Summary
While cell signaling devotees tend to think of the endoplasmic reticulum (ER) as a Ca2+ store, those who study protein synthesis tend see it more as site for protein maturation, or even degradation when proteins do not fold properly. These two worldviews collide when inositol 1,4,5-trisphosphate (IP3) receptors are activated, since in addition to acting as release channels for stored ER Ca2+, IP3 receptors are rapidly destroyed via the ER-associated degradation (ERAD) pathway, a ubiquitination- and proteasome-dependent mechanism that clears the ER of aberrant proteins. Here we review recent studies showing that activated IP3 receptors are ubiquitinated in an unexpectedly complex manner, and that a novel complex composed of the ER membrane proteins SPFH1 and SPFH2 (erlin 1 and 2) binds to IP3 receptors immediately after they are activated and mediates their ERAD. Remarkably, it seems that the conformational changes that underpin channel opening make IP3 receptors resemble aberrant proteins, which triggers their binding to the SPFH1/2 complex, their ubiquitination and extraction from the ER membrane and finally, their degradation by the proteasome. This degradation of activated IP3 receptors by the ERAD pathway serves to reduce the sensitivity of ER Ca2+ stores to IP3 and may protect cells against deleterious effects of over-activation of Ca2+ signaling pathways.
1.1 IP3 receptors and their activation
IP3 receptors are large (~2,700 amino acid) ER membrane proteins which form tetrameric channels that govern the release of Ca2+ stored within the ER lumen of vertebrate cells (Figure 1) [1–3]. They are named for their ability to bind to and be opened by the second messenger IP3, which is generated at the plasma membrane in response to cell surface receptor activation. Thus, IP3 receptors are pivotal in signaling pathways that couple extracellular hormones, neurotransmitters and growth factors to increases in cytoplasmic Ca2+ concentration and the regulation of Ca2+-dependent events (e.g. secretion, fertilization, apoptosis and gene expression). There are three closely-related IP3 receptor homologs in mammals (IP3R1, IP3R2 and IP3R3), that form both homo- and heterotetramers, and which have slightly different properties and markedly different tissue distributions. IP3R1 appears to be expressed ubiquitously, while IP3R2 and IP3R3 have more sporadic distributions. Largely for this reason, IP3R1 has received the most attention, culminating in the development of several 3-dimensional models of the IP3R1 tetramer (Figure 1A) [1].
Figure 1. IP3 receptor structure and activation.
A. Cryo-electron microscopy image of tetrameric IP3R1 purified from mouse cerebellum (modified from reference 8). The scale bar =100Å, and the region thought to span the ER membrane and contain the channel pore is indicated by the double lines. Models obtained by other groups are broadly similar to the image shown, but are not identical [1]. B. Model of channel opening; for clarity, only two IP3R1 subunits are shown (modified from reference 6; see text for description). The domains indicated are the SD (yellow, amino acids 1–223), deletion of which creates a protein that binds IP3, but which cannot not form functional channels; the LBD (orange and red, amino acids 224–575), which is composed of 2 halves linked by a putative hinge; the channel domain (blue, amino acids 2276–2749), which contains 6 TM helices linked by 3 lumenal loops and 2 cytosolic loops, and a coiled-coil (CC) region that participates in tetramer assembly; and the intervening coupling domain (green, amino acids 576–2275), which contains several regulatory sites. The channel pore is formed by TM helices 5 and 6 and the intervening lumenal loop [2,6,7]. The arrows indicate the putative movements that occur after IP3 binding that cause channel opening.
Channel opening is a complex process involving the binding of both IP3 and Ca2+ (which are co-agonists) to multiple subunits within the IP3 receptor tetramer, and while it is clear that IP3 binds to and alters the conformation of the ligand-binding domain (LBD), the sites and effects of Ca2+ binding remain controversial (Figure 1B) [1–3]. Although the atomic structures of the LBD and the adjacent suppressor domain (SD) have been solved [3,4] the structure of the remainder of the protein, including the pore, is undefined. Further, and somewhat disappointingly, the 3-dimensional models of IP3 receptor tetramers (Figure 1A) are not detectibly affected by IP3 binding, and although Ca2+ does have an effect on conformation [5], it is not yet clear how this relates to receptor activation [1]. Thus, channel opening has yet to be visualized. This has encouraged the building of models to explain channel opening, based on the effects of mutagenesis, the mapping of which parts of IP3 receptors interact with each other, and molecular modeling onto better-defined K+ channels. The current idea [1–4,6,7] is that IP3 binding to the LBD causes its two parts to close together around a putative hinge, that this moves the SD away from the cytoplasmic loop between transmembrane (TM) helices 4 and 5 with which the SD normally interacts, and that this causes reorganization of the pore-forming sequences, and Ca2+ flow (Figure 1B).
