Summary
The actin-based motility of the intracellular pathogen Listeria monocytogenes relies on ActA, a bacterial factor mimicking the activity of host cell nucleation-promoting factors of the WASP/WAVE family. The activity of WASP and WAVE is tightly regulated in cells. However, it is not known whether the activity of ActA is regulated upon L. monocytogenes infection. Here, we used an RNAi-based genetic approach in combination with computer-assisted image analysis to investigate the role of host factors in L. monocytogenes spread from cell to cell. We showed that the host cell serine/threonine kinase CK2 is required for efficient actin tail formation. We demonstrated that, similar to WASP and WAVE, the affinity of ActA for the ARP2/3 complex is regulated by CK2-mediated phosphorylation. We also demonstrated the importance of this regulatory mechanism in a mouse model of infection. Our work suggests that ActA is a bacterial virulence factor that not only displays a structural mimic of the VCA domain of WASP/WAVE family members, but also co-opted CK2 as the host cell factor regulating its activity, a form of mimicry that we refer to as regulatory mimicry. We present comparative evidence supporting the notion that unrelated pathogens displaying actin-based motility may have evolved a similar strategy.
Introduction
The ability of various intracellular pathogens to spread from cell to cell without having to be exposed to the extra-cellular environment is a fundamental aspect of their pathogenic properties. To accomplish this important part of their infectious cycle, various bacterial pathogens have mastered the art of manipulating the host cell cytoskeleton (Gouin et al., 2005; Stevens et al., 2006). Listeria monocytogenes is a Gram-positive bacterium that displays the ability to invade mammalian epithelial cells and propagate into several epithelial structures, including the intestinal, feto-placental and blood-brain barriers (Schlech, 1997). During intra-cytosolic growth, L. monocytogenes polymerizes host actin at one pole of the bacterial surface, allowing the bacteria to move through the cytosol via actin-based motility (Lambrechts et al., 2008). When the bacteria encounter the plasma membrane, the formation of membrane protrusions from an infected cell into an adjacent cell leads to the formation of a bacteria-containing double membrane vacuole in the receiving cell. The bacteria escape this secondary vacuole and resume intra-cytosolic growth (Tilney and Portnoy, 1989).
L. monocytogenes actin-based motility relies on ActA (Brundage et al., 1993; Domann et al., 1992; Kocks et al., 1992), a bacterial factor that mimics the nucleation-promoting activity of WASP/WAVE family members (Goley and Welch, 2006; Pollard, 2007; Takenawa and Suetsugu, 2007) by recruiting the ARP2/3 complex to the bacterial surface (Welch et al., 1997; Welch et al., 1998).
The nucleation promoting activity of WASP/WAVE family members relies on their VCA domain (V (verprolin) region (or WH2 domain), C (central) region and the A (acidic) region). The VCA domain promotes nucleation by bringing together the first G-actin of a new filament and the ARP2/3 complex. The V region interacts with G-actin and the C and A regions cooperate to recruit the ARP2/3 complex (Machesky and Insall, 1998; Marchand et al., 2001; Rohatgi et al., 1999). The C region is characterized by the presence of basic and hydrophobic residues that are essential for interaction with the ARP2/3 complex (Marchand et al., 2001; Panchal et al., 2003). In agreement with the importance of these basic residues in the C region of WASP, the R477K and K476E mutations (Fillat et al., 2001; Kolluri et al., 1995) have been identified in patients displaying the Wiskott-Aldrich syndrome (WAS), a form of immunodeficiency originally linked to mutations in the N-terminal part of WASP (Derry et al., 1994). The A region is characterized by the presence of a trytophane residue embedded in a stretch of acidic residues. The exact function of this region is still unclear, but the tryptophane residue is essential for interaction with the ARP2/3 complex (Marchand et al., 2001).
Deletion analysis of ActA showed that the structural motif essential for interaction with the ARP2/3 complex in vitro and actin tail formation in vivo is a short stretch of basic residues located in the N-terminal part of the protein (Lasa et al., 1997; Skoble et al., 2000; Zalevsky et al., 2001), a region that display similarities with the C region of WASP/WAVE VCA domain, including a stretch of basic residues (Bi and Zigmond, 1999).
In agreement with the importance of the conserved basic residues, introduction of the corresponding mutation in the N-terminal region of ActA (amino-acid position 146–150) completely abolished actin tail formation as well as the recruitment of the ARP2/3 complex at the bacterial surface in vivo (Pistor et al., 2000). ActA does not display a region equivalent to the A region of WASP/WAVE in the vicinity of the conserved basic residues (C region). Instead, mapping and mutational analysis identified an acidic region harboring a key tryptophane residue located in the N-terminal domain of ActA (aminoacid position 41–45) (Boujemaa-Paterski et al., 2001; Skoble et al., 2000). Thus, similar to WASP and WAVE, two regions of ActA, displaying similarities with the C and A regions of the VCA domain of WASP and WAVE, cooperates for binding the ARP2/3 complex.
The nucleation-promoting activity of WASP/WAVE family members is tightly regulated in cells. For instance, all WASP family members harbor an auto-inhibitory GBD domain that binds either to the C region of the VCA domain or the Rho-family GTPases (Goley and Welch, 2006; Pollard, 2007). WAVE family members do not display the auto-inhibitory GBD domain. However, their nucleation-promoting activity is also tightly regulated through interaction with binding partners and post-translational modifications (Takenawa and Suetsugu, 2007). Interestingly, recent studies have shown that the VCA domain of both WASP and WAVE 2 is subject to CK2-mediated phosphorylation, which increases their affinity for the ARP2/3 complex (Cory et al., 2003; Pocha and Cory, 2009). Although ActA was shown to be necessary and sufficient to promote the nucleation activity of the ARP2/3 complex in vitro (Welch et al., 1997), it is unclear whether host cell processes may be required to regulate the activity of ActA in vivo. Interestingly, ActA was shown to be highly phosphorylated upon infection in mammalian cells (Brundage et al., 1993), presumably by host cell kinase(s). However, the functional relevance of ActA phosphorylation in infected cells is unknown. Here, we show that, similar to WASP and WAVE, the affinity of ActA for the ARP2/3 complex is regulated by CK2-mediated phosphorylation. We discuss the implications of our findings with respect to the general mechanisms regulating the activity of nucleation-promoting factors and the evolution of functional mimicry by pathogens that hijack host cell cytoskeleton components.
