTT, 50 mM CHES pH 9.5, for 10 min at 37 °C; the reaction was stopped with 10 µL 1M KOH, and neutralized with 8.4 µL 1M HCl. A 1.5 µL sample of the reaction mixture was spotted on a silica gel plate, and resolved with a 5:2 (v/v) solution of methanol and ammonium hydroxide. The chromatogram was developed for 1 hr; the plate was allowed to dry and stained with 4 mg/mL ninhydrin in 95% EtOH, then heat dried [19]. The Rf values of the compounds were calculated using equation 3.
| (3) |
Mass spectrometry analysis
100 µg of PAD was subjected to SDS-PAGE. The band corresponding to PAD was then excised for protein fingerprinting analysis using matrix assisted laser desorption/ time of flight (MALDI/TOF) at the Proteomic Facility at Temple University School of Medicine. The Autoflex program from the MALDI/TOF/TOF (Bruker Daltonics) uses Swiss-Prot for peptide mass finger printing.
MALDI/TOF linear mode was used to determine the molecular mass of the enzyme. A sample (1 mg/mL) of enzyme was transferred from the storage buffer to 0.1% TFA solution by Sephadex G-50 column centrifugation to remove salts before mass spectrometric analysis [20]. A 4 µL aliquot of sample was mixed with 4 µL of matrix (10 mg of 3, 5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid), 50% acetonitrile and 0.05% TFA). The matrix-PAD mixture (4 µL) was transferred to an aluminum plate, air-dried and analyzed. The spectrum of the ionized protein is the average of 30 laser hits on the matrix-protein mixture. The molecular mass standards for mass spectrometry were 100 pmoles of bovine serum albumin (66430.09 Da) and 100 pmoles of aldolase (39212.28 Da) (Sigma, MSCAL1).
Metal analysis
To determine if PAD requires exogenous metals for catalysis, assays were carried out in the presence of NaCl, KCl, MnCl2, MgCl2, NiCl2, ZnCl2, CuSO4, CoCl2, FeSO4, EGTA or EDTA at concentrations of 1 and 10 mM. PAD was further analyzed for metal content after gel filtration chromatography through a Sephadex G-50 centrifuge column that had been equilibrated with deionized, distilled Chelex-100 treated H2O. A 500 µg sample of PAD was denatured by boiling for 10 min in a sealed microfuge tube, and then centrifuged at 14000 rpm for 5 min to pellet the denatured protein. The supernatant, pellet and intact PAD samples were analyzed for metal content by inductively coupled plasma-emission spectrometry at the Chemical Analysis Laboratory at the University of Georgia.
DNA and Protein Sequence
Truncated and full-length PAD-pCRT7/ NT-TOPO clones were sent for DNA sequencing at the DNA Sequencing Facility of the University of Pennsylvania. The results were later compared to the database from the P. gingivalis genome sequencing project [13]. A BLAST search was also performed with the nucleotide sequence and the deduced amino acid sequence to identify possible homologues. For protein sequencing, 100 µg of PAD was loaded into three wells of an SDS-PAGE gel and transferred electrophoretically to a 0.45 micron PVDF membrane (Millipore) at 100 V for 1 hr. The membrane was stained with Coomassie blue, and then destained with 10% acetic acid and 10% methanol. The bands that corresponded to PAD were excised and sent to the Protein Facility of the Iowa State University, where the samples were subjected to 4 cycles of Edman degradation.
Quantitative amino acid analysis
To assess possible auto-catalyzed deimination of protein arginyl residues, quantitative amino acid analysis was performed on several PAD samples as a function of post-purification storage times. The protein samples were transferred from the storage buffer to 1% acetonitrile solution by column centrifugation (Sephadex G-50) to remove salts and buffer. The samples were placed in microcentrifuge tubes and sent to AAA Service Laboratory, Inc., Damascus, OR for hydrolysis and quantitative amino acid analysis.
Results
Expression and localization of PAD
The truncated and full-length forms of PAD were successfully cloned from P. gingivalis into pCRT7/TOP10F’. The plasmid that yielded the correct band sizes after restriction enzyme digestion and the correct DNA sequence for recombinant, truncated PAD was designated pCRT7/mPAD and the full-length encoding plasmid was designated pCRT7/PAD. pCRT7/mPAD and pCRT7/PAD were transformed into BL21 Star (DE3)pLysS to evaluate PAD expression. Although PAD was expressed in E. coli, the expressed protein was only detected following Western blot analysis using antibody against the Xpress epitope. The protein concentrated from the medium, lysate from non-induced cells, IPTG-induced cell lysate, and the pellet containing the cell debris was tested for PAD expression. The amount of PAD found in the pellet from induced cells was insignificant compared to the PAD found in the supernatant from the sonication step (data not shown). The non-induced cell lysate and the protein recovered following ultra-filtration of the media showed neither expression of PAD nor any enzymatic activity. Protein expression was optimal when induced with 0.5 mM IPTG at 37 °C for 6 hr.
The expression of PAD was quite modest, thus multiple approaches were explored to increase the levels of expressed protein. First, IPTG-induction of PAD was carried out at 25 °C, 30 °C, and 37 °C; the change in temperature only affected the period of time required for the cells to reach the optimal density for induction. Second, in the preparation of the inoculating cell culture, cells were grown in LB medium supplemented with 34 µg/mL chloramphenicol and 100 µg/mL carbenicillin. Before dilution of the overnight culture into the fresh medium, the cells were centrifuged and washed with LB medium to ensure that minimal secreted β-lactamase was transferred with the cells to the fresh medium. Once the cells were incubated in the fresh pre-warmed 37 °C medium for 45 min, 34 µg/mL chloramphenicol and 100 µg/mL ampicillin were added. These steps were taken as contamination control to ensure better growth of cells and the optimum IPTG-induction. Protein misfolding, a possible reason for poor expression of PAD, was addressed by overexpressing a variety of exogenous protein chaperones, DnaK-DnaJ-GrpE and GroES-GroEL (pG-KJE8 from TAKARA), in the host prior to expression of PAD. The presence of the chaperones in BL21(DE3)pLysS did not improve the expression of PAD as reflected in Western blots of the cell lysate or activity measurements (data not shown). The plasmid-containing cells appeared very small under the phase contrast microscope compared to host cells without mPAD/pCRT7 plasmid, but upon testing for toxicity, the cells were able to grow into colonies in the presence of 1 mM IPTG on an LB plate supplied with proper antibiotics.