1.2 IP3 receptor down-regulation
In 1991 it was discovered in mammalian cell lines that in response to activation of certain IP3-generating cell surface receptors, IP3 receptors are “down-regulated”, i.e. there is a rapid and dramatic decline in cellular IP3 receptor content [9]. Typically, this decline is >50%, with half-maximal effect at 30–60 minutes [10–14], but is particularly marked in αT3-1 anterior pituitary cells, in which gonadotropin-releasing hormone (GnRH) receptor activation down-regulates IP3R1 by ~70%, with half-maximal effect at ~15 minutes [15,16]. Subsequently, it was shown that down-regulation is mediated by an increase in the rate of IP3 receptor degradation [10,11], that it occurs for all IP3 receptor types and is specific, since other ER and signaling proteins are not simultaneously affected [11,12,15,16], that it reduces the sensitivity of ER Ca2+ stores to IP3 [9,12,13], and that it occurs in a wide range of mammalian cell lines in vitro [9–16], in various primary cultures [10,17,18], in mouse oocytes after fertilization [19,20], and in rat pancreas in vivo [18]. Thus, IP3 receptor down-regulation appears to be a widespread homeostatic process – cells adapt to persistent activation of IP3-dependent signaling pathways by reducing the level of the channels that respond to IP3 [14]. This provides cells with a mechanism to limit increases in cytoplasmic Ca2+ concentration and may, thus, protect against deleterious effects of over-activation of Ca2+ signaling pathways (e.g. that which may occur during certain neuropathologies [21], acute pancreatitis [22], and cholestasis [23]), or may serve to oppose processes that depend upon IP3 receptor-induced Ca2+ mobilization from the ER (e.g. apoptosis) [24]. Interestingly, IP3 receptor down-regulation is clearly evident in a rodent model of acute pancreatitis [18] and appears to be a final common event in bile duct epithelia of patients with cholestasis [23].
Initially, the mechanism by which activated IP3 receptors were down-regulated was unclear [14]. It then emerged that activated IP3 receptors are substrates for the ubiquitin-proteasome pathway (UPP); i.e. they are first tagged with the 76 amino acid protein ubiquitin, and are then degraded by the proteasome [12,25].
2.1 The UPP
The UPP is currently the focus of intense interest, since it is now known to be the major route of protein degradation in eukaryotic cells and mediates the selective destruction of many important proteins, including signaling pathway proteins and regulators of the cell cycle and transcription [26]. In addition, it is responsible for “quality control” in the ER; i.e., the selective degradation of misfolded proteins, and of unused subunits of multimeric protein complexes, in a process known as ER-associated degradation (ERAD) (Figure 2) [27].
Figure 2. Simplified model of the ERAD pathway.
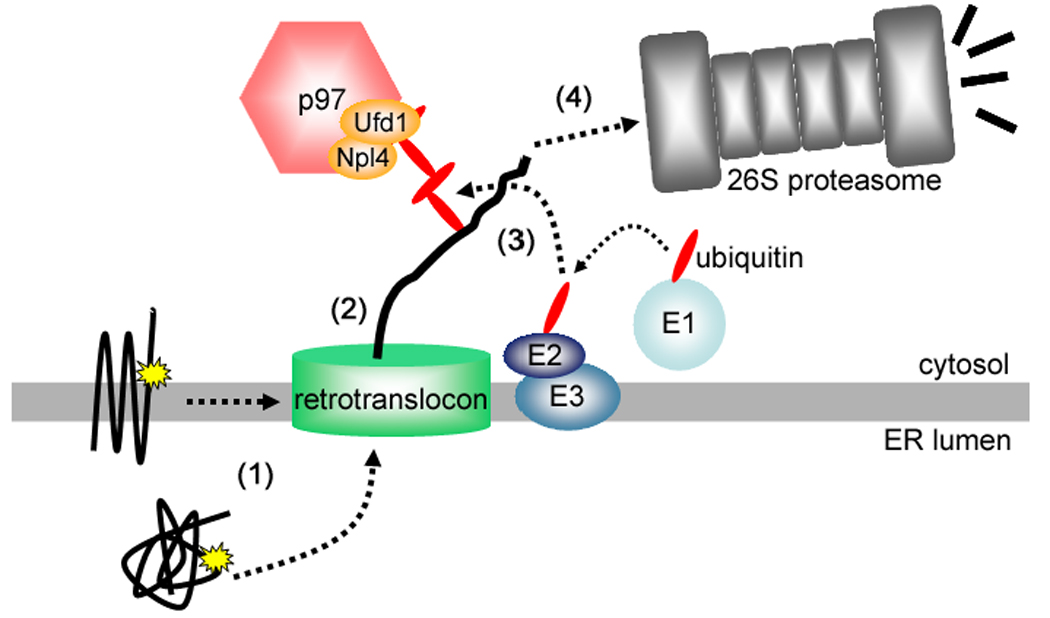
ERAD of membrane or luminal proteins containing some kind of aberration (yellow stars) can be thought of as a 4-step process consisting of (1) recognition, (2) retrotranslocation, (3) ubiquitination, and (4) proteasomal degradation (see text for details).