Results
CK2 is required for L. monocytogenes spread from cell to cell
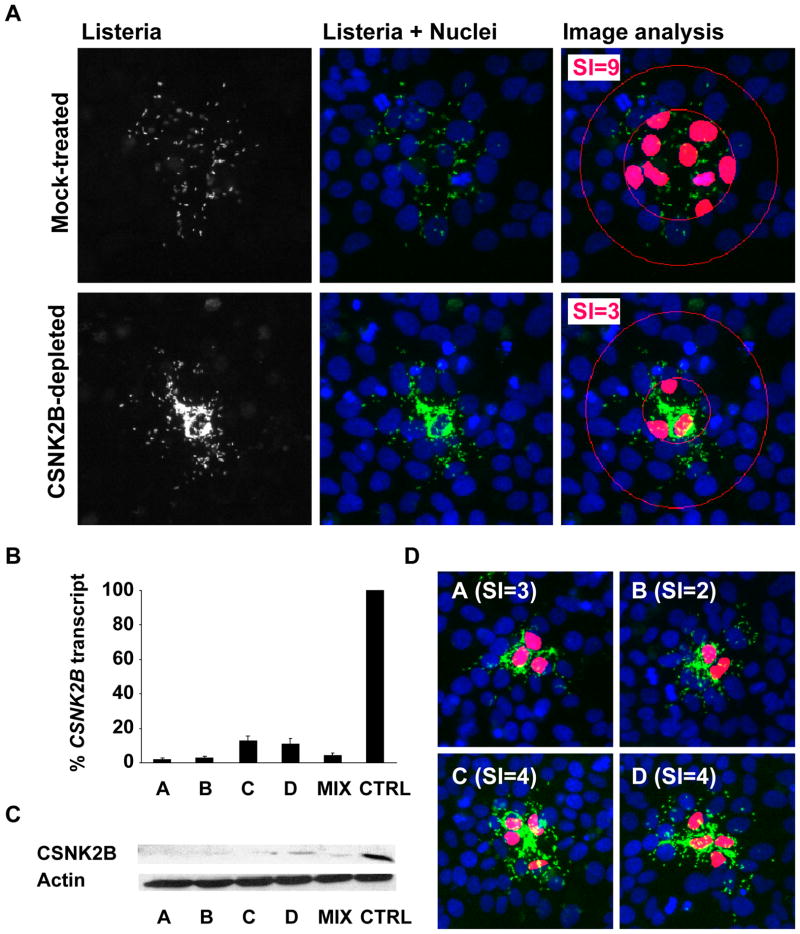
We recently conducted an RNAi screen for host factors involved in L. monocytogenes spread from cell to cell (Swiss et al., unpublished) and identified CSNK2B as a strong candidate gene. CSNK2B encodes the regulatory subunit of Casein Kinase 2 (CK2), a pleitropic serine/threonine kinase catalyzing the phosphorylation of more than 300 substrates in eukaryotic cells (Pinna, 2002). To confirm the involvement of CSNK2B in L. monocytogenes spread from cell to cell, we used a pool of 4 redundant siRNA duplexes and silenced the expression of CSNK2B (see Experimental Procedures). In mock-treated cells, GFP-expressing L. monocytogenes formed a characteristic spreading pattern across the monolayer of cells (Figure 1A, Mock-treated). In stark contrast, the bacteria accumulated in the center of a given infection foci in CK2-depleted cells, suggesting that the vast majority of the bacteria failed to spread to the neighboring cells (Figure 1A, CSNK2B-depleted). To quantify these phenotypes, we used computer-assisted image analysis to determine the spreading index (SI) as the number of cells harboring 75% of the total number of bacteria in a given infection foci (Figure S1). When wild-type cells were infected for 8 hours, the spreading index was 9 +/−2 (Figure 1A, Mock-treated, Image analysis, SI=9). In agreement with a role for CSNK2B in L. monocytogenes spread, the spreading index dropped to 3 +/−1 in CSNK2B-depleted cells (Figure 1A, CSNK2B-depleted, Image analysis, SI=3).
Figure 1. CK2 is required for L. monocytogenes spread from cell to cell.
(A) Representative images of HeLa 229 cells (Nuclei, blue channel) infected with GFP-expressing L. monocytogenes (Listeria, green channel) in mock-treated and CK2-depleted cells. The spreading index (SI) values and +/− SD were determined as described in Figure S1 by analyzing at least 12 foci of infection for each condition tested in three independent experiments.
(B and C). Efficiency of the silencing reagents. Cells were transfected with individual CSNK2B siRNA duplexes (A, B, C and D) or a pool of the four silencing reagents (MIX) and compared to mock-treated cells (CTRL) for silencing efficiency at the transcript level (B) and at the protein level (C).
(D) Cell-to-cell-spreading phenotype in CK2-depleted cells using individual siRNA duplexes as described in (B) and (C) and quantified as shown in (A).
To further validate the specific involvement of CSNK2B, we tested the 4 redundant silencing reagents individually. We confirmed their ability to silence CSNK2B expression at the transcript and protein levels (Figure 1B and Figure 1C). We also determined their corresponding spreading index and confirmed their ability to confer the cell-to-cell spreading defect (Figure 1D). These results ruled out the possibility of an off-target effect and established that the observed cell-to-cell spreading phenotype is specific to CSNK2B depletion.
CK2 is a tetrameric enzyme displaying 2 redundant catalytic sub-units, CSNK2A1 and CSNK2A2, and two regulatory subunits, CSNK2B. We next tested whether CK2 alpha and/or CK2 alpha’ may be involved in L. monocytogenes cell-to-cell spread. siRNA treatment targeting either CSNK2A1 or CSNK2A2 did not result in a significant spreading defect (Figure S2). However, the simultaneous silencing of both CSNK2A1 and CSNK2A2 expression conferred a spreading defect phenotype similar to the one observed upon CSNK2B silencing (Figure S2). These results indicate that the CK2 holoenzyme is involved in L. monocytogenes cell-to-cell spread and that CSNK2A1 and CSNK2A2 isoforms probably play a (redundant) role as catalytic subunits.
CK2 is required for actin tail formation
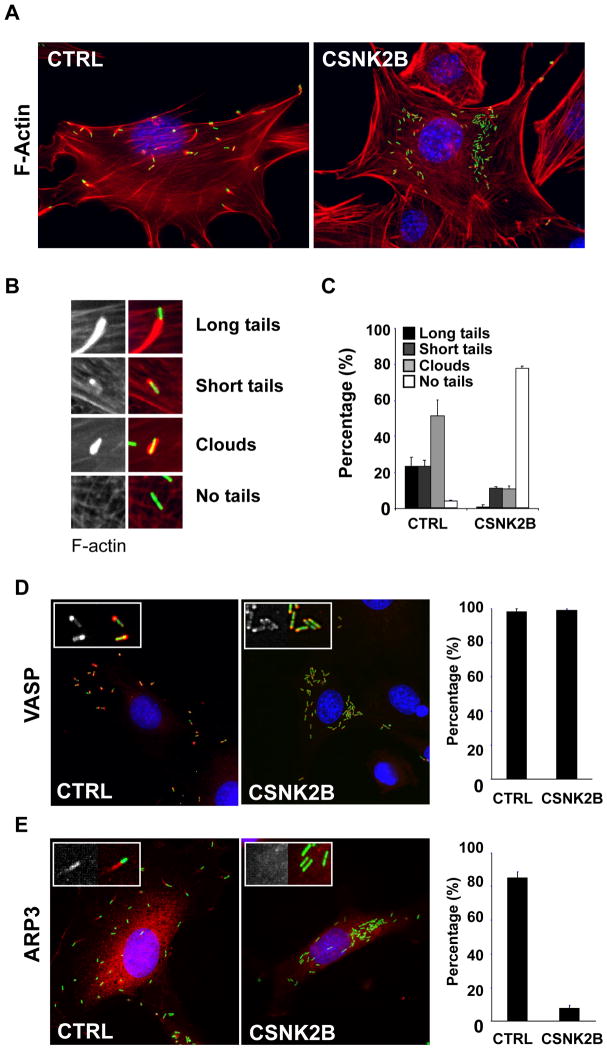
To further analyze the role of CK2 in L. monocytogenes spread, we analyzed the formation of actin tails in mock-treated and CK2-depleted cells (Figure 2A). We scored bacteria that displayed an actin tail (Figure 2B, long and short tails), or were surrounded by F-actin (Figure 2B, clouds), or were not associated with F-actin at all (Figure 2B, no tails). In mock-treated cells, more than 90% of the bacteria were found in association with polymerized actin (Figure 2A, CTRL and Figure 2C, CTRL). In CK2-depleted cells, the bacteria grew as micro-colonies, next to the nucleus (Figure 2A), and more than 70% of the bacteria were not associated with polymerized actin (Figure 2C). The rare bacteria that were associated with polymerized actin displayed very short tails or clouds and were often located next to the plasma membrane. Similar results were obtained in various cells types (Table S1). Altogether, these results indicate that the CK2 kinase is involved in L. monocytogenes cell-to-cell spread through the regulation of actin tail formation.
Figure 2. CK2 is required for actin tail formation and recruitment of the ARP2/3 complex.
(A) Representative images of mock-treated (CTRL) and CSNK2B-depleted (CSNK2B) Swiss 3T3 cells infected with wild-type L. monocytogenes (green) and stained for DNA (blue) and F-actin (red).
