Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) plays a pivotal role in multiple neurologic diseases by mediating caspase-independent cell death, which has recently been designated parthanatos to distinguish it from other forms of cell death such as apoptosis, necrosis and autophagy. Mitochondrial apoptosis-inducing factor (AIF) release and translocation to the nucleus is the commitment point for parthanatos. This process involves a pathogenic role of poly (ADP-ribose) (PAR) polymer. It generates in the nucleus and translocates to the mitochondria to mediate AIF release following lethal PARP-1 activation. PAR polymer itself is toxic to cells. Thus, PAR polymer signaling to mitochondrial AIF is the key event initiating the deadly crosstalk between the nucleus and the mitochondria in parthanatos. Targeting PAR-mediated AIF release could be a potential approach for the therapy of neurologic disorders.
Keywords: AIF, poly(ADP-ribose), PAR, PARP-1, calpain, parthanatos, cell death
1. Introduction
Apoptosis-inducing factor (AIF) is a mitochondrial flavoprotein contributing to both cell life and death (Boujrad, et al., 2007, Krantic, et al., 2007, Modjtahedi, et al., 2006). Under physiological conditions, AIF maintains mitochondrial structure (Cheung, et al., 2006) and plays an essential role in oxidative phosphorylation (Joza, et al., 2005, Vahsen, et al., 2004). Conversely, under pathological conditions, AIF is a key mediator of caspase-independent cell death. Although the mechanism of how AIF contributes to cell death is obscure, one pivotal event is that mitochondrial AIF translocates to nucleus, where it induces chromatin condensation and large-scale DNA fragmentation (≈ 50 kb) leading to cell death (Susin, et al., 1999).
AIF is a key factor that mediates poly (ADP-ribose) polymerase-1 (PARP-1)-dependent cell death (Yu, et al., 2002). PARP-1-mediated cell death differs from other forms of cell death, such as apoptosis, necrosis and autophagy (Table 1). It causes phosphatidylserine flipping onto the outer plasma membrane, dissipation of mitochondrial membrane potential, chromatin condensation and large DNA fragmentation (Delettre, et al., 2006, Delettre, et al., 2006, Susin, et al., 1999, Wang, et al., 2004, Yu, et al., 2002). However, unlike apoptosis, it does not cause apoptotic body formation or small scale DNA fragmentation. Moreover, PARP-1-induced cell death cannot be rescued by pan-caspase inhibitors, such as z-VAD-fmk and boc-aspartyl-fmk (BAF) (Yu, et al., 2002). Although PARP-1-mediated cell death shows loss of membrane integrity similar to necrosis, it does not induce cell swelling (Wang, et al., 2004, Yu, et al., 2002). It also clearly differs from autophagy, which involves autophagic vacuoles formation and lysosomal degradation (Edinger and Thompson, 2004, Kroemer, et al., 2005). These observations suggest that PARP-1-mediated cell death is unique compared with apoptosis, necrosis and autophagy. To distinguish from other forms of cell death, PARP-1-mediated cell death is named as parthanatos, after poly(ADP-ribose) (PAR) polymer, which is a product of PARP-1 activation and thanatos, which is the Greek personification of death and mortality (Harraz, et al., 2008).
Table 1.
Differential features of apoptosis, necrosis, autophagy and parthanatos
| Apoptosis | Necrosis | Autophagy | Parthanatos | |
|---|---|---|---|---|
| Membrane | Apoptotic bodies; Blebbing; PS externalization | Disrupted, Loss of integrity; Blebbing, | Formation of double- membrane bound autophagosomes | Loss of integrity; PS externalization |
| Cytoplasm | Morphologically intact; Condensation | Vacuolation of the cytoplasm Disrupted; Cell swelling | Autophagic vacuoles, Lysosomal degradation | Condensation |
| Mitochondria | Depolarization; cyt c release to cytosol; | Loss of mitochondria ultrastructure | Degradation | Depolarization; AIF release to nucleus; |
| Nuclei | chromatin condensation; shrink | Morphologic changes; chromatin digestion | − | Chromatin condensation; shrink |
| DNA | DNA fragmentation (DNA ladder) | DNA hydrolysis (smear) | − | Large DNA fragmentation (≈50 kb) |
| ATP | Energy-dependent | No energy requirement | Energy-dependent | Energy-independent |
| Key mediators | Subset of caspases | − | Atg6/Beclin-1 | PARP activation; PAR polymer formation; Caspase-independent |
| Propidium iodide | − | + | + | + |
| Annexin V | + | − | − | + |
| Inflammatory | − | + | − | − |
| Neurologic Disorders | Excitotoxicity, ischemia, stroke, trauma, AD, PD, ALS, HD, ataxias | Excitotoxicity, stroke, ischemia, AD, PD, ALS, HD, epilepsy | AD, PD, ALS, HD, prion disease | Excitotoxicity, stroke, ischemia, PD |
AD, Alzheimer’s disease; ALS, Amyotrophic lateral sclerosis; Cyt c, cytochrome c; HD, Huntington’s disease; PD, Parkinson’s disease; PS, phosphatidylserine.