The canonical summary of the UPP is that substrates for degradation are first polyubiquitinated (tagged with ubiquitin chains linked via ubiquitin’s lysine 48 (K48) residue) and then processed by the proteasome, a 26S, multi-subunit protease composed of a 20S proteolytic core and two 19S regulatory caps, that can recognize polyubiquitinated proteins and unfold and degrade them [26–28]. This is certainly an over-simplification, however, since it is now clear that some UPP substrates can be modified with ubiquitin conjugates other than K48-linked chains and that the proteasome can recognize a variety of ubiquitin conjugates [29]. Nevertheless, ubiquitination is the key step in targeting a protein for proteasomal degradation, and much is known about its enzymology [26–29]. It is achieved through the hierarchical action of 3 enzymes, termed ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (Ubc or E2), and ubiquitin-protein ligase (E3). While there is only one or two E1(s), there are dozens of E2s, and hundreds of E3s. In summary, E1-activated ubiquitin is transferred to an E2, and with the guidance of an E3, the ubiquitin moiety is coupled to the ε-amino group of a lysine residue in the substrate through an isopeptide bond. The process can conclude with the addition of just one ubiquitin moiety (causing monoubiquitination), or a polyubiquitin chain can be formed by multiple rounds of ubiquitination; the C-terminus of incoming ubiquitin moieties are isopeptide bonded to lysine residues in the already attached ubiquitin. Originally, ubiquitin’s K48 was thought to be the only lysine used for chain synthesis, but it is now emerging that ubiquitin’s other lysines (K6, K11, K27, K29, K33 and K63) can also be used [29]. Ubiquitination of a substrate is triggered by variety of signals (e.g. phosphorylation or the exposure of hydrophobic patches) [30,31], but interestingly, there is no amino acid consensus sequence that facilitates the selection of particular lysines [32]. Rather, it is often the case that multiple lysines within a certain region are ubiquitinated, indicating that when a substrate and an E2 are juxtaposed, any accessible lysine within that region will be ubiquitinated [30]. Structural features appear to influence selection of lysines, since a systematic analysis of 135 ubiquitination sites in yeast proteins showed that ubiquitin was preferentially added to lysines in surface–exposed peptide loops, as compared to α-helices or β-sheets [32]. Other findings support this view; e.g. the EGF receptor is ubiquitinated at 6 lysines clustered in the kinase domain, all of which are located on exposed surfaces [33].
2.2 ERAD
In addition to being a Ca2+ store, the ER is of course, also the synthesis site of membrane and secreted proteins, which account for ~1/3 of all proteins [27]. It has emerged in recent years that a sophisticated system, ERAD, exists in eukaryotes for the disposal of proteins that do not fold properly or which cannot find their normal binding partners (Figure 2) [27]. Intriguingly, some ER-resident proteins that are stable under normal conditions are also processed in this manner, the prototype being 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), the rate limiting enzyme in sterol synthesis, which is targeted for ERAD when sterols are in excess [34,35].
How substrates are recognized for ERAD (Figure 2, step 1) is not well understood, and it seems that recognition can be prompted in a variety of ways – either by generic signals (e.g. surface exposed hydrophobic patches), or by specific recognition factors (e.g. Insigs, which mediate HMGR degradation) [30,31,34,35]. Even more mysterious is how substrates are “retrotranslocated” from the ER lumen or membrane (step 2) to the ubiquitination machinery in the cytosol; several routes have been proposed (e.g. a proteinaceous “retrotranslocon”, or a lipid-based pore), but as yet, there is no consensus [27,31]. Much more is known about the enzymes that mediate ERAD substrate ubiquitination (step 3) [31]. In yeast, Hrd1p, an ER membrane E3, mediates the degradation of proteins with misfolded lumenal and membrane domains, while Doa10, another ER membrane E3, mediates the degradation of ER membrane proteins with misfolded cytosolic domains [31,36]. A similar, but more complex, situation appears to exist in mammals, where two Hrd1p homologues (Hrd1 and gp78) and a putative Doa10 homologue (TEB4) [30,31], co-exist with several additional ER membrane-located ligases, possibly with more specialized roles (e.g. RMA1/RNF5 and Kf-1) [31,37,38]. Ubiquitination occurs subsequent to, or simultaneously with retrotranslocation, and a cytosolic complex composed of the ATPase p97 and its polyubiquitin-binding co-factors, Ufd1p and Npl4p, couples ATP hydrolysis to substrate extraction, although precisely how, is not yet clear [27,30]. Finally, (step 4) polyubiquitinated substrates are recruited to the 26S proteasome via shuttle proteins that bind both ubiquitin and the 19S cap, or by interacting directly with intrinsic subunits of the 19S cap that contain ubiquitin-binding motifs [27,29,39]. It is probable that some of the aforementioned steps are integrated, since multiprotein complexes that carry out more than one step are being defined; e.g. the complex centered around Hrd1p contains proteins that recognize, polyubiquitinate, and even perhaps retrotranslocate ERAD substrates [27,36], and proteasomes are found at the cytoplasmic face of the ER and could provide some of the motive force for retrotranslocation [27].
2.3 Are IP3 receptors ERAD substrates ?
Evidence that IP3 receptors are UPP substrates came from experiments showing that IP3 receptors are polyubiquitinated, and that proteasome inhibitors block their down-regulation [12,16,18,25,40]. Obviously, their location in the ER immediately suggested that IP3 receptors could be targeted by the ERAD pathway and subsequent studies supported this view – an E2 that ubiquitinates IP3 receptors is ubc7 [40], an enzyme implicated in both yeast and mammalian ERAD pathways [31,36], and the p97-Ufd1-Npl4 complex mediates the extraction of ubiquitinated IP3 receptors from the ER membrane [41]. The identity of the E3 that catalyses IP3 receptor ubiquitination remains to be resolved, however [42]. This intersection between IP3 receptors and the ERAD pathway raises several fascinating questions; notably, why and how are IP3 receptors selected for ERAD, at what sites are IP3 receptors ubiquitinated and with what conjugates, and how are tetrameric IP3 receptor complexes dissembled and delivered to the proteasome ? The remainder of this review describes the progress that has been made in answering these questions, and highlights the issues that remain to be resolved.