(B) Scored actin tail patterns. The left images show F-actin only and the right images show F-actin (red) and the bacteria (green).
(C) Histogram illustrating the quantification of scored actin tail patterns in mock-treated (CTRL) and CSNK2B-depleted (CSNK2B) cells. Values represent the mean ± SD of three independent experiments.
(D and E) Images illustrating the recruitment of VASP (D) and ARP3 (E) in mock-treated (CTRL) and CK2-depleted (CSNK2B) Swiss 3T3 cells. For each image, the left inset shows the corresponding immuno-staining only (VASP or ARP3) and the right inset shows immuno-staining (red) and bacteria (green). The histograms on the left illustrate the quantification of the scored recruitment. Values represent the mean ± SD of three independent experiments.
CK2 is required for recruitment of the ARP2/3 complex
Two main host actin cytoskeleton regulators are involved in ActA-mediated actin tail formation. VASP binds a domain in ActA displaying proline-rich regions, and enhances the actin tail elongation process (Lasa et al., 1995; Niebuhr et al., 1997; Smith et al., 1996). The ARP2/3 complex nucleates actin polymerization at the bacterial cell surface (Welch et al., 1998), and binds a domain in ActA displaying limited similarities to the C region of the VCA domain of WASP (Pistor et al., 2000; Skoble et al., 2000; Zalevsky et al., 2001). We first tested whether the actin tail formation defect observed in CK2-depleted cells may be related to a defect in VASP recruitment. In control cells, VASP was recruited to the cell surface of more than 95% of the bacteria and displayed a polar localization (Figure 2D, CTRL). In CK2-depleted cells, VASP recruitment was indistinguishable from the situation observed in control cells (Figure 2D, CSNK2B). These observations suggest that CK2 does not play a major role in VASP recruitment. We next tested the role of CK2 in the recruitment of the ARP2/3 complex. In control cells, the ARP2/3 complex co-localized with bacteria, as well as the actin tails (Figure 2E, CTRL). With some rare exceptions, the ARP2/3 complex did not co-localize with the bacteria in CK2-depleted cells (Figure 2E, CSNK2B). These data indicate that CK2 is required for efficient recruitment of the ARP2/3 complex to the bacterial cell surface in vivo.
The C region of ActA is phosphorylated by CK2 in vitro
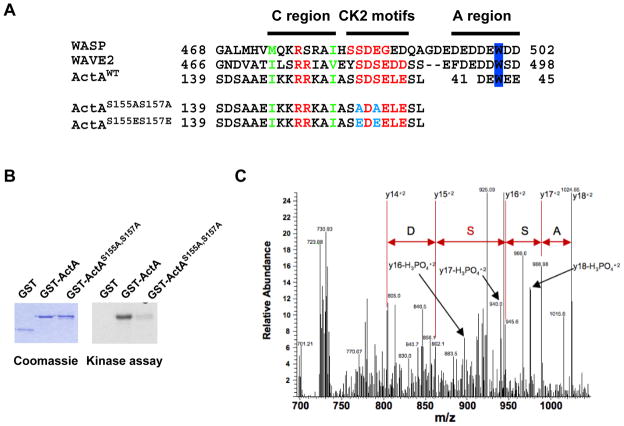
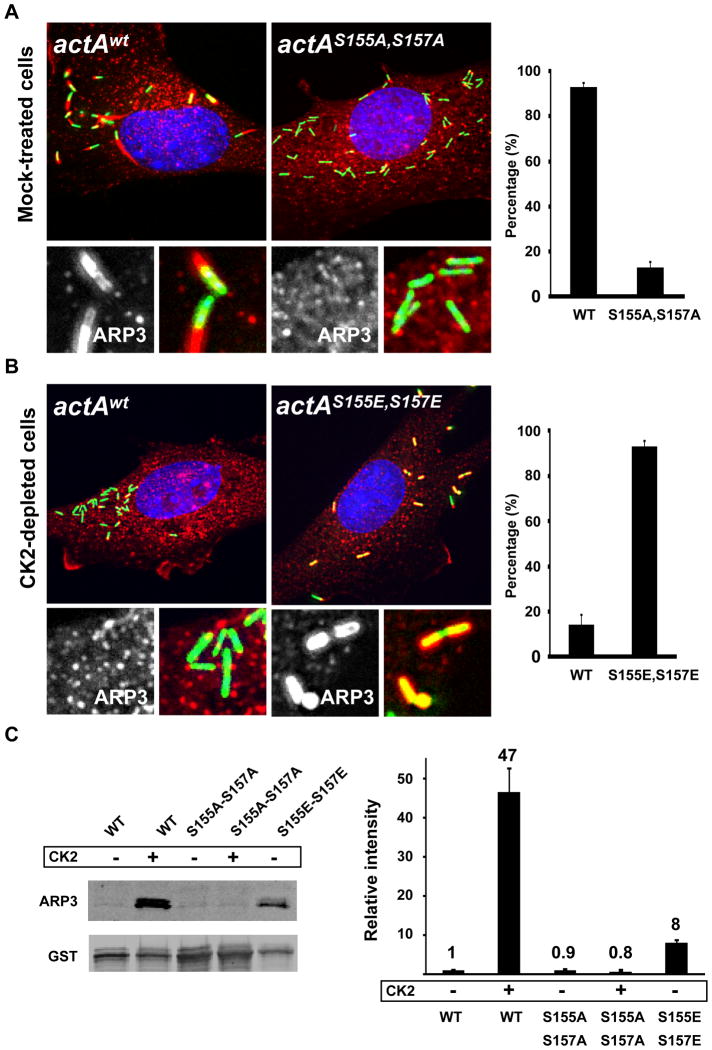
WASP/WAVE family members interact with the ARP2/3 complex through the C and A regions of their VCA domain (Machesky and Insall, 1998; Rohatgi et al., 1999). It was suggested that the N-terminal domain of ActA displays a stretch of basic residues (146-KKRRK-150) similar to the C region of WASP and WAVE (Bi and Zigmond, 1999) (see Figure 3A, C region). Structure/function analyses demonstrated that this short stretch of basic residues in ActA is essential for binding the ARP2/3 complex in vitro and for actin tail formation in vivo (Lasa et al., 1997; Pistor et al., 2000; Skoble et al., 2000). In addition, mapping and mutational analysis demonstrated the existence of a region in ActA equivalent to the A region of WASP/WAVE (Boujemaa-Paterski et al., 2001; Skoble et al., 2000) (see Figure 3A, A region). Interestingly, recent studies have shown that WASP and WAVE 2 are subject to CK2-mediated phosphorylation in the vicinity of their C region, which increases their affinity for the ARP2/3 complex (Cory et al., 2003; Pocha and Cory, 2009). Our comparative sequence analysis of WASP, WAVE 2 and ActA clearly indicated the presence of putative CK2 phosphorylation sites in the corresponding region of ActA (Figure 3A, SXXE/D). We used a kinase assay to show that the N-terminal region of ActA corresponding to amino acids 114 to 194 was phosphorylated by CK2 in vitro (Figure 3B, GST-ActA). The introduction of the S155A-S157A mutations in the corresponding N-terminal region of ActA (Figure 3A, ActAS155A, S157A) strongly decreased CK2-mediated phosphorylation, suggesting that residues S155 and S157 are potential targets for CK2 phosphorylation (Figure 3B, GST-ActAS155A, S157A).
Figure 3. CK2 phosphorylates the ARP2/3 complex binding domain of ActA.
(A) Alignment of the peptide sequence of WASP, WAVE 2, ActA (ActAWT) and ActA mutated versions (ActAS155A,S157A and ActAS155E,S157E) showing the C region, the identified CK2 phosphorylation motifs and the A region (position 41–45 in ActA) with the conserved W residue.