The roles of AIF and parthanatos have been widely implicated in some neurologic diseases. Recently, new progress has been made to elucidate the mechanism of mitochondrial AIF release in different forms of cell death. Our group found that nonprotein PAR polymer functions as a cell death signal and plays a pivotal role in mitochondrial AIF release in parthanatos (Andrabi, et al., 2006, Yu, et al., 2006). Here, we review biological properties and functions of AIF in neurologic diseases and the mechanism of mitochondrial AIF release.
2. AIF properties and mitochondrial location
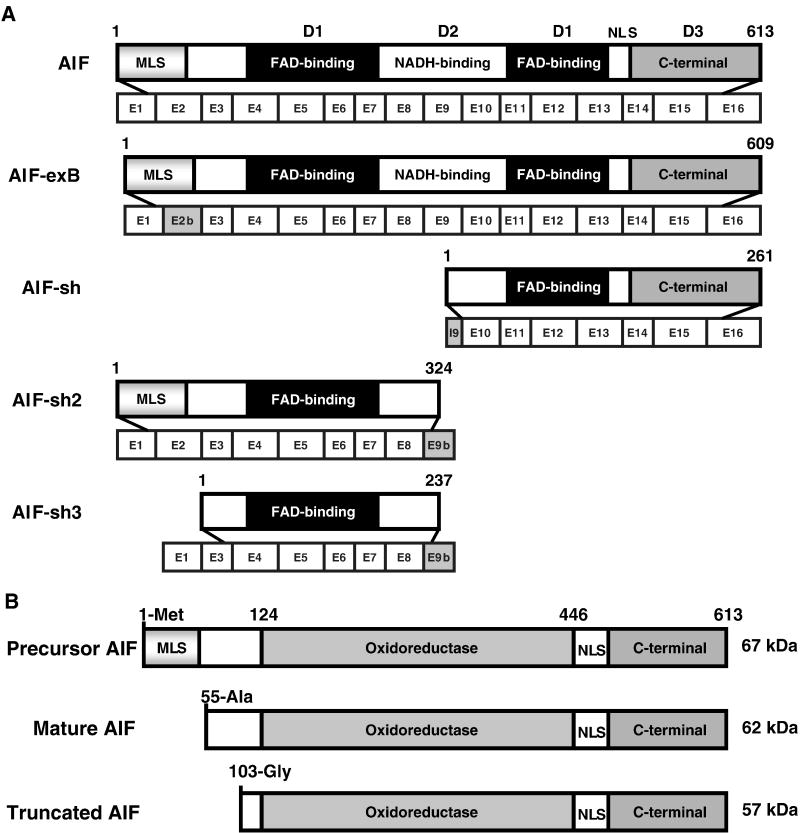
AIF, which was initially identified from mouse liver mitochondria following permeability transition pore opening, comprises 16 exons and is located on chromosome X (Susin, et al., 1999). It is synthesized as a 67 kDa-precursor in the cytoplasm and imported into mitochondria. It contains a predicted mitochondrial localization sequence (MLS) in its N-terminus (Fig. 1). During mitochondria import, AIF is processed to the mature 62 kDa form by cleavage at Met53/Ala54 (Figure 1B) (Otera, et al., 2005). Although previously, mature AIF was thought to exist as a 57 kDa soluble species (Susin, et al., 1999), now evidence from different laboratories clearly indicates that the mature AIF is 62 kDa (Cao, et al., 2007, Otera, et al., 2005).
Figure 1.