3.1 IP3 receptor ubiquitination is surprisingly complex
A fundamental unanswered question for most UPP substrates concerns where they are ubiquitinated and with what, and only recently, with the advent of mass spectrometry-based technologies [33,43–45], has it been possible to address this question. Application of this approach to IP3R1 isolated from GnRH-stimulated αT3-1 cells, showed that at least 11 of IP3R1’s 167 lysines can be sites of ubiquitination (Figure 3A), that of the attached ubiquitin moieties, at least ~40% are monoubiquitin, with the vast majority of the remainder being roughly evenly divided between K63- and K48-linked chains (Figure 3B), and that on average, IP3R1 subunits are ubiquitinated at ~6 lysines with a total of ~ 8 ubiquitin moieties (Figure 3C) [46]. Interestingly, all of the identified ubiquitination sites are found in the cytosolic coupling domain and are likely in exposed regions, as they are found within or adjacent to binding sites for modifiers, or close to surface-exposed loops (Figure 3A). As more of the IP3R1 structure is defined, it will be fascinating to see how these sites are arranged in 3-dimensions. Certainly, the proximity of the ubiquitinated lysines to regulatory sites also raises the possibility (as yet untested) that ubiquitination might play a role in the acute regulation of channel function, in addition to triggering proteasomal degradation.
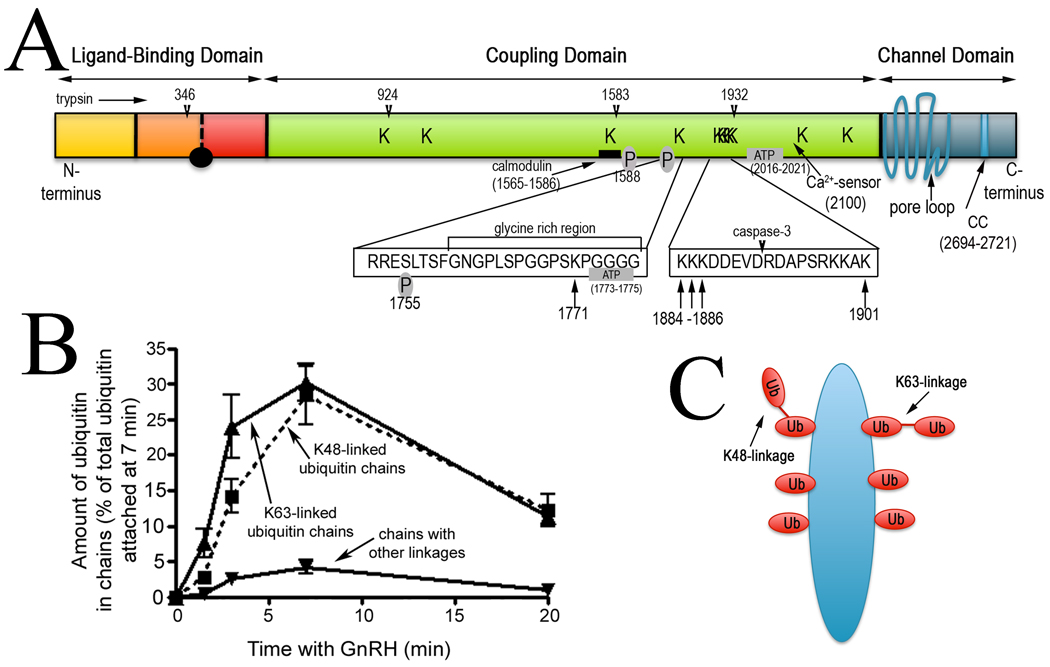
Figure 3. Ubiquitination sites and ubiquitin chain linkages on IP3R1.
A A schematic representation of mouse IP3R1, with the 11 ubiquitination sites (K916, 962, 1571, 1771, 1884, 1885, 1886, 1901, 1924, 2118 and 2257) indicated by “Ks” in the main diagram or by arrows in the expanded regions. Also indicated are sites at which trypsin preferentially cleaves IP3R1 and which are thought to be surface-exposed loops (arrowheads), a glycine rich region, a caspase-3 cleavage site, ATP- and calmodulin-binding sites, the “Ca2+-sensor”, and sites of PKA-mediated phosphorylation [2]. Note that the ubiquitination sites are near exposed loops, or regulatory sites. B Relative amounts of ubiquitin in chains linked via K48 or K63 or other lysines. C Depicted is a single IP3R1 subunit ubiquitinated in a manner that approximates to the average modification; the subunit is tagged at 6 sites with a total of 8 ubiquitin moieties, with 1 ubiquitin moiety containing a K48 linkage and 1 containing a K63 linkage.
Given the canonical view of the UPP, the accumulation of so much monoubiquitin and K63-linked ubiquitin on activated IP3R1 is surprising. K48 and K63-linked chains have very different structures [29], and while K48-linked chains clearly signal for proteasomal degradation, K63-linked chains are generally considered to be involved in other events, such as regulation of the NF-κB pathway, receptor enodocytosis and DNA repair [29]. However, substrates modified with K63-linked chains are rapidly cleaved by the proteasome, at least in vitro [47], indicating that the K63 linkages on IP3R1 may also signal for degradation. Likewise, while monoubiquitination is generally thought to influence protein trafficking [29], recent work shows that certain proteasome subunits have a high affinity for monoubiquitin [39]. Clearly, the roles of monoubiquitin and the different chain types in IP3 receptor regulation need to be resolved experimentally. Likewise, it needs to be determined whether K48 and K63 linkages are found in the same (mixed-linkage) chains, or are segregated into different chains, and precisely which enzymes govern the accumulation of the different ubiquitin conjugates. Interestingly, it has recently become apparent that the same questions are applicable to other UPP substrates, since many other proteins (e.g. the EGF receptor, cyclin B1, RIP1 and IRAK1) are also modified with a mixture of monoubiquitin and K48- and K63-linked chains [33,45,48].