(B) Kinase assay with purified CK2 and GST fusion proteins displaying the wild-type C region of ActA (GST-ActA) or the S155A-S157A mutated version (GST-ActAS155A,S157A).
(C) MS/MS analysis of peptide spanning residues 151–170. The arrows indicate the MS/MS doubly charged y fragment ions for residues 153 to 156 (ASS*D). The y14+2 to the y18+2 ions plus the y16-H3PO4 +2 to the y18-H3PO4 +2 ions demonstrate that Ser 155 is phosphorylated.
The C region of ActA is phosphorylated in vivo
We next used mass spectrometric approaches to specifically detect ActA phosphorylation in vivo. ActA was immuno-precipitated from infected cell extracts and subjected to SDS electrophoresis (Brundage et al., 1993). The peptides resulting from in-gel digest of ActA and its phosphorylated products (Brundage et al., 1993) were fractionated into three pools and were analyzed by nano-LC tandem mass spectrometry for identification (Table S2). The first analyzed fraction represented 20% of the total protein digest (Figure S3A). The remaining 80% was enriched for phosphopeptides with TiO2 affinity matrix and both the enriched (Figure S3B) and the flow-through fractions (Figure S3C) were analyzed by LC-MS/MS. The data showed the enrichment of phosphopeptide 151AIASSDSELESLTYPDKPTK170 (harboring Ser155 and Ser157) as revealed by the presence of the triply charged phosphorylated ion (m/z 744.68, Figure S3A and Figure S3B) and the absence of the unmodified triply charged form of the peptide ion (m/z 718.03, Figure S3B and Figure S3C). The analysis of MS/MS spectrum revealed the presence of the doubly charged y fragment ions for residues 153 to 156 (-ASSD-, Figure 3C). The y14+2 to the y18+2 ions and the y16-H3PO4+2 to the y18-H3PO4+2 ions demonstrated that Ser155 is phosphorylated (Figure 3C). We also conducted MS analysis in CK2-depleted cells for the non-phosphorylated version of the phosphopeptide of interest (m/z 718.0), and two (multiply charged) version of the detected phosphopeptide of interest (m/z 744.7, 1116.5) containing S155 and S157. The data show a consistent decrease for the signal corresponding to the phosphorylated AIASSDSELESLTYPDKPTK peptide ions (Table S3). These results suggest that the relative level of phosphorylation of ActA on S155 is reduced in CK2-depleted cells.
ActA phosphorylation regulates the actin tail formation process
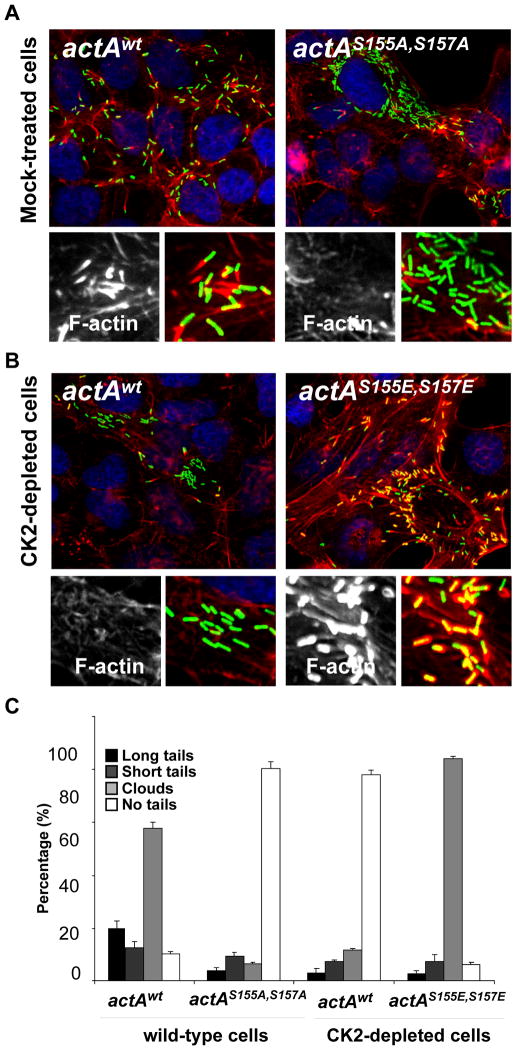
To test the functional relevance of S155 and S157 phosphorylation upon infection, we introduced the single S155A and double S155A, S157A mutations by allelic exchange at the actA locus in the L. monocytogenes chromosome (see Experimental Procedures). The mutant bacteria displaying the actAS155A allele displayed no striking actin tail formation defect (data not shown). However, and similar to the situation observed in CSNK2B-depleted cells infected with wild-type bacteria, infection of mock-treated cells with mutant bacteria displaying the actAS155A, S157A allele revealed a strong spreading defect (Figure 4A, spreading index = 4+/−2, n=12), as well as a strong defect in actin tail formation (Figure 4A and Figure 4C, actAwt and actAS155A,S157A). The actAS155A, S157A mutant bacteria were as infectious as wild-type and the expression level of ActAS155A,S157A was similar to the expression level of ActAwt in infected cells (Figure S4). Moreover, the mutant bacteria expressing ActAS155A, S157A recruited VASP as efficiently as wild-type bacteria upon infection (Figure S5), indicating that ActAS155A, S157A was properly expressed at the bacterial cell surface. These results strongly suggested that phosphorylation on S155 and S157 is required for actin tail formation. In agreement with this assumption, we also showed that the introduction of the phospho-mimic S155E-S157E mutations in actA could partially rescue the actin polymerization defect observed in CK2-depleted cells (Figure 4B and Figure 4C, actAwt and actAS155E, S157E). Similar to ActAS155A, S157A, ActAS155E, S157E was properly expressed at the bacterial cell surface (Figure S4 and Figure S5). We noted that the vast majority of the mutant bacteria were associated with massive accumulation of F-actin, forming clouds or short tails, but failed to form long tails (Figure 4B and Figure 4C, actAS155E, S157E), a defect also observed in wild-type cells infected with the actAS155E, S157E mutant bacteria (data not shown). Despite the formation of actin clouds and short tails, the bacteria displaying the phospho-mimic S155E-S157E mutations in actA did not spread properly (Figure 4B, spreading index = 4+/−2, n=12). These data indicate that, low level of phosphorylation, as mimicked by the S155A-S157A mutations, as well as high level of phosphorylation, as mimicked by the S155E-S157E mutations, result in actin tail formation defects, suggesting that the phosphorylation level of S155 and S157 must be tightly regulated in vivo in order to achieve wild-type tail formation and efficient cell-to-cell spread.
Figure 4. ActA phosphorylation regulates actin tail formation and cell-to-cell spread.
(A and B). Cells infected with GFP-expressing bacteria (green) and stained for DNA (blue) and F-actin (red). The high-magnification images displayed at the bottom of each image show F-actin only (left) and F-actin (red) and bacteria (green) (right). The corresponding spreading index values and +/− SD were determined by image analysis of at least 12 foci of infection in three independent experiments as described in Figures S1: actAwt in mock-treated cells, SI= 9+/−3; actAS155A-S157A in mock-tretaed cells, SI= 4+/−2; actAwt in CK2-depleted cells, SI= 3+/−1 and actAS155E-S157E in CK2-depleted cells, SI= 3+/1. (C) Histogram scoring actin tail formation as presented in A and B. Values represent the mean ± SD of three independent experiments.