The possibility of poor translation was also addressed by transforming pCRT7/mPAD into Rosetta (DE3)pLysS. Rosetta (DE3)pLysS (Novagen) contains a plasmid encoding argU, argW, glyT,ileX, leuW, metT, proL, thrT, thrU, and tyrU, which allows expression of genes encoding tRNAs for rare arginine codons, AGA, AGG, and CGA; the glycine codon, GGA; the isoleucine codon, AUA; the leucine codon, CUA; and the proline codon, CCC. These codons are rarely used in E. coli. No improvement in PAD expression was observed following SDS-PAGE and Western blot analysis (data not shown).
Purification of truncated PAD
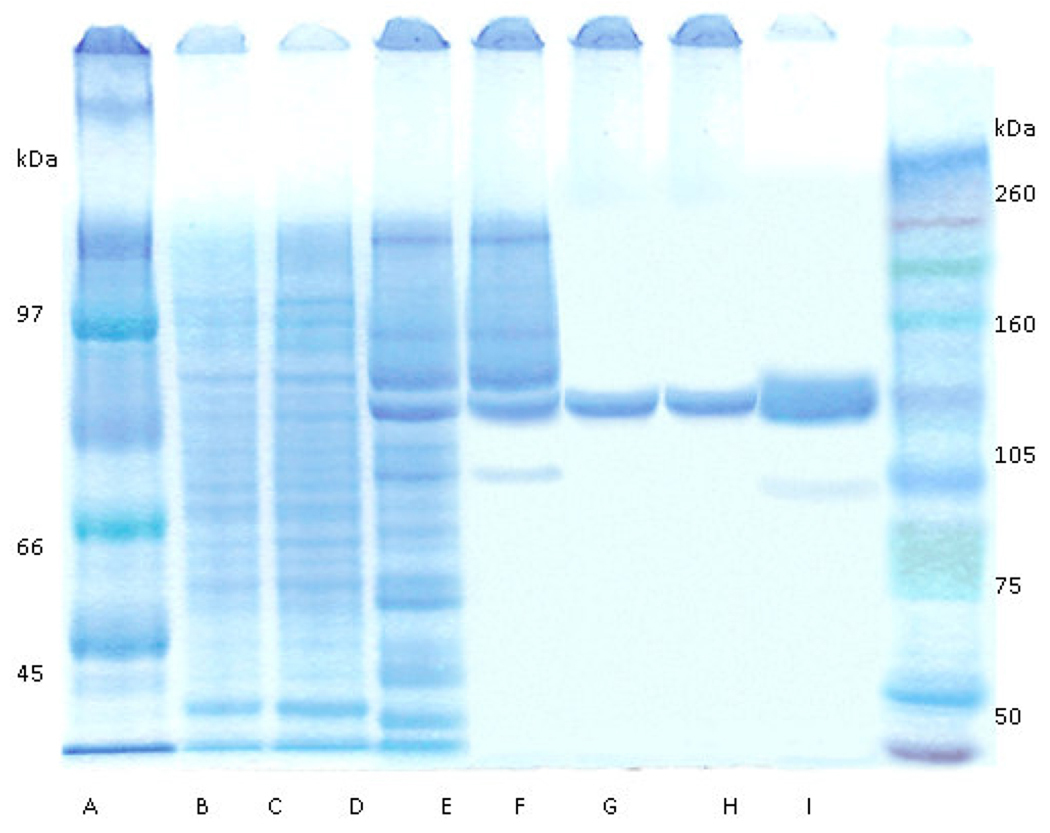
The supernatant from low speed centrifugation of lysate from cells concentrated from 12 L of culture was mixed with 15 mL of Ni+-NTA resin and eluted as described in above. The band representing the enzyme was visible following Coomassie staining of an SDS-PAGE gel, but contaminating proteins were readily visible on the gel after this step (Figure 1). The ultra-filtration step yielded concentrated PAD and eliminated many of the smaller proteins. After gel filtration chromatography and a second ultra-filtration step for concentration, the enzyme appeared to be approximately 90% pure (Figure 1), with recovery of 51% activity (Table 1). The purification steps permitted the isolation of approximately 1.75 mg of PAD per liter of culture. The purification of recombinant truncated PAD is summarized in Table 1.
Figure 1.
Coomassie-stained SDS-PAGE of truncated PAD at various stages of purification. A, low range molecular mass marker (Amersham) B, 5 µg lysate; C, 5 µg supernatant after centrifugation; D, 5 µg Ni+-NTA elute; E, 5 µg 1st ultrafiltration retentate; F, 5 µg pooled gel filtration fractions with enzyme activity; G, 5 µg 2nd ultrafiltration retentate; H, 25 µg 2nd ultrafiltration retentate; I, full range molecular mass markers (Amersham).
Table 1.
Purification of PADa
| Fraction | Volume (mL) |
Protein (mg) |
Activity (µmol/min) |
Specific activity (Unit/mg)b |
Yield (%) |
|---|---|---|---|---|---|
| Clarified extract | 86 | 20,000 | 6.9 | 3.45×10−4 | 100 |
| Ni+-NTA eluate | 8.5 | 65 | 4.3 | 0.066 | 62 |
| 1st ultrafiltation retentate | 1 | 61 | 3.9 | 0.063 | 57 |
| Gel filtration peak | 10.5 | 22 | 3.7 | 0.168 | 54 |
| 2nd ultrafiltration retentate | 1 | 20 | 3.5 | 0.175 | 51 |
The enzyme reaction was carried out as described in Materials and Methods, using 100 µM NαBA as substrate and 50 µg of PAD.