3.2 Special delivery
To contemplate how ubiquitinated IP3 receptors might be degraded by the proteasome is quite daunting. To enter the catalytic core of the proteasome, proteins must first be unfolded [27,28], yet IP3 receptor subunits have 6 TM domains and in their native state are tightly associated into tetramers ~1MDa in size (Figure 1A). Two new pieces of data appear to speak to this issue. First, in contrast to many model ERAD substrates [49,50], IP3 receptors are not released into the cytosol prior to degradation [25], and second, IP3 receptor subunits are not fragmented prior to complete degradation, suggesting that they are consumed in one step [12,25,46]. Together, these data suggest a mechanism like that shown in Figure 2, whereby subunits of activated IP3 receptors are “fed” into the proteasome as the peptide is extracted from the ER membrane, with the assistance of ubiquitin-binding factors, like the p97-Npl4-Ufd1 complex. But how can individual IP3 receptor subunits be extracted ? Analysis of IP3 receptor ubiquitination may provide a clue, since not all subunits in an IP3 receptor tetramer are ubiquitinated and sub-tetrameric IP3 receptor complexes form as IP3 receptor degradation proceeds [46], suggesting that individual subunits are selectively ubiquitinated, extracted and degraded. Since IP3 receptors are ubiquitinated in the coupling domain which is normally exposed to the cytosol [46], and depletion of p97 causes ubiquitinated IP3 receptors to accumulate in the ER membrane [41], it appears that ubiquitination is the event that triggers IP3 receptor extraction, rather than vice versa.
3.3 The SPFH1/2 complex and selection of activated IP3 receptors for ERAD
Several proteins, including p97, associate with IP3 receptors in an activation-dependent manner [41] and most recently it has been demonstrated that SPFH1 and SPFH2 (erlin 1 and erlin 2; see Section 3.4) [51], also have this property [42,52]. Figure 4A–C shows the essential features of these two proteins – that they associate rapidly with IP3 receptors in a manner that precedes maximal IP3 receptor ubiquitination and association of p97, that they are type II ER membrane glycoproteins, and that they oligomerize into a huge, ~2MDa complex. RNA interference shows that depletion of this complex inhibits IP3 receptor ubiquitination and degradation [42,52], indicating that it mediates an early step in IP3 receptor ERAD. The structural data provide tantalizing hints about function – the open ring-shape immediately raises the possibility that the SPFH1/2 complex forms some kind of pore in the ER membrane, or that it could completely or partially encircle IP3 receptor tetramers. Indeed, the membrane and intralumenal region of IP3R1 has a diameter of ~100Å (Figure 1A), which could easily fit within the ~125Å cavity in the luminal domain of the SPFH1/2 complex. Interestingly, the IP3 receptor channel pore-forming sequences include an intraluminal loop located between the fifth and sixth TM domains that contains the pore helix and selectivity filter and is thought to undergo some kind of rearrangement upon IP3 receptor activation [1–3,7]. This loop is already known to interact with ERp44 [53] and chromogranins [54], and could also provide a docking site for the luminal domain of the SPFH1/2 complex. Overall, the most likely scenario (Figure 4D) is that the SPFH1/2 complex is a recognition factor that binds to the luminal regions of activated IP3 receptors, and triggers IP3 receptor ubiquitination in the coupling domain by recruiting the appropriate E2 and E3. Interestingly, the SPFH1/2 complex also associates with other proteins undergoing ERAD; e.g. CFTRΔF508 [55] and the α1D-adrenergic receptor [56], and affects the stability of some model ERAD substrates [52]. Thus, while the SPFH1/2 complex is clearly essential for IP3 receptor ERAD, it may also play a role in the degradation of other substrates.
Figure 4. The SPFH1/2 complex and its role in IP3 receptor ERAD.
A. SPFH1 and SPFH2 associate very rapidly with activated IP3R1, as observed when αT3-1 cells are exposed to GnRH, and anti-IP3R1 immunoprecipitates are probed for ubiquitin, p97, SPFH1 and SPFH2. B. The basic molecular architecture of SPFH1 and SPFH2 is shown, with the N-terminal TM domains indicated by black boxes, the SPFH domains by gray boxes, the glycans by the asterisks, and the locations of the various motifs by amino acid number. C. A 3-dimensional reconstruction of the SPFH1/2 complex, determined at a resolution of ~33Å and contoured at a volume corresponding to a calculated molecular mass of ~2MDa (scale bar = 100Å). Putative side (1), top (2) and bottom (3) views are shown, with the membrane-spanning and lumenal regions indicated by arrows and arrowheads, respectively. D. Summary model of how IP3 receptor ERAD occurs. Upon binding of the co-agonists IP3 and Ca2+, IP3 receptor tetramers undergo a conformational change that both opens the Ca2+ channel and triggers the association of the SPFH1/2 complex, which recruits the E2 and E3 enzymes that catalyze the ubiquitination of IP3 receptors in the coupling domain. Ubiquitinated receptors are then extracted from the membrane and delivered to the proteasome through the action of the cytosolic p97-Ufd1-Npl4 complex.