CK2 phosphorylation regulates interaction of ActA with the ARP2/3 complex
We next tested whether the actin tail formation defects observed with the mutant bacteria expressing ActAS155A, S157A may be related to a defect in the recruitment of the ARP2/3 complex. In mock-treated cells, the ARP2/3 complex co-localized with bacteria, as well as the actin tails (Figure 5A, actAWT). The ARP2/3 complex did not co-localize with the bacteria expressing ActAS155A, S157A (Figure 5A, actAS155A, S157A), indicating that CK2 phosphorylation on S155-S157 is required for efficient recruitment of the ARP2/3 complex to the bacterial cell surface in vivo. Accordingly, the introduction of the phospho-mimic S155E-S157E mutations in actA could partially rescue the ARP2/3 complex recruitment defect observed in the CK2-depleted cells infected with wild-type bacteria (Figure 5B, actAwt and actAS155E, S157E). We next tested in vitro whether CK2-mediated phosphorylation on S155-S157 may affect the affinity of ActA for the ARP2/3 complex. To this end, we purified the N-terminal domain of ActA (1–194) as a GST fusion protein and used a pull-down assay to analyze the interaction of ActA with purified ARP2/3 complex. In vitro phosphorylation of GST-ActAWT with purified CK2 dramatically increased the interaction with the ARP2/3 complex (Figure 5C, GST-ActAWT and GST-ActAWT + CK2). In agreement with our in vivo observations, the introduction of the S155A-S157A mutation completely abolished the effect of the CK2-mediated phosphorylation (Figure 5C, GST-ActAS155A-S157A and GST-ActAS155A-S157A + CK2). Also in agreement with our in vivo observations, the introduction of the phospho-mimic S155E-S157E mutations also increased the interaction of ActA with the ARP2/3 complex (Figure 5C, GST-ActAS155E-S157E), although the increase was not as dramatic as the one conferred by phosphorylation on S155 and S157 (8-fold versus 47-fold, Figure 5C). Altogether, our in vivo and in vitro analyses demonstrate that CK2-mediated phosphorylation of ActA on S155 and S157 regulates its interaction with the ARP2/3 complex.
Figure 5. ActA phosphorylation regulates ARP2/3 complex binding.
(A and B). Mock-treated and CK2-depleted cells infected with GFP-expressing bacteria (green) and stained for DNA (blue) and ARP3 (red). The high-magnification images displayed at the bottom of each image show ARP3 only (left) and ARP3 (red) and bacteria (green) (right). The histograms on the right illustrate the quantification of the scored recruitment.
(C) Interaction of wild-type (WT) and mutant (S155A-S157A and S155E-S157E) GST fusion proteins with purified ARP2/3 complex and western blot analysis (left panel) and quantification of three independent experiments (right panel). ARP3 signals were normalized with the GST values and the normalized signal for WT was given an arbitrary value of 1 (Relative intensity). Values represent the mean ± SD of three independent experiments.
ActA phosphorylation affects Listeria monocytogenes infection
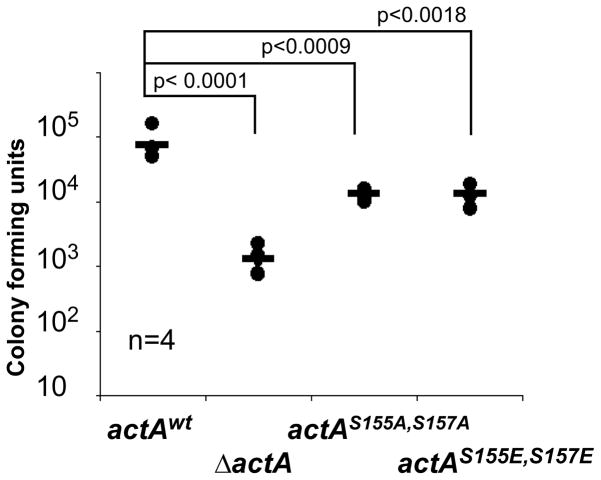
We further tested the importance of the S155-S157 phosphorylation of ActA in a mouse model of infection. As a measure of the infection process, we found that the ability of the strains harboring the S155A-S157A or S155E-S157E actA alleles to colonize the spleen was clearly attenuated, displaying an intermediate phenotype, as compared to spleen colonization by the wild-type strain and the actA deletion mutant strain (Figure 6). We conclude that the regulation of actin-tail formation through the CK2-mediated phosphorylation of ActA constitutes an important aspect of L. monocytogenes infection.
Figure 6. ActA phosphorylation affects L. monocytogenes infection.
Data showing CFUs (Colony-Forming Units) harvested from the spleen of 3 mice infected with wild-type (WT), ActA mutant (ΔActA), actAS155A, S157A and actAS155E, S157E strains. Two-tailed Student’s t-tests were performed. The experiment was repeated twice with similar results.
Conservation of CK2 phosphorylation motifs in nucleation-promoting factors
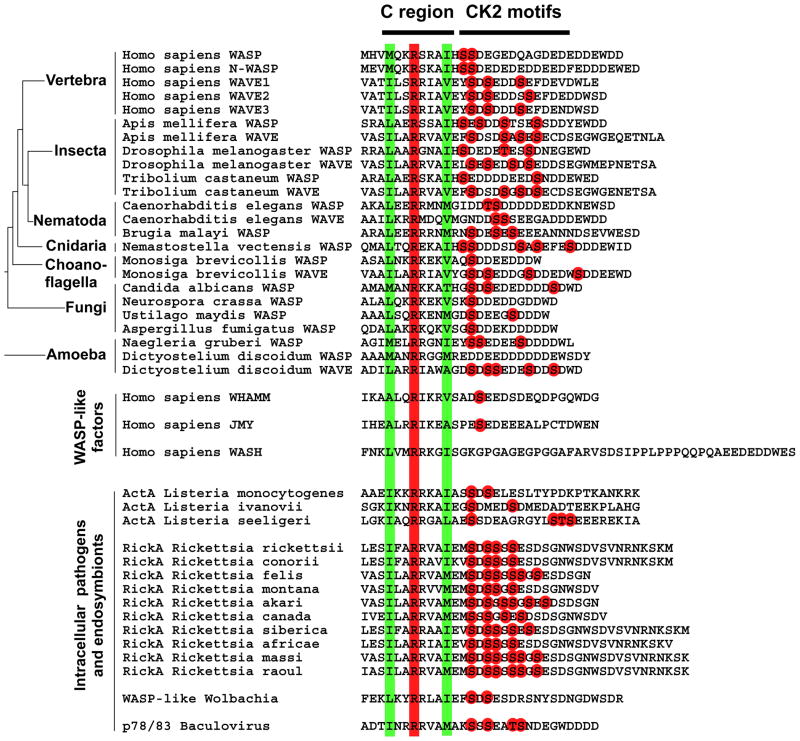
Structural analysis of the C region of WASP revealed the presence of conserved hydrophobic and charged residues predicted to form an amphipathic helix interacting with the ARP2/3 complex and the WASP auto-inhibitory domain (Panchal et al., 2003). We conducted comparative sequence analysis showing that the C region of all WASP/WAVE family members examined, including WASP-like factors produced by bacterial and viral pathogens, such as Listeria spp., Rickettsia spp., Wolbachia spp. and Baculoviruses, displays a signature that includes a perfectly conserved arginine residue surrounded by two hydrophobic residues (Figure 7). Importantly, with the exception of WASH (Linardopoulou et al., 2007), all WASP/WAVE-like family members examined also displayed putative CK2 phosphorylation sites (S/TXXD/E) in the vicinity of the C region (Figure 7). Our analysis therefore supports the notion that, as shown for ActA in this study, unrelated virulence factors displaying nucleation-promoting activity, such as RickA (Gouin et al., 2004; Jeng et al., 2004) and p78/83 (Goley et al., 2006), may also be regulated by CK2-mediated phosphorylation (Figure 7). To the best of our knowledge, ActA, and potentially RickA and p78/83, constitutes the first example of virulence factors that not only evolved as a functional mimicry of a host cell activity, but also precisely coopted the host cell mechanism modulating this activity.
Figure 7. Peptide sequence alignment showing the presence of CK2 phosphorylation motifs in the vicinity of the C region of WASP/WAVE family members.
The conserved hydrophobic and arginine residues in the C region are boxed in green and red, respectively. The red dots indicate putative CK2 phosphorylation sites (pS/TXXD/E).