Units, µmoles/min at 37 °C
Citrulline colorimetric assays were performed to determine enzymatic activity after each purification step. Some of the contaminants in the lysate interfered with the colorimetric assay, which was more reliable later in the purification procedure. The concentration of PAD was quantified spectrophotometrically using a calculated extinction coefficient [21] of 81180 M−1cm−1 at 280 nm; therefore the absorbance of 1 mg/mL of truncated PAD is 1.33 ± 0.07.
Purification of the full-length form of PAD
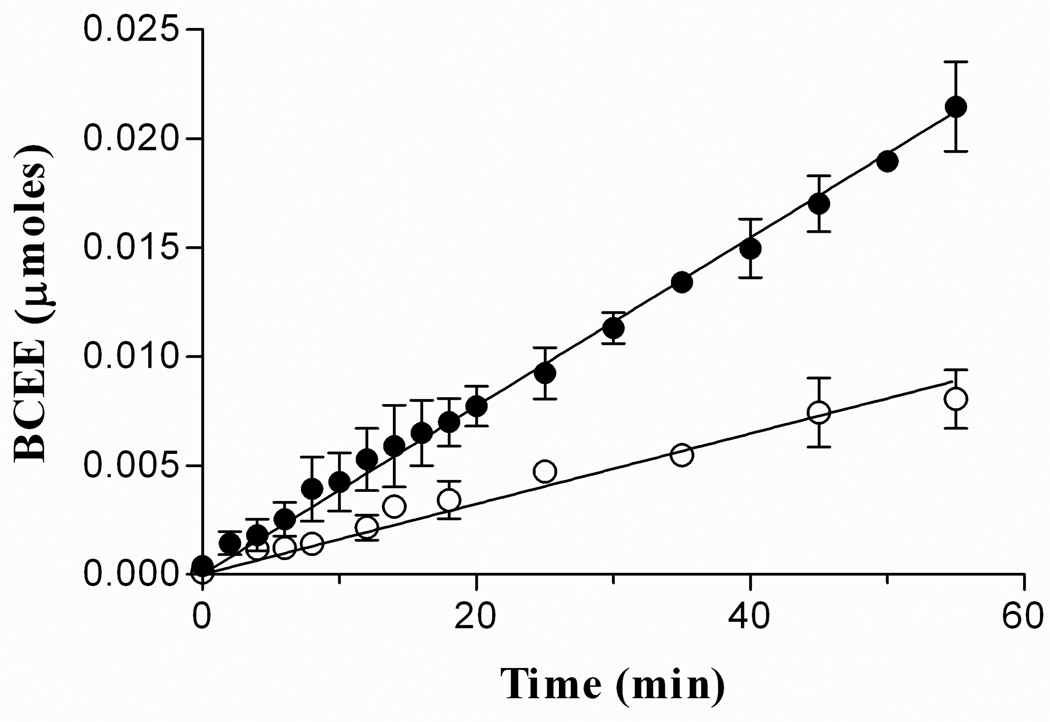
The enzyme collected from P. gingivalis growth media had a deletion of 43 amino acids from the N-terminus when compared to the predicted amino acid sequence [14]. The 43 amino acids missing from the enzyme could have been removed during export from the cell, a process normally coupled to proteolysis, or perhaps proteases found in the medium cleaved some or all of the 43 amino acids from PAD. Recombinant full-length PAD was purified and treated under the same conditions as the truncated form of the enzyme. Expression of the enzyme was determined by Western blot analysis (data not shown) and the activity by colorimetric citrulline assay. The full-length PAD only yielded 0.5 mg of protein per liter of cell culture. The protein precipitated within the first day after purification even at 25 °C in the presence of FMN, DTT or β-mercaptoethanol (data not shown). Immediately after purification, full-length PAD only had 40% activity relative to the truncated form (Figure 2). As a consequence, the truncated form was used for all further experiments.
Figure 2.
Activities of truncated and full-length PAD as a function of time. 4 µM of the enzymes (truncated and full-length PAD) were pre-incubation at 37 °C in 50 mM CHES/HCl pH 9.5 and 10 mM DTT; the reaction was started with the addition of 20 mM BAEE. The slopes of the graph show that the full-length form of PAD (○) has 40% activity of truncated form of PAD (●). The error bars in each point represent the mean ± S.D. of the results from 4 independent experiments.
Both the truncated and full-length forms of recombinant PAD have 36 extra amino acids at the N-terminus. The removal of the fused poly-His and Xpress epitope tags by enterokinase, leaving only four extra N-terminal amino acid residues (Asp-Pro-Thr-Leu), did not change PAD activity or stability (data not shown).