3.4 SPFH domain-containing proteins
SPFH1 and SPFH2 belong to a family of ~100 mammalian proteins that contain an “SPFH” domain, an ~250 amino acid motif named because of minor sequence similarities in the proteins Stomatin, Prohibitin, Flotillin (reggie), and HflC/K [51]. SPFH domain-containing proteins share some similarities, including localization to cholesterol-rich, detergent-resistant membranes (DRMs), and assembly into large (>1MDa) oligomeric structures [51]. To date, however, no universal function has been attributed to the SPFH domain, and SPFH domain-containing proteins have distinct subcellular localizations and roles [51]. For example, stomatin and stomatin-related proteins regulate the function of epithelial sodium channels of the ENaC/degenerin family at the plasma membrane in a cholesterol-dependent manner [57–59], and the prohibitins are found primarily in the mitochondrial inner membrane, where they carry out a variety of functions, including the regulation of membrane protein stability [60]. Intriguingly, two plasma membrane-associated SPFH domain-containing proteins, MEC-2 from C. elegans and the mammalian stomatin-like protein, podocin, directly bind cholesterol via their SPFH domains, and it is possible that all SPFH domain-containing proteins share this property [61]. Cholesterol binding undoubtedly relates to the localization of these proteins to DRMs, and suggests that they either help recruit cholesterol and other lipids to membrane microdomains, or are drawn to DRMs by cholesterol where they play other roles [51]. Interestingly, although the cholesterol content of intracellular membranes is low [62], SPFH1 and SPFH2 localize to putative ER-derived DRMs in a cholesterol-dependent manner [63], and evidence that IP3 receptors relocate to lipid raft-like microdomains after cell stimulation, raises the possibility that the SPFH1/2 complex and activated IP3 receptors might interact in DRMs [64].
4. Conclusions and perspectives
That a fraction of activated IP3 receptors are hived off for ERAD is both surprising and intriguing. The cell is inactivating IP3 gated channels in a very radical manner – by degradation as opposed to a reversible modification. This could represent a finely tuned mechanism to suppress Ca2+ signaling. Alternatively, it could be more by accident than design – it is possible that during the activation process, IP3 receptors “accidentally” expose regions (e.g. hydrophobic patches) that makes them resemble aberrant proteins and allows for recognition by the ERAD pathway. Whether other ion channels are similarly subject to activation-dependent ubiquitination has yet to be defined, but significantly there currently are no reports that ryanodine receptors, which are also ER-located Ca2+ channels [2], become ubiquitinated upon activation. The challenges now are to better define the role of the SPFH1/2 complex in IP3 receptor regulation, to identify the enzymes that control ubiquitin conjugation to IP3 receptors, to define the functions of the various ubiquitin conjugates, and to establish how ubiquitinated receptors are delivered to the proteasome. Addressing these challenges will teach us much about IP3 receptor processing and about the ERAD pathway in general.
Acknowledgements
The authors wish to apologize to those whose work was omitted due to space constraints, and wish to thank the National Institutes of Health, the Pharmaceutical Research and Manufacturers of America Foundation, and the American Heart Association for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Taylor CW, da Fonseca PCA, Morris EP. IP3 receptors: the search for structure. Trends Biochem. Sci. 2004;29:210–219. doi: 10.1016/j.tibs.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Foskett JK, White C, Cheung K-H, Mak D-OD. Inositol trisphosphate receptor Ca2+ release channels. Physiol. Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikoshiba K. IP3 receptor/Ca2+ channel: from discovery to new signaling concepts. J. Neurochem. 2007;102:1426–1446. doi: 10.1111/j.1471-4159.2007.04825.x. [DOI] [PubMed] [Google Scholar]
- 4.Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M. Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor. J. Mol. Biol. 2007;373:1269–1280. doi: 10.1016/j.jmb.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 5.Hamada K, Terauchi A, Mikoshiba K. Three-dimensional rearrangements within inositol 1,4,5-trisphosphate receptor by calcium. J. Biol. Chem. 2003;278:52881–52889. doi: 10.1074/jbc.M309743200. [DOI] [PubMed] [Google Scholar]
- 6.Schug ZT, Joseph SK. The role of the S4–S5 linker and C-terminal tail in inositol 1,4,5-trisphosphate receptor function. J. Biol. Chem. 2006;281:24431–24440. doi: 10.1074/jbc.M604190200. [DOI] [PubMed] [Google Scholar]
- 7.Schug ZT, da Fonseca PC, Bhanumathy CD, Wagner L, 2nd, Zhang X, Bailey B, Morris EP, Yule DI, Joseph SK. Molecular characterization of the inositol 1,4,5-trisphosphate receptor pore-forming segment. J. Biol. Chem. 2008;283:2939–2948. doi: 10.1074/jbc.M706645200. [DOI] [PubMed] [Google Scholar]