Discussion
In this study, we demonstrate that a host cell serine/threonine kinase, CK2, is required for efficient cell-to-cell spread of the intracellular pathogen L. monocytogenes. We show that the CK2-mediated phosphorylation of the ARP2/3-binding domain of ActA is required for proper actin tail formation and regulates the affinity of ActA for the ARP2/3 complex. These findings have implications for our general understanding of the mechanisms regulating host cell activities and the evolution of microbial factors mimicking these host cell activities.
Host cell nucleation-promoting factors of the WASP/WAVE family bind and activate the ARP2/3 complex through the C and A regions of their VCA domain (Goley and Welch, 2006; Pollard, 2007). Biochemical studies have suggested that the C region of WASP, N-WASP and WAVE harbors a conserved arginine residue (position 477 in WASP, Figure 6) and several conserved hydrophobic residues folding into an amphipathic helix that constitute the structural motif interacting with the auto-inhibitory GBD domain, as well as the ARP2/3 complex (Panchal et al., 2003). Our comparative analyses of the C region of WASP/WAVE family members and the corresponding C region found in nucleation-promoting factors expressed by intracellular microbial pathogens revealed the strict conservation of the arginine residue and two surrounding hydrophobic residues (Figure 7). This conservation is however less apparent for the newly identified WASP-like factors, including WHAMM (Campellone et al., 2008) and JMY (Zuchero et al., 2009) (Figure 7). We propose that this signature constitutes the minimal structural motif required for interaction with the ARP2/3 complex with the C region, and as such, constitutes the structural basis for the functional mimicry displayed by the C region of ActA (Figure 7).
We present genetic and biochemical evidence showing that phosphorylation of the C region of ActA on S155 and S157 is important for the recruitment of the ARP2/3 complex. Both sites qualify as bone fide CK2 phosphorylation sites (consensus sequence SXXD/E). We also present compelling evidence suggesting that S155 is a direct target for CK2 phosphorylation in vitro and in vivo. However, we were not able to detect S157 phosphorylation in vivo. Thus, although very likely, the actual involvement of CK2 in S157 phosphorylation remains to be demonstrated.
Similar to ActA, the CK2-mediated phosphorylation of the C region of WASP and WAVE 2 (see Figure 3A) was shown to increase the affinity of the VCA domain for the ARP2/3 complex (Cory et al., 2003; Pocha and Cory, 2009). It is noteworthy that the vast majority of CK2 phosphorylation sites are serine/threonine residues surrounded by acidic residues from position −1 to +4, with an absolute conservation in position +3 (S/TXXE/D) (Pinna, 2002). Interestingly, the acidic residues surrounding S155 and S157 were identified as important for ActA activity in a systematic charged-to-alanine mutational analysis of the N-terminal region of ActA (see (Lauer et al., 2001), mutant 48, 155-SDSELE > 155-SASALA). Taking all these observations together, we propose a model in which the negative charges provided by the acidic residues surrounding the phosphorylation sites contribute to the affinity of the C region for the ARP2/3 complex (Figure 3A). In this model, the addition and removal of phosphate groups allows to modulate the interaction with the ARP2/3 complex. Accordingly, the in vitro phosphorylation of ActA on S155 and S157 dramatically increased the affinity of ActA for the ARP2/3 complex (Figure 5C). Moreover, the introduction of the S155A-S157A mutations in ActA led to a strong defect in actin tail formation and recruitment of the ARP2/3 complex in vivo (Figure 5A). Furthermore, the introduction of the phospho-mimic mutation S155E-S157E increased the affinity of ActA for the ARP2/3 complex in vitro (Figure 5C) and the corresponding mutant strain could recruit the ARP2/3 complex in CK2-depleted cells (Figure 5B). However, the affinity of the phosphorylated wild-type protein was significantly higher than the affinity of the phosphomimic mutant. In addition, the rescue of actin tail formation in CK2-depleted cells was only partial, since the bacteria failed to form long tails (Figure 4C). These results may indicate that, in addition to the negative charges, the phosphate groups may provide additional features that contribute to the affinity of the C region for the ARP2/3 complex. It is also possible that, in vivo, in order to achieve proper actin tail formation, the level of ActA phosphorylation must be tightly regulated through phosphorylation/de-phosphorylation cycles, perhaps suggesting the existence of a phosphatase activity that modulates the phosphorylation state of the C region in vivo. Thus, L. monocytogenes may provide a tractable system to understand the exact role of the CK2-mediated phosphorylation and to identify additional factors regulating the activity of the C region of WASP/WAVE family members.
With the exception of the conserved residues in the C region and putative CK2 phosphorylation sites, unrelated microbial factors displaying nucleation-promoting activities, such as ActA in Listeria spp., RickA in Rickettsia spp. (Gouin et al., 2004; Jeng et al., 2004) and p78/83 in Baculoviruses (Goley et al., 2006; Machesky et al., 2001), do not display extensive amino-acid sequence similarities with WASP/WAVE family members. Thus, it is likely that these microbial mimics of WASP and WAVE were generated by convergent evolution (Stebbins and Galan, 2001). In this context, it is remarkable that ActA, and potentially RickA and p78/83, co-opted a regulatory mechanism similar to WASP and WAVE. This probably indicates that the CK2-mediated phosphorylation represents a unique solution to the problem of regulating the cellular activity of nucleation-promoting factors displaying a C region. We propose that ActA defines a novel type of bacterial mimicry, that we refer to as regulatory mimicry, whereby a pathogen has not only evolved a structural mimic of a host cell activity, such as the VCA domain and its role in the recruitment of the ARP2/3 complex, but has also precisely co-opted the host cell machinery that regulates this host cell activity, such as the serine/threonine kinase CK2 and its role in regulating the affinity of the VCA domain for the ARP2/3 complex.
Experimental Procedures
Bacterial and eukaryotic cell growth conditions
L. monocytogenes strain 10403S was grown overnight in BHI (Difco) at 30°C without agitation prior to infection. HeLa 229 cells (ATCC #CCL-2.1) and Swiss 3T3 (ATCC #CCL 92) cells were grown in DMEM (Invitrogen) supplemented with 10% FBS (Gibco) at 37°C in a 5% CO2 incubator.
Listeria monocytogenes infection
Cells were infected (MOI=20) with Listeria monocytogenes strain (10403S) expressing GFP under the control of a IPTG-inducible promoter (Agaisse, unpublished). The plates were centrifuged at 1000 rpm for 5 minutes and internalization of the bacteria was allowed to proceed for 1 hour at 37° before gentamicin (50 μM final) and IPTG (4 mM final) were added in order to kill the remaining extra-cellular bacteria and visualize internalized bacteria. The cells were incubated at 37° for 6–8 hours and then fixed in PBS 4% formaldehyde at 4 °C overnight. The cells were stained with Hoechst (Molecular Probes, Inc.), washed in PBS and kept at 4° until imaging.
CSNK2B silencing and validation procedure
Cells were transfected by reverse transfection with Dharmafect1 and individual CSNK2B siRNA (A, B, C and D, 50 nM final) or a pool of the four silencing reagents (12.5 nM each, 50nM total) and incubated for 72hrs on coverslips in a 24-well plate format. The sequences targeted by the individual siRNA duplexes were as follows: A: CAACCAGAGUGACCUGAUU; B: GACAAGCUCUAGACAUGAU C: CAGCCGAGAUGCUUUAUGG; D: GCUCUACGGUUUCAAGAUC For real-time PCR analysis, total RNA and first-strand cDNA synthesis was performed using the TaqMan Gene Expression Cells-to-Ct Kit (Applied Biosystems) as recommended by the manufacturer with the addition of DNaseI for the removal of unwanted genomic DNA. CSNK2B mRNA levels were determined by quantitative real-time PCR using the LightCycler 480 SYBR Green 1 Master Kit and LightCycler 480 instrument (Roche Biochemicals, Indianapolis, IN).