Molecular mass analysis
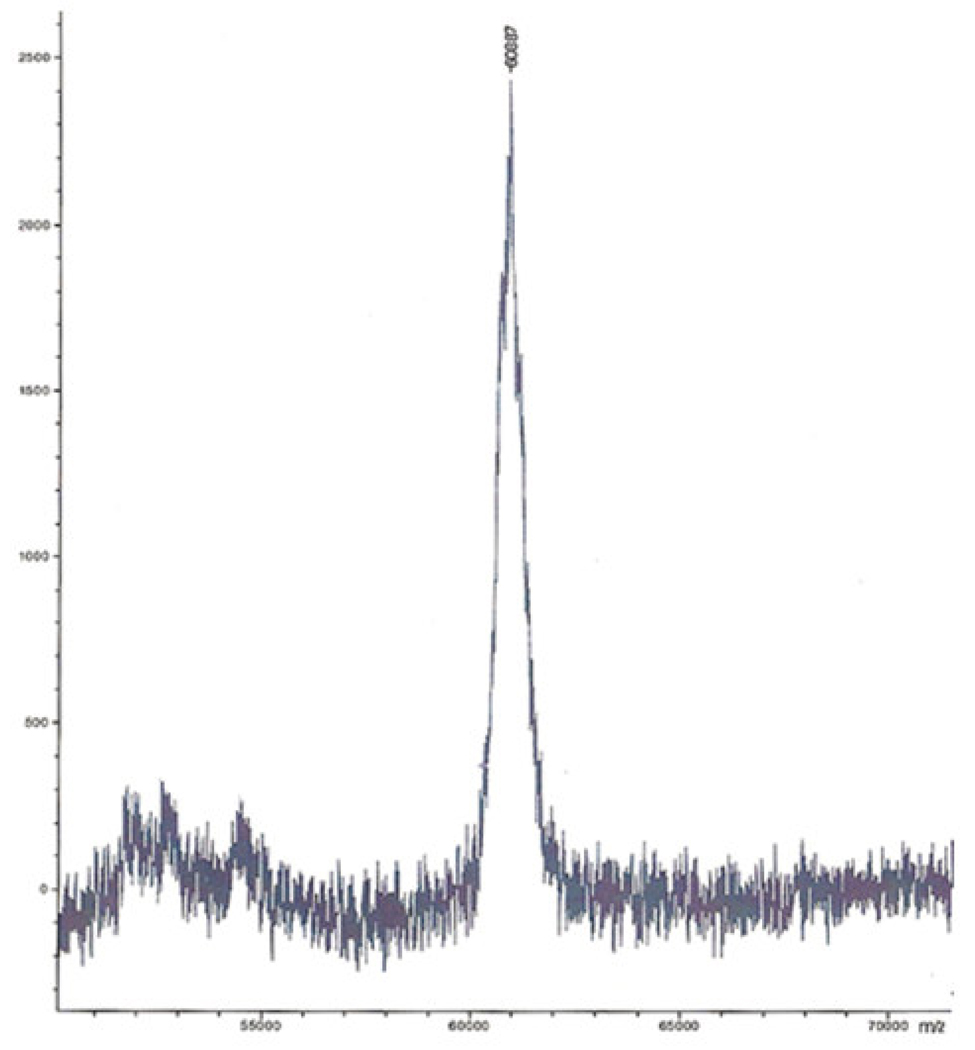
Purified recombinant truncated PAD (missing 43 N-terminal amino acid residues but including Xpress and poly-His tags) migrated as a single band on SDS-PAGE. The molecular weight calculated from SDS-PAGE was approximately 51 kDa (Figure 1). The molecular weight for truncated PAD was also calculated from a standard curve generated by gel filtration chromatography with proteins of known molecular weight as described in Materials and Methods. Elution of the active enzyme from the calibrated gel filtration column indicated a mass of approximately 53 kDa. The experimental mass determined by linear mode MALDI/TOF analysis was 60885±3 Da (Figure 3), which is consistent with the calculated molecular mass of truncated PAD with its poly-His and Xpress epitope tags (60884.24 Da). The calculated mass of poly-His and Xpress epitope tags is 3969.69 Da.
Figure 3.
MALDI/TOF, linear mode analysis of truncated PAD. The spectrum shows a mass for PAD at 60887 Da, which is consistent with the calculated mass of truncated PAD (60884.24 Da) that includes the mass of the poly-His and Xpress epitope tags. The spectrum represents the average of 30 laser hits (mass ionization).
Sequence analysis
MALDI/TOF/TOF of trypsin-digested recombinant truncated PAD was used to analyze and identify PAD by SWISS protein peptide mass finger printing. The result of the database analysis demonstrated a protein match to the P. gingivalis PAD; the same result was obtained by BLAST analysis performed on the DNA sequence and the protein sequence derived from PAD [22]. A match was also obtained for the peptide (1810.00 Da) that contained the N-terminal amino acids of the truncated PAD (AFQETNPPAGPVR) and the C-terminal amino acid residues of the tags. The sequence of entire tag is MRGSHHHHHHG MASMTGGQQM GRDLYDDDDKDPTL, and that of the tryptic peptide is DPTLAFQETNPPAGPVR.
MALDI/TOF/TOF protein finger printing was also used to identify the other proteins that eluted from the Ni+-NTA column, but which were subsequently separated during gel filtration chromatography. This analysis indicates that that one of the proteins was E. coli bifunctional polymyxin resistance ArnA protein and the other was E. coli glucosamine-fructose-6-phosphate aminotransferase.
Amino terminal sequencing of recombinant truncated PAD was carried out through 4 cycles of Edman degradation, and gave the sequence: Met-Arg-Gly-Ser for the major amino acid HPLC peak for each cycle (major peak represents 80% of the sample). This sequence matches that expected for the N-terminal sequence of the poly-His tag fused to the truncated PAD.
Product analysis
The accuracy of the colorimetric citrulline assay was confirmed using a variety of controls to ensure that the colored product generated by the action of PAD on L-arginine was indeed L-citrulline. TLC on silica gel was used to confirm substrate depletion and product formation by PAD. A final concentration of 10 mM citrulline, and 10 mM arginine were used as markers. The Rf value for L-arginine was 0.33 and the Rf value for L-citrulline was 0.86, giving a clear separation of these two compounds. The results showed citrulline formation proportional to the amount of arginine added to the enzyme reaction (data not shown).
A series of experiments was performed to ensure that the results observed in the citrulline colorimetric assay were due to enzyme activity. In the first set of experiments, linear rates of product formation were observed with the substrates L-arginine, NαBA, and BAEE. Second, the reaction rate was compared using 50 µg and 100 µg of PAD, incubated for 20 min in the presence of 150 µM NαBA; the expected doubling of rate was observed (data not shown). Third, a portion of the enzyme was boiled for 10 min to ensure the denaturation of the enzyme, and cooled for another 10 min at room temperature. The activity of the boiled PAD was compared to the activity of the enzyme incubated at 25 °C, 37 °C, and 55 °C. No product formation was observed in reaction mixtures containing the PAD sample that had been boiled. Additionally, the optimum activity of the enzyme was observed at 55 °C (data not shown), which is in agreement with previous work [14]. Finally, substrate saturation was observed using BAEE over the range of 5 to 50 mM, yielding a Km of 18 mM (data not shown).
pH profile
PAD activity was measured using the colorimetric citrulline assay over a pH range from 5.0 to 12.0 with a 50 mM mixed buffer system (MES, MOPS, CAPS, HEPES, CAPSO, BISTRIS, CHES) in the presence of 100 µM of NαBA. In agreement with previous work [14], optimum activity is observed at pH 9.5, with 38.5% activity remaining at pH 6 and 3.4% activity remaining at pH 11.0 (data not shown). The pH data suggest that the catalytic activity of the enzyme is likely dependent on the ionization states of active site residues. A thorough analysis of the pH dependence of kinetic parameters will be presented elsewhere.