- 8.Sato C, Hamada K, Ogura T, Miyazawa A, Iwasaki K, Hiroaki Y, Tani K, Terauchi A, Fujiyoshi Y, Mikoshiba K. Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. J. Mol. Biol. 2004;336:155–164. doi: 10.1016/j.jmb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 9.Wojcikiewicz RJH, Nahorski SR. Chronic muscarinic stimulation of SH-SY5Y neuroblastoma cells suppresses inositol 1,4,5-trisphosphate action: Parallel inhibition of inositol 1,4,5-trisphosphate-induced Ca2+ mobilization and inositol 1,4,5-trisphosphate binding. J. Biol. Chem. 1991;266:22234–22241. [PubMed] [Google Scholar]
- 10.Wojcikiewicz RJH, Furuichi T, Nakade S, Mikoshiba K, Nahorski SR. Muscarinic receptor activation down-regulates the type I inositol 1,4,5-trisphosphate receptor by accelerating its degradation. J. Biol. Chem. 1994;269:7963–7969. [PubMed] [Google Scholar]
- 11.Wojcikiewicz RJH. Type I, II and III inositol 1,4,5-trisphosphate receptors are unequally susceptible to down-regulation and are expressed in markedly different proportions in different cell types. J. Biol. Chem. 1995;270:11678–11683. doi: 10.1074/jbc.270.19.11678. [DOI] [PubMed] [Google Scholar]
- 12.Bokkala S, Joseph SK. Angiotensin II-induced down-regulation of inositol trisphosphate receptors in WB rat liver epithelial cells: Evidence for the involvement of the proteasome pathway. J. Biol. Chem. 1997;272:12454–12461. doi: 10.1074/jbc.272.19.12454. [DOI] [PubMed] [Google Scholar]
- 13.Tovey SC, de Smet P, Lipp P, Thomas D, Young KW, Missiaen L, De Smedt H, Parys JB, Berridge MJ, Thuring J, Holmes A, Bootman MD. Calcium puffs are generic InsP3-activated elementary calcium signals and are downregulated by prolonged hormonal stimulation to inhibit cellular calcium responses. J. Cell Sci. 2001;114:3979–3989. doi: 10.1242/jcs.114.22.3979. [DOI] [PubMed] [Google Scholar]
- 14.Wojcikiewicz RJH. Regulated ubiquitination of proteins in GPCR-initiated signalling pathways. Trends Pharmacol. Sci. 2004;25:35–41. doi: 10.1016/j.tips.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Willars GB, Royall JE, Nahorski SR, El-Gehani F, Everest H, McArdle CA. Rapid down-regulation of the type I inositol 1,4,5-trisphosphate receptor and desensitization of gonadotropin-releasing hormone-mediated Ca2+ responses in αT3-1 gonadotropes. J. Biol. Chem. 2001;276:3123–3129. doi: 10.1074/jbc.M008916200. [DOI] [PubMed] [Google Scholar]
- 16.Wojcikiewicz RJH, Xu Q, Webster JM, Alzayady K, Gao C. Ubiquitination and proteasomal degradation of endogenous and exogenous inositol 1,4,5 trisphosphate receptors in αT3-1 anterior pituitary cells. J. Biol. Chem. 2003;278:940–947. doi: 10.1074/jbc.M206607200. [DOI] [PubMed] [Google Scholar]
- 17.Lee B, Gai W, Laychock SG. Proteasomal activation mediates down-regulation of inositol 1,4,5-trisphosphate receptor and calcium mobilization in rat pancreatic islets. Endocrinol. 2001;142:1744–1751. doi: 10.1210/endo.142.5.8150. [DOI] [PubMed] [Google Scholar]
- 18.Wojcikiewicz RJH, Ernst SA, Yule DI. Secretagogues cause the ubiquitination and down-regulation of inositol 1,4,5 trisphosphate receptors in rat pancreatic acinar cells. Gastroenterology. 1999;116:1194–1201. doi: 10.1016/s0016-5085(99)70023-5. [DOI] [PubMed] [Google Scholar]
- 19.Parrington J, Brind S, De Smedt H, Gangeswaran R, Lai AF, Wojcikiewicz RJH, Carroll J. Expression of inositol 1,4,5 trisphosphate receptors in mouse oocytes and early embryos: the type I isoform is upregulated in oocytes and down-regulated after fertilization. Dev. Biol. 1998;203:451–461. doi: 10.1006/dbio.1998.9071. [DOI] [PubMed] [Google Scholar]
- 20.Jellerette T, He CL, Wu H, Parys JB, Fissore RA. Down-regulation of the inositol 1,4,5-trisphosphate receptor in mouse eggs following fertilization or parthenogenetic activation. Dev. Biol. 2000;223:238–250. doi: 10.1006/dbio.2000.9675. [DOI] [PubMed] [Google Scholar]
- 21.Verkhratsky A. Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiol. Rev. 2005;85:201–279. doi: 10.1152/physrev.00004.2004. [DOI] [PubMed] [Google Scholar]
- 22.Ward JB, Petersen OH, Jenkins SA, Sutton R. Is an elevated concentration of acinar cytosolic free ionized calcium the trigger for acute pancreatitis ? Lancet. 1995;346:1016–1019. doi: 10.1016/s0140-6736(95)91695-4. [DOI] [PubMed] [Google Scholar]
- 23.Pusl T, Nathanson MH. The role of inositol 1,4,5-trisphosphate receptors in the regulation of bile secretion in health and disease. Biochem. Biophys. Res. Commun. 2004;322:1318–1325. doi: 10.1016/j.bbrc.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 24.Rong YP, Distelhorst CW. Bcl-2 protein family members: versatile regulators of calcium signaling in cell survival and apoptosis. Annu. Rev. Physiol. 2008;70:73–91. doi: 10.1146/annurev.physiol.70.021507.105852. [DOI] [PubMed] [Google Scholar]
- 25.Oberdorf JA, Webster JM, Zhu CC, Luo SG, Wojcikiewicz RJH. Down-regulation of types I, II and III inositol 1,4,5 trisphosphate receptors is mediated by the ubiquitin / proteasome pathway. Biochem. J. 1999;339:453–461. [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: Implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 27.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nature Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nature Rev. Drug Disc. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda F, Dikic I. Atypical ubiquitin chains: New molecular signals. EMBO reports. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Rev. Mol. Cell Biol. 2008;9:679–689. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell Dev. Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catic A, Collins C, Church GM, Ploegh HL. Preferred in vivo ubiquitination sites. Bioinformatics. 2004;20:3302–3307. doi: 10.1093/bioinformatics/bth407. [DOI] [PubMed] [Google Scholar]