For Western Blot analysis, cells were transfected with the corresponding silencing reagents in a 24-well plate format and cells were lysed directly in Laemmli sample buffer. The anti-CSNK2B antibody (1:200) was obtained from Santa Cruz (sc-20710). The anti-actin antibody (1:10,000) was obtained from Sigma (A2066). The anti-rabbit -HRP conjugated secondary antibody (1:10,000) was obtained from Jackson ImmunoResearch (111-035-14).
Immunofluorescence procedures
Swiss 3T3 cells were seeded on glass coverslips, infected with L. monocytogenes and fixed in PBS 4% para-formaldehyde. The cells were permeabilized and stained in PBS/0.05% Saponin/1%BSA containing the appropriate primary antibody for 2 hours at room temperature, washed 5 times in PBS/0.05% Saponin/1%BSA before addition of secondary antibodies in PBS/0.05% Saponin/1%BSA for 1 hour at room temperature. The samples were then mounted onto glass slides using FluoroGuard Antifade Reagent (Bio-Rad). Rabbit monoclonal anti-VASP antibody (1:100) was obtained from Cell Signaling (Clone 9A2). Rabbit polyclonal anti-Arp3 antibody (1:100) was obtained from Upstate (07-272). Alexa-fluor 488 secondary antibody (1:1,000), Alexa-fluor 568 conjugated phalloidin (1:500) and Hoechst (1:500) were obtained from Invitrogen.
ActA mutagenesis
The S155A-S157A double mutation (TCG>GCA and AGT>GCA, respectively) was introduced using the overlapping primers 5S155A-S157A (AGAAGAAAAGCCATAGCGTCGGCAGATGCAGAGCTTGAAAGCCTTACTTAT) and 3S155A-S157A (ATAAGTAAGGCTTTCAAGCTCTGCATCTGCCGACGCTATGGCTTTTCTTCT). The construct was generated by overlap PCR using the flanking 5′ primer ActA5Eco (GAAGAATTCGCGCTTCAACAAGCAGCG) with 3S155A-S157A and 3′ primer ActA3BamHI (GGAGGATCCCACTTATCAGAGCCGGCGCGC) with 5S155A-S157A. The two overlapping fragment were gel purified and used as template for PCR amplification using ActA5Eco and ActA3BamHI as flanking primers. The resulting 1.6 kb PCR fragment encoding the corresponding S155AS157A mutation was digested by EcoRI and BamHI and cloned into pMAD (Arnaud et al., 2004). The resulting plasmid, pMAD-ACTAS155AS157A, was introduced into L. monocytogenes 10403S by electroporation and the gene replacement was conducted as previously described (Arnaud et al., 2004). The resulting mutant strain (10403S actAS155A-S157A) was verified by PCR amplification using chromosomal DNA of the corresponding strain and direct DNA sequencing of the PCR product. The actAS155E-S157E mutant strain was generated similarly by generating plasmid pMAD-ACTAS155ES157E using the primers 5S155E-S157E (AGAAGAAAAGCCATAGCGTCGGAAGATGAAGAGCTTGAAAGCCTTACTTAT) and 3S155E-S157E (ATAAGTAAGGCTTTCAAGCTCTTCATCTTCCGACGCTATGGCTTTTCTTCT).
In vitro kinase assay
The DNA fragment corresponding to amino acids 114 to 194 of ActA was PCR-amplified from chromosomal DNA using primers 5GST1Eco (GAAGAATTCTAGTGGCTATAAATGAAGAGGCTTCA) and 3GSTH3 (AAGAAGCTTCTGCATGCTAGAATCTAAGTCACTTTC), digested with EcoRI and HindIII and cloned into pGEX-KG (Amersham Pharmacia Biotech). The S155A-S157A double mutation was amplified using 5GST1Eco and 3GSTH3 as primers and pMAD-ActAS155AS157A as template. Both constructs were verified by DNA sequencing. The corresponding fusion proteins were produced in E. coli BL21 (Invitrogen). The bacteria were grown at 30°C until OD600= 0.5 and expression was induced by adding 0.5 mM IPTG for 1 hour. After re-suspension of the bacteria in PBS and disruption of the bacterial cell wall by sonication, the fusion proteins were purified using Gluthathione Sepharose 4B (GS4B) (Amersham Pharmacia Biotech). To conduct the kinase assay, fusion proteins attached to the GS4B beads were washed and re-suspended in kinase buffer (20 mM Tris-HCl pH 7.5, 10 mM KCl, 10 mM MgCl2, 100 μM ATP and 200 μM sodium orthovanadate) supplemented with 10 μCi (γ-32P)ATP and 50 U CK2 (New England Biolabs, Inc) and incubated for 10 min at 30°C. GS4B beads were washed three times in PBS, re-suspended in 30 μl Laemmli buffer (Sigma) and subjected to SDS-PAGE analysis. The gel was stained with Coomassie brillant blue, dried and incorporated 32P was detected using autoradiography.
Mass Spectrometric Analysis
In gel Protein Digestion
Coomassie stained gel bands corresponding to ActA Protein isolated in 1 dimensional polyacrylamide gel were subjected to in situ enzymatic digestion. The gel plugs were washed with 250μl 50% acetonitrile/50% water for 5 minutes followed by 250μl of 50mM ammonium bicarbonate/50% acetonitrile/50% water for 30 minutes. One final wash was preformed using 10mM ammonium bicarbonate/50% acetonitrile/50% water for 30 minutes. After washing, the gel plugs were dried in a Speedvac and rehydrated with 0.1μg of modified trypsin (Promega) per (approximately) 15mm3 of gel in 15 μl 10mM ammonium bicarbonate. Samples were digested at 37 °C for 16 hours.
Titanium dioxide enrichment
The digest was acidified with 0.5% TFA, 50% acetonitrile. Top Tips (Glygen Corp.) were prepared by washing 3 times with 40μl of each100% acetonitrile, followed by 0.2M sodium phosphate pH 7.0, and 0.5% TFA, 50% acetonitrile. Washes were spun through into an eppendorf tube at 2,000 rpm for 1 minute. The acidified digest supernatant was loaded into the TopTip, spun at 1,000 rpm for 1 minute, and then 3,000 rpm for 2 minutes. Gel pieces were rinsed with 40μl 0.5% TFA, 50% acetonitrile, with the supernatant transferred to the Top Tip and the spin repeated. The Top Tip was then washed with 40μl 0.5% TFA, 50% acetonitrile and the spin repeated. The flow through from these washes were saved and analyzed by LC-MS/MS as below. Phosphopeptides were eluted from the TopTip with 30μl 28% ammonium hydroxide (3 times). Both the flow through and eluted fractions were dried in speedvac, and re-dried in 40 μl water. Samples were dissolved in 3μl 70% formic acid, vortexed, diluted with 7μl 0.1% TFA, spun and transferred to LC-MS/MS vials where 5μl was injected.
LC-MS/MS
The tandem mass spectral analysis was performed with a Thermo Fischer Scientific LTQ-Orbitrap mass spectrometer equipped with a Waters nano-Acquity UPLC system. Peptides/phosphopeptides were loaded onto a Waters Symmetry® C18 180μm × 20mm trap column and separated through a 1.7 μm, 75 μm × 250 mm nanoAcquity™ UPLC™ column (35°C) immediately connected to the electrospray ionization source. Trapping was performed at 15μl/min, 99% Buffer A (100% water, 0.1% formic acid) for 1 min. Peptide separation was performed at 300 nl/min with Buffer A: 100% water, 0.1% formic acid and Buffer B: 100% CH3CN, 0.075% formic acid. A linear gradient (51 minutes) was run with 5% buffer B at initial conditions, 50% B at 50 minutes, and 85% B at 51 minutes. MS was acquired in the LTQ-Orbitrap using 1 microscan, and a maximum inject time of 900microseconds. A precursor mass spectral acquisition was taken in the orbitrap followed by four data dependant MS/MS acquisitions in the ion trap. MS3 scans were also obtained if neutral loss of a phosphate group was detected corresponding to m/z differences of 98.0, 49.0, and 32.7. The data was searched using the Mascot (Matrix Science) database search algorithm against the NCBInr protein database.