Stability
The purified enzyme showed no appreciable loss of activity when quick frozen and stored at −80 °C or when stored at room temperature (25 °C) in the presence of 1 mM DTT or 0.5 mM β-mercaptoethanol for 2 weeks. However, when the purified enzyme was stored at −20 °C or 4 °C, aggregation of the protein occurred. The aggregation was readily apparent in the microcentrifuge tube, and upon centrifugation a pellet formed. The Coomassie stained gel of both the supernatant and pellet from a 32 day old sample showed that approximately 75% of PAD was in the pellet. Additionally, this result was confirmed by activity measurements. PAD completely aggregates at 4 °C after 30 min of incubation, but aggregated PAD displayed variable activity for up to one week after re-suspension with storage buffer (data not shown). These results suggest that either aggregation does not inactivate PAD or that inactivation is reversible in the presence of substrate. The aggregation of PAD also occurred when the enzyme concentration was greater than 60 mg/mL. The enzyme at 13 mg/mL remained active at pH 7.0, 8.0 and 9.5 for several days at room temperature.
Cofactors
Based on the earlier observation that FMN stabilized the enzyme [14], the purification of PAD was carried out in the presence and in the absence of FMN in all buffers; results were compared to test the difference in stability and activity of the enzyme at every step. There was no improvement in enzyme stability or activity with the addition of FMN at concentrations in the range of 1 µM to 10 mM, and the addition of FAD, FMN or NADPH at concentrations in the range of 5–25 µM did not change the rate of the reaction (data not shown). Furthermore, no evidence for tightly bound enzyme-associated cofactors was found in a spectrophotometric scan from 260 nm to 600 nm.
Metal analysis
Since it is known that some members of the guanidino modifying superfamily, of which PAD is proposed to be a member, are metalloenzymes or need metal ions for catalysis [23, 24], PAD was evaluated for the presence of tightly bound metals or metal ion activation. The results of metal analysis from the Chemical Analysis Laboratory at the University of Georgia revealed no significant quantities of Ca++, Cd++, Co++, Cr++, Cu++, Fe++, K+, Mg++, Mn++, Mo+6, Na+, Ni++, or Zn++ tightly associated with PAD. PAD was assayed in the presence of 0.1 mM, 1 mM, and 10 mM concentrations of various metal ions (Na+, K+, Mn++, Mg++, Ni+, Zn++, Cu++, Co++, Fe++), as well as EGTA and EDTA to test the effect of metal on the reaction. There was no significant change in the enzyme rate when the metals or the chelating reagents were present in the reaction mixture (data not shown), except for Zn++ and Cd++, which were inhibitory. At 1 mM Zn++, the enzyme activity was inhibited by 26%, while 53% inhibition was observed in the presence of 1 mM Cd++. These analyses suggest that PAD is not a metalloenzyme and does not require exogenous metals for catalysis.
Kinetic analysis
Substrate specificity studies were conducted to map the active site and determine requirements for catalytic activity. Kinetic parameters for the enzyme in the presence of potential substrates are summarized in Table 2. Modification of the α-amino group of arginine with the bulky hydrophobic benzoyl group (Nα-benzoyl-L-arginine) provides a significant reduction in Km, with a halving of kcat compared to L-arginine. Michaelis-Menten analysis provides a Km value of 10 µM for Nα-benzoyl-L-arginine. However, because of the high concentrations of enzyme required for assays and the relatively low Km value determined for this compound, the concentration of substrate is not significantly greater than the concentration of enzyme and Michaelis-Menten analysis may not apply. The kinetic data for this substrate were also analyzed by the method of Dixon as outlined in Segel [25] for conditions of high enzyme concentration where a significant fraction of the total substrate may be bound to the enzyme. This analysis yielded a range of 15–20 µM for Km. For Nα-benzoyl-L-arginine there is thus a 33–66-fold increase in catalytic efficiency relative to L-arginine. This trend is extended to the peptide substrate bradykinin in which a 50-fold decrease of Km is accompanied by 5-fold decrease of kcat compared to L-arginine. Similarly, the tripeptide P-F-R had a kcat/Km approximately 10-fold larger than the corresponding value for L-arginine.
Table 2.