- 33.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem. Rev. 2009;109:1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho P, Goder V, Rapoport T. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 37.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 38.Maruyama Y, Yamada M, Takahashi K, Yamada M. Ubiquitin ligase Kf-1 is involved in the endoplasmic reticulum-associated degradation pathway. Biochem. Biophys. Res. Commun. 2008;374:737–741. doi: 10.1016/j.bbrc.2008.07.126. [DOI] [PubMed] [Google Scholar]
- 39.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster JM, Tiwari S, Weissman AM, Wojcikiewicz RJH. Inositol 1,4,5 trisphosphate receptor ubiquitination is mediated by mammalian Ubc7, a component of the Endoplasmic Reticulum-Associated Degradation pathway, and is inhibited by chelation of intracellular Zn2+ J. Biol. Chem. 2003;278:38238–38246. doi: 10.1074/jbc.M305600200. [DOI] [PubMed] [Google Scholar]
- 41.Alzayady KJ, Panning MM, Kelley GG, Wojcikiewicz RJH. Involvement of the p97-Ufd1-Npl4 Complex in the Regulated Endoplasmic Reticulum-associated Degradation of Inositol 1,4,5-Trisphosphate Receptors. J. Biol. Chem. 2005;280:34530–34537. doi: 10.1074/jbc.M508890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearce MMP, Wormer DB, Wilkens S, Wojcikiewicz RJH. An endoplasmic reticulum (ER) membrane complex composed of SPFH1 and SPFH2 mediates the ER-associated degradation of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 2009;284:10433–10445. doi: 10.1074/jbc.M809801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 44.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteaomics. Nat. Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi S. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 46.Sliter D, Kirkpatrick DS, Alzayady K, Kubota K, Gygi SP, Wojcikiewicz RJH. Mass spectral analysis of type I inositol 1,4,5-trisphosphate receptor ubiquitination. J. Biol. Chem. 2008;283:35319–35328. doi: 10.1074/jbc.M807288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 48.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 49.Carlson EJ, Pitonzo D, Skach WR. p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J. 2006;25:4557–4566. doi: 10.1038/sj.emboj.7601307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Pearce MMP, Wang Y, Kelley GG, Wojcikiewicz RJH. SPFH2 mediates the ERAD of IP3 receptors and other substrates in mammalian cells. J. Biol. Chem. 2007;282:20104–20115. doi: 10.1074/jbc.M701862200. [DOI] [PubMed] [Google Scholar]
- 53.Higo T, Hattori M, Nakamura T, Natsume T, Michikawa T, Mikoshiba K. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell. 2005;120:85–98. doi: 10.1016/j.cell.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 54.Kang J, Kang S, Yoo SH, Park S. Identification of residues participating in the interaction between an intraluminal loop of inositol 1,4,5-trisphosphate receptor and a conserved N-terminal region of chromogranin B. Biochim. Biophys. Acta. 2007;1774:503–509. doi: 10.1016/j.bbapap.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, et al. Hsp90 Cochaperone Aha1 Downregulation Rescues Misfolding of CFTR in Cystic Fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 56.Lyssand JS, DeFino MC, Tang XB, Hertz AL, Feller DB, Wacker JL, Adams ME, Hague C. Blood pressure is regulated by an alpha1D-adrenergic receptor/dystrophin signalosome. J. Biol. Chem. 2008;283:18792–18800. doi: 10.1074/jbc.M801860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price MP, Thompson RJ, Eshcol JO, Wemmie JA, Benson CJ. Stomatin Modulates Gating of Acid-sensing Ion Channels. J. Biol. Chem. 2004;279:53886–53891. doi: 10.1074/jbc.M407708200. [DOI] [PubMed] [Google Scholar]
- 58.Wetzel C, Hu J, Riethmacher D, Benckendorff A, Harder L, Eilers A, Moshourab R, Kozlenkov A, Labuz D, Caspani O, Erdmann B, Machelska H, Heppenstall PA, Lewin GR. A stomatin-domain protein essential for touch sensation in the mouse. Nature. 2006;445:206–209. doi: 10.1038/nature05394. [DOI] [PubMed] [Google Scholar]
- 59.Goodman MB, Ernstrom GG, Chelur DS, O'Hagan R, Yao CA, Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 60.Merkwirth C, Langer T. Prohibitin function within mitochondria: Essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Huber TB, Schermer B, Muller RU, Hohne M, Bartram M, Calixto A, Hagmann H, Reinhardt C, Koos F, Kunzelmann K, et al. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. USA. 2006;103:17079–17086. doi: 10.1073/pnas.0607465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prinz W. Cholesterol trafficking in the secretory and endocytic systems. Semin. Cell Dev. Biol. 2002;13:197–203. doi: 10.1016/s1084-9521(02)00048-4. [DOI] [PubMed] [Google Scholar]
- 63.Browman DT, Resek ME, Zajchowski LD, Robbins SM. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 2006;119:3149–3160. doi: 10.1242/jcs.03060. [DOI] [PubMed] [Google Scholar]
- 64.Weerth SH, Holtzclaw LA, Russell JT. Signaling proteins in raft-like microdomains are essential for Ca2+ wave propagation in glial cells. Cell Calcium. 2007;41:155–167. doi: 10.1016/j.ceca.2006.06.006. [DOI] [PubMed] [Google Scholar]