In vitro protein binding assay
Plasmids pGEX-GST3, pGEX-GST3S155AS157A and pGEX-GST3S155ES157E were generated using oligonucleotides 5GST3 (GAAGAATTCGTGGGATTAATTAGATTTATGCGTGCG) and 3GSTH3 (AAGAAGCTTCTGCATGCTAGAATCTAAGTCACTTTC) as primers, and chromosomal DNA, pMAD-ACTAS155AS157A and pMAD-ACTAS155ES157E as templates, respectively. The corresponding BL21 strains were grown at 37°C to an O.D. of 0.5 before addition of IPTG (1mM final concentration) and further incubation for one hour at 30°C. Bacteria were subsequently harvested in 1ml PBS supplemented with 10μl of a protease inhibitor cocktail (Sigma) and lysed by sonication. The GST fusion proteins were immobilized on 200 μl of glutathione-Sepharose beads (GE Healthcare) for 2hrs at 4°C. CK2 treated beads were incubated at 30°C for 20 minutes in CK2 reaction buffer (20 mM Tris-HCl [pH7.5], 50 mM KCl, 10 mM MgCl2) supplemented with 200 μM ATP and 50U of purified CK2 enzyme (New England Biosciences). Untreated and CK2-treated beads were washed once in XB (10mM HEPES [pH 7.6], 100 mM KCl, 1 mM MgCl, 0.1 mM EDTA, 1 mM DTT) and subsequently incubated with 5 μM purified bovine Arp2/3 complex (Cytoskeleton) in XB plus 0.1% (v/v) Tween-20, 0.2 mM ATP, and 0.5 mg/ml BSA (50 n M) at 4°C for 2hrs. Beads were subsequently washed twice in 150 μl of XB plus 0.1% (v/v) Tween-20 and 0.2 mM ATP. Beads were resuspended in 40μl of Laemelli sample buffer, boiled for 10 mins and subjected to western blot analysis. Membranes were incubated in blocking buffer (PBS plus 5% dry non-fat milk) and probed with either rabbit polyclonal Anti-Arp3 (1:500) obtained from Millipore or rabbit polyclonal Anti-GST (1:250) obtained from Santa Cruz Biotechnology. IRDye800 conjugated anti-rabbit IgG (Rockland, Gilbertsville, PA) were used as secondary antibodies. Signal intensities were quantified using the Odyssey infrared image system (LiCor).
Mouse infection
Groups of four, eight-week-old female BALB/c mice were infected intra-peritoneally with 5×104 bacteria of a given genotype. Four days post-infection, the animals were euthanized, spleens were collected, homogenized in PBS and the number of colony forming-units (CFUs) was determined on BHI agar plates.
Supplementary Material
Acknowledgments
We thank Dan Portnoy for the generous gift of the ActA antibody. We thank Jorge Galan for suggesting the mouse experiments. We thank Neal Gliksman for helping with image analysis. We thank Isabelle Derré, Ana-Maria Dragoi, Jorge Galan and Craig Roy for critical reading of the manuscript. This work was supported by the Mallinckrodt foundation (H.A.), the National Institutes of Health training Grant T32 AI-07640 (R.C.), Northeast Biodefense Center Grant U54 AI-057158 for Proteomics Core (K.L.S. and E.E.G.) and Grant R01 AI-073904 (H.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Zigmond SH. Actin polymerization: Where the WASP stings. Curr Biol. 1999;9:R160–163. doi: 10.1016/s0960-9822(99)80102-x. [DOI] [PubMed] [Google Scholar]
- Boujemaa-Paterski R, Gouin E, Hansen G, Samarin S, Le Clainche C, Didry D, Dehoux P, Cossart P, Kocks C, Carlier MF, et al. Listeria protein ActA mimics WASp family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochemistry. 2001;40:11390–11404. doi: 10.1021/bi010486b. [DOI] [PubMed] [Google Scholar]
- Brundage RA, Smith GA, Camilli A, Theriot JA, Portnoy DA. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc Natl Acad Sci U S A. 1993;90:11890–11894. doi: 10.1073/pnas.90.24.11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory GO, Cramer R, Blanchoin L, Ridley AJ. Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Mol Cell. 2003;11:1229–1239. doi: 10.1016/s1097-2765(03)00172-2. [DOI] [PubMed] [Google Scholar]
- Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wachter M, Wuenscher M, Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. Embo J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillat C, Espanol T, Oset M, Ferrando M, Estivill X, Volpini V. Identification of WASP mutations in 14 Spanish families with Wiskott-Aldrich syndrome. Am J Med Genet. 2001;100:116–121. doi: 10.1002/ajmg.1228. [DOI] [PubMed] [Google Scholar]
- Goley ED, Ohkawa T, Mancuso J, Woodruff JB, D’Alessio JA, Cande WZ, Volkman LE, Welch MD. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science. 2006;314:464–467. doi: 10.1126/science.1133348. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, Li R, Cossart P. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Jeng RL, Goley ED, D’Alessio JA, Chaga OY, Svitkina TM, Borisy GG, Heinzen RA, Welch MD. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kolluri R, Shehabeldin A, Peacocke M, Lamhonwah AM, Teichert-Kuliszewska K, Weissman SM, Siminovitch KA. Identification of WASP mutations in patients with Wiskott-Aldrich syndrome and isolated thrombocytopenia reveals allelic heterogeneity at the WAS locus. Hum Mol Genet. 1995;4:1119–1126. doi: 10.1093/hmg/4.7.1119. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Gevaert K, Cossart P, Vandekerckhove J, Van Troys M. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 2008;18:220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Lasa I, David V, Gouin E, Marchand JB, Cossart P. The amino-terminal part of ActA is critical for the actin-based motility of Listeria monocytogenes; the central proline-rich region acts as a stimulator. Mol Microbiol. 1995;18:425–436. doi: 10.1111/j.1365-2958.1995.mmi_18030425.x. [DOI] [PubMed] [Google Scholar]
- Lasa I, Gouin E, Goethals M, Vancompernolle K, David V, Vandekerckhove J, Cossart P. Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. Embo J. 1997;16:1531–1540. doi: 10.1093/emboj/16.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P, Theriot JA, Skoble J, Welch MD, Portnoy DA. Systematic mutational analysis of the amino-terminal domain of the Listeria monocytogenes ActA protein reveals novel functions in actin-based motility. Mol Microbiol. 2001;42:1163–1177. doi: 10.1046/j.1365-2958.2001.02677.x. [DOI] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Insall RH, Volkman LE. WASP homology sequences in baculoviruses. Trends Cell Biol. 2001;11:286–287. doi: 10.1016/s0962-8924(01)02009-8. [DOI] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. Embo J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- Pinna LA. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- Pistor S, Grobe L, Sechi AS, Domann E, Gerstel B, Machesky LM, Chakraborty T, Wehland J. Mutations of arginine residues within the 146-KKRRK-150 motif of the ActA protein of Listeria monocytogenes abolish intracellular motility by interfering with the recruitment of the Arp2/3 complex. J Cell Sci. 2000;113(Pt 18):3277–3287. doi: 10.1242/jcs.113.18.3277. [DOI] [PubMed] [Google Scholar]
- Pocha SM, Cory GO. WAVE2 is regulated by multiple phosphorylation events within its VCA domain. Cell Motil Cytoskeleton. 2009;66:36–47. doi: 10.1002/cm.20323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Schlech WF., 3rd Listeria gastroenteritis--old syndrome, new pathogen. N Engl J Med. 1997;336:130–132. doi: 10.1056/NEJM199701093360211. [DOI] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.