Kinetic constants for PAD substrates
| Substrate | Km (µM) |
Relative Vmax (%) |
kcat (sec−1) |
kcat/Km (µM−1sec−1) |
|---|---|---|---|---|
| L-Arginine | (1.2 ± 0.5)×103 | 100 | 0.22 | 1.9×10−4 |
| Nα-Benzoyl-L-arginine (Nα-BA) | 10–20 | 54.5 | 0.12 | (6–12)×10−3 |
| D-Arginine | (1.9 ± 0.5)×103 | 26.4 | 0.058 | 3.1×10−5 |
| BK (R-P-P-G-F-S-P-F-R) | 21 ± 1 | 18.8 | 0.043 | 2.1×10−3 |
| BK antagonist (K-R-P-P-G-F-S-P-L) | 33 ±1 | 3.1 | 6.8×10−3 | 2.1×10−4 |
| P-R-F | 70 ± 1 | 3.4 | 7.5×10−3 | 1.1×10−4 |
| P-F-R | 89 ± 1 | 78.2 | 0.17 | 1.9×10−3 |
| L-Canavanine | (6 ± 1)×103 | 2.5 | 5.2×10−3 | 9.3×10−7 |
| L-Arginine hydroxamate | (6.9 ± 0.5)×103 | 68.2 | 0.15 | 2.2×10−5 |
| Benzoyl arginine ethyl ester (BAEE) | (18 ± 1)×103 | 8.1 | 0.019 | 9.9×10−7 |
| Agmatine | (31 ± 1)×103 | 3.2 | 7.0×10−3 | 2.3×10−7 |
| Homoarginine | (4.7 ± 0.8)×103 | 50 | 0.11 | 2.4×10−5 |
| Nω-Hydroxy-L-arginine | (1.5 ± 0.5)×103 | 2.1 | 4.6×10−3 | 3.0×10−6 |
| Nω,Nω-Dimethyl-L-arginine | >1×106 a | <0.0017 |
No activity detected with 100mM Nω,Nω-dimethyl-L-arginine. Products from Nω-hydroxy-L-arginine and Nω, Nω-dimethyl-L-arginine have been previously measured using the citrulline colorimetric assay [33].
In contrast, removal of the carboxyl group or the addition of hydrophobic bulk at that site was deleterious. Agmatine, which lacks a carboxylate group, was a very poor substrate, with Km about 30-fold higher than L-arginine and kcat 30-fold lower, for a catalytic efficiency 900-fold lower than that of L-arginine. The ethyl ester of N-α-benzoyl-L-arginine showed a Km about 20-fold greater than L-arginine with a kcat about 7-fold lower. The peptide P-R-F had a 15-fold lower Km but 30-fold slower kcat. Arginine hydroxamate, which adds bulk to carboxylate but preserves its negative charge, is bound about 5-fold weaker, but yields a kcat similar to that of L-arginine. Modification of the guanidino group, as in Nω-hydroxyl-L-arginine, resulted in a 50-fold decrease in kcat and a Km comparable to that of L-arginine. Nω, Nω-dimethyl-L- arginine gave no detectable activity.
The general length and conformation of the substrate were also important. Homoarginine, with a side chain extended by one carbon, bound about 4-fold weaker than arginine, and showed a 2-fold lower kcat. The D form of arginine was less tightly bound than L-arginine, and kcat was about 4-fold lower.
Auto-catalyzed Deimination of PAD
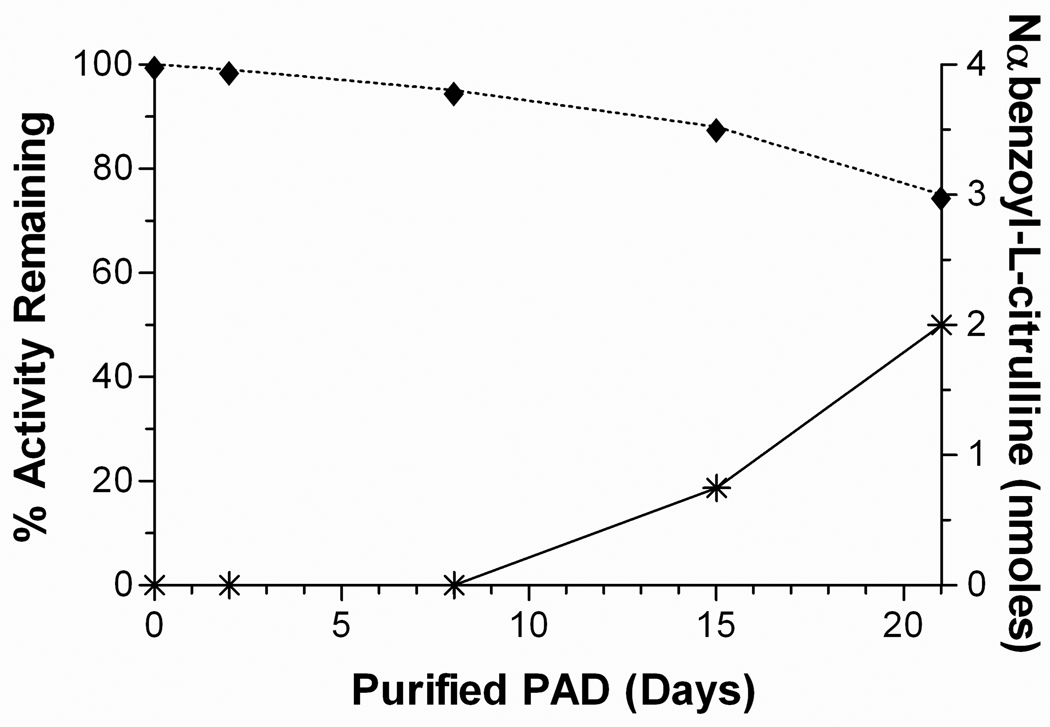
Purified PAD that had been stored for various periods of time was analyzed for self-deimination of arginyl residues. Acid hydrolysis and quantitative amino acid analysis showed a decrease in the number of arginyl residues as a function of time. Freshly purified PAD had 18 arginine residues, which is in agreement with the number predicted from the DNA sequence of truncated PAD with its poly-His and Xpress epitope tags; at the end of 21 days, this value had decreased to 16. These results are consistent with those obtained by colorimetric determination of citrulline residues (Figure 4) where the presence of protein-citrullinyl residues was directly proportional with the disappearance of arginine from the protein. Figure 4 also shows that the activity of the enzyme decreases as a function of self-deimination; Western blot analysis verified that the enzyme had not undergone proteolysis over the 21 day period (data not shown).
Figure 4.
PAD self-deimination. Measurement of citrulline formation (*) in solution with 0.9 nmoles of PAD as a function of time (storage periods after purification). PAD activity decreases as a function of time (♦).
Discussion
We have successfully expressed and purified two recombinant proteins, a truncated form and a full-length form of peptidylarginine deiminase from P. gingivalis in BL21 Star (DE3)pLysS E. coli. The expression of PAD was not as robust as generally observed in protein overexpression systems, but provided sufficient protein for activity studies. Reasons for poor expression are not clear; perhaps PAD interferes with its own expression by deiminating arginine residues in proteins necessary for translation.
Protein and DNA sequencing along with the mass spectrometry results confirmed that the isolated protein was the PAD from P. gingivalis. Further confirmation was provided by the citrulline colorimetric assay and TLC; the product detected by both assays was citrulline. NH4+, the other expected product, was detected using the coupled spectrophotometric assay.
The enzyme was purified and stored at room temperature. PAD has 8 cysteine residues, therefore 1 mM DTT or 0.5 mM β-mercaptoethanol was used to maintain the thiols in the enzyme in the reduced state [26]. The oxidation state of the secreted enzyme is unknown but we do note that the concentration of oxygen in solution is 0.2 mM and PAD is an extracellular enzyme. The enzyme stability improved after purification. The data obtained from inclusion of FMN in the storage buffer and in the enzyme reaction mixture showed no change in the enzyme kinetic parameters or improvement of the enzyme stability, despite the fact that an FMN requirement has been reported for PAD stability [14]. The origin of this discrepancy is unclear, and may result from the inclusion of N-terminal tags in the recombinant protein. The same results were observed when other redox factors such as NADPH and FADH were tested as PAD cofactors. The removal of the poly-His and Xpress epitope tags by enterokinase did not affect the enzyme activity or its stability, thus all the experiments were done with a tagged enzyme.
Metal analysis results showed that PAD does not require exogenous metal for catalysis nor it is a metalloenzyme. This is one of the enzyme characteristics that differentiate the P. gingivalis PAD from mammalian PAD4, a Ca++ dependent metalloenzyme [23, 27] and ADI. Although no metal ions have been identified in crystal structures of ADI [24], the inclusion of 60 mM KCl to assay mixtures results in a 6-fold increase in initial velocity, while the addition of 20 mM MgCl2 results in an 8-fold increase in velocity [28,29]. The P. gingivalis PAD is inhibited by zinc and cadmium, a result consistent with the proposed involvement of a nucleophilic cysteine residue, as demonstrated for other members of the GME superfamily [16, 23, 28].
The full-length and truncated forms of PAD cloned in E. coli were compared to determine whether the cleavage of the first 43 amino acids at the N-terminus might be essential for catalysis or enzyme stability. The full-length PAD is very unstable compared to the truncated form of PAD. The full-length form precipitates within the first day of purification even at 25 °C in the presence of DTT or β-mercaptoethanol; under these conditions the truncated form retains 100% activity for up to three weeks. The full-length PAD has lower levels of expression in the host cell (0.5 mg/L of culture), and immediately after purification, it only has 40% of the activity of the truncated form. Thus, the truncated form of the enzyme was used for the studies reported in this work. The instability and the low activity of the full-length PAD may provide rationale for truncation of the enzyme during or after secretion. Although there have been publications that suggest that E. coli can secrete eukaryotic proteins [30], in this study, E. coli secreted neither the full-length nor the truncated form of PAD into the medium.
Kinetic studies with L-arginine, L-arginine analogs and peptides with arginine at different locations showed that PAD prefers arginine residues at the carboxyl terminus. Based on the kinetic constants obtained for arginine and its analogs as substrates for PAD, it seems that the carboxylate group of the arginine plays an important role in determining the rate of catalysis. The results with the tripeptides Pro-Arg-Phe and Pro-Phe-Arg as well as bradykinin and bradykinin antagonist demonstrate that the position of the arginine does not significantly affect the substrate Km, but that a C-terminal arginine enhances the rate of product formation. Interestingly, the previous report for this enzyme indicated that only the C-terminal arginine of bradykinin was deiminated [14], while the current results indicate that the internal residue of bradykinin antagonist is deiminated by the enzyme. Mammalian PAD4 shows similar substrate specificity, with the rate of catalysis dependent on substrate structure and the position of arginine in the peptide sequence. Furthermore, for PAD4 the residues neighboring the arginyl substrate are important in determining substrate activity [31]. Although PAD has a higher specificity for C-terminal arginine residues, quantitative amino acid analysis showed that PAD undergoes self-deimination even though the closest arginine residue to the C-terminus is 8 residues away [13]. Our results are consistent with previous observations [14] showing that approximately 4.0 to 6.5 mol of citrulline per mol of PAD (depending on the age of the sample) were observed after acid hydrolysis of PAD and citrulline determination.
According to a Conserved Domain Database for protein classification (CDD), the predicted active site of PAD, generated by alignment of sequences for the related enzymes ADI, DDHA and AT, contains amino acids Asp 130, Asp 187, His 236, Asp 238 and Cys 351 [32]. The next step will be to establish whether Cys 351, or some other cysteine residue, acts as a nucleophile to initiate the enzymatic reaction and form a covalent intermediate as has been shown for other members of this enzyme superfamily, while the remaining predicted active site residues mediate multiple proton transfers. These results will provide significant insight into the catalytic mechanism of PAD and form the basis for future inhibitor design.
Acknowledgments
We thank Dr. R. Reczkowski for his help in designing the clones, Drs. C. Grubmeyer, W. E. Masker, and D. R. Soprano for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by National Institutes of Health Grant GM067788 to D.E.A. and graduate research fellowship 42-0509-151 to D.E.A. from the Commonwealth Universal Research Enhancement Program of the Pennsylvania Department of Health.
Abbreviations used: NαBA, N-α-benzoyl arginine; BAEE, N-α-benzoyl arginine ethyl ester; BK, bradykinin; CAPS, 3-(cyclohexylamino)-1-propanesulfonic acid; CAPSO, 3-(Cyclohexylamino)-2-hydroxy-1-propanesulfonic; CHES, 2-(N-cyclohexylamino)-ethanesulfonic acid; BISTRIS, bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane; BSA, bovine serum albumin; DNase I, deoxyribonuclease I; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; FADH2, reduced flavin adenine dinucleotide; FMN, flavin mononucleotide; FPLC, fast protein liquid chromatography system; HEPES, N-(2-hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid); IPTG, isopropyl β-D-thiogalactoside; MES, 2-(N-morpholino)ethanesulfonic acid; MOPS, 3-(N-morpholino)propane sulfonic acid; NADPH, reduced nicotinamide adenine dinucleotide phosphate; PMSF, phenylmethylsulfonyl fluoride; RNase A, ribonuclease A; TLC, thin-layer chromatography; Tris, tris(hydroxymethyl)aminomethane.
References
- 1.Carlsson J, Hofling JF, Sundqvist GK. J. Med. Microbiol. 1984;18:39–46. doi: 10.1099/00222615-18-1-39. [DOI] [PubMed] [Google Scholar]
- 2.Lambe DW, Jr, Ferguson KP, Mayberry WR. Can. J. Microbiol. 1982;28:367–374. doi: 10.1139/m82-056. [DOI] [PubMed] [Google Scholar]
- 3.Renvert S, Pettersson T, Ohlsson O, Persson GR. J. Periodontology. 2006;77:1110–1119. doi: 10.1902/jop.2006.050336. [DOI] [PubMed] [Google Scholar]
- 4.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Infection and Immunity. 1999;67:5792–5798. doi: 10.1128/iai.67.11.5792-5798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Angelis M, Mariotti L, Rossi J, Servili M, Fox PF, Rollan G, Gobbetti M. Appl. Environ. Microbiol. 2002;68:6193–6201. doi: 10.1128/AEM.68.12.6193-6201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi N. Oral Microbiol Immunol. 2003;18:109–113. doi: 10.1034/j.1399-302x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 7.Griffen AL, Lyons SR, Becker MR, Moeschberger ML, Leys EJ. J. Clin. Microbiol. 1999;37:4028–4033. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- 9.Niederman R, Brunkhorst B, Smith S, Weinred RN, Ryder MI. Arch. Oral Biol. 1990;35 Suppl:205S–209S. doi: 10.1016/0003-9969(90)90159-8. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi N, Saito K, Schachtele CF, Yamada T. Oral Microbiol Immunol. 1997;12:323–328. doi: 10.1111/j.1399-302x.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 11.Imamura T, Potempa J, Pike RN, Travis J. Infect. Immun. 1995;63:1999–2003. doi: 10.1128/iai.63.5.1999-2003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozarov EV, Dorn BR, Shelburne CE, Dunn WA, Jr, Progulske-Fox A. Arterioscler. Thromb. Vasc. Biol. 2005;25:e17–e18. doi: 10.1161/01.ATV.0000155018.67835.1a. [DOI] [PubMed] [Google Scholar]
- 13.Nelson K, Fleishmann R, DeBoy R, Paulsen I, Fouts D, Eisen J, Daugherty S, Dodson R, Durkin A, Gwinn M, Haft D, Kolonay J, Nelson W, White O, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin J, Duncan M, Dewhirst F, Fraser C. J. Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGraw WT, Potempa J, Fraley D, Travis J. Infect. Immun. 1999;67:3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirai H, Blundell TL, Mizuguchi K. Trends Biochem. Sci. 2001;26:465–468. doi: 10.1016/s0968-0004(01)01906-5. [DOI] [PubMed] [Google Scholar]
- 16.Shirai H, Mokrab Y, Mizuguchi K. Proteins. 2006;64:1010–1023. doi: 10.1002/prot.20863. [DOI] [PubMed] [Google Scholar]
- 17.Knipp M, Vasak M. Anal. Biochem. 2000;286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 18.Özer N. Biochem. Med. 1985;33:367–371. doi: 10.1016/0006-2944(85)90012-2. [DOI] [PubMed] [Google Scholar]
- 19.TLC procedure. Organic Chemistry Lab Course. 2003 Orgchem.colorado.edu/hndbksupport/TLC/TLC.html.
- 20.Penefsky HS. Methods Enzymol. 1979;55:527–530. doi: 10.1016/0076-6879(79)56050-9. [DOI] [PubMed] [Google Scholar]
- 21.Gill SC, Von Hippel PH. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 22.Ye J, McGinnis S, Madden LT. Nucleic Acids Res. 2006;34:W6–W9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 24.Galkin A, Lu X, Dunaway-Mariano D, Herzberg O. J. Biol. Chem. 2005;280:34080–34087. doi: 10.1074/jbc.M505471200. [DOI] [PubMed] [Google Scholar]
- 25.Segel IH. Enzyme Kinetics. New York: John Wiley and Sons; 1975. pp. 72–77. [Google Scholar]
- 26.Cleland W. Biochemistry. 1963;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 27.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Nat. Struct. Mol. Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 28.Lu X, Li L, Wu R, Feng X, Li Z, Yang H, Wang C, Guo H, Galkin A, Herzberg O, Mariano PS, Martin BM, Dunaway-Mariano D. Biochemistry. 2006;45:1162–1172. doi: 10.1021/bi051591e. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Li L, Feng X, Wu Y, Dunaway-Mariano D, Engen JR, Mariano PS. J. Am. Chem. Soc. 2005;127:16412–16413. doi: 10.1021/ja056226p. [DOI] [PubMed] [Google Scholar]
- 30.Hartl F, Wiedmann M. Curr. Biol. 1993;3:86–89. doi: 10.1016/0960-9822(93)90161-g. [DOI] [PubMed] [Google Scholar]
- 31.Nomura K. Arch. Biochem. Biophys. 1992;293:362–369. doi: 10.1016/0003-9861(92)90407-n. [DOI] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, Anderson BJ, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. Nucleic Acids Res. 2005;33(Database Issue):D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone EM, Schaller TH, Bianchi H, Person MD, Fast W. Biochemistry. 2005;44:13744–13752. doi: 10.1021/bi051341y. [DOI] [PubMed] [Google Scholar]