A. Structure of five different human AIF isoforms. Full-length AIF is encoded by a 16-exon X chromosome gene giving rise to a protein precursor composed of 613 amino acids (Susin, et al., 1999). AIF-exB is an isoform with 609 amino acids generated by the alternative usage of exon 2b instead of exon 2 (Loeffler, et al., 2001). AIF and AIF-exB both contain three functional domains: FAD-binding domains D1 (black), NADH-binding domain D2 (white) and C-terminal domain D3 (gray), and a mitochondrial localization sequence (MLS) at the N-terminus and a nuclear localization sequence (NLS) at the C-terminus. Short AIF (AIFsh) contains C-terminal AIF encoded by exons 10–16, lacking of MLS. AIFsh2 and AIFsh3 are generated by the alternative splicing of the exon 9b (Delettre, et al., 2006). AIFsh2 contains the MLS and the oxidoreductase domain, but lacks the C-terminus of AIF. AIFsh3 has a similar structure as AIFsh2 with the splicing of exon 2, which leads to the loss of MLS. B. Graphic representation of precursor AIF, mature AIF and truncated AIF.
AIF is thought to be primarily located in the intermembrane space of mitochondria (Susin, et al., 1999). Otera et al. showed that the mature AIF is produced by cleaving off the N-terminal 52 amino acids, but not the N-terminal 101 amino acids. Therefore it contains a hydrophobic transmembrane segment (amino acid residues 66–84). Presumably mature AIF is a type-I inner membrane protein with the N-terminus exposed to the matrix and the C-terminal portion to the intermembrane space (Otera, et al., 2005). If this is the case, AIF has to be processed into a soluble form by removing the hydrophobic transmembrane segment after cell death stimulation. Thus, compared to cytochrome c (cyt c), AIF would take a longer time to translocate to nucleus. However, AIF release occurs earlier than that of cyt c in certain cell death paradigms, such as parthanatos (Wang, et al., 2004, Yu, et al., 2002). Considering that AIF is a much larger protein than cyt c, the relative kinetics of AIF translocation to the nucleus versus cyt c release from mitochondria is puzzling. One possible explanation could be that a second pool of AIF exists in mitochondria. For instance, perhaps a pool of AIF is localized at the outer mitochondrial membrane. Upon PARP-1 activation, AIF quickly responds to the cell death signal and translocates from the mitochondria to the nucleus as an uncleaved form. It is reasonable to speculate that different mechanisms might be involved in mitochondrial AIF release due to its localization. Therefore, it is important to further study the localization of AIF at the submitochondrial level.
The crystal structure of AIF reveals that AIF is comprised of three putative functional domains: 1) a N-terminal FAD binding domain, 2) a central NADH binding domain and 3) a C-terminal domain (Figure 1A) (Mate, et al., 2002, Susin, et al., 1999). The first two domains compose the oxidoreductase part of AIF, which confers electron transfer activity to the protein (Delettre, et al., 2006, Miramar, et al., 2001). The C-terminal domain mainly contributes to the caspase-independent cell death properties of AIF (Delettre, et al., 2006). AIF is ubiquitously expressed in different tissues contributing to both cell life and death. So far, four isoforms of human AIF have been identified: AIF-exB, AIFsh, AIFsh2 and AIFsh3 (Figure 1A) (Delettre, et al., 2006, Delettre, et al., 2006, Loeffler, et al., 2001). AIF-exB contains an alternative exon 2b instead of the original exon 2, which does not affect AIF-exB mitochondrial import and function (Loeffler, et al., 2001). AIFsh (AIF short) is a cytosolic protein comprising seven exons derived from exons 10 to 16 (Delettre, et al., 2006). Although it lacks the N-terminal oxidoreductase domain, AIFsh still provokes chromatin condensation and large-scale DNA fragmentation, supporting that the oxidoreductase part of AIF is not necessary for the induction of cell death (Delettre, et al., 2006). AIFsh2 is a newly identified mitochondrial spliced isoform with NADH oxidase activity (Delettre, et al., 2006). It contains the AIF MLS and the oxidoreductase domain, but lacks the C-terminus. Therefore, AIFsh2 cannot translocate to nucleus to trigger chromatin condensation and DNA fragmentation. Under physiological conditions, AIFsh2 mRNA is absent in normal brain tissue, but it is expressed in neuroblastoma-derived cells. AIFsh3 has a similar structure as AIFsh2 with the splicing of exon 2, which leads to the loss of MLS. The expression of AIF-exB, AIFsh, AIFsh2 and AIFsh3 is regulated independently from the expression of AIF. However, it still remains unknown what events lead to differential splicing of AIF and the role of these different AIF isoforms in cell death and survival needs further investigation.
3. AIF functions and AIF release mechanisms
3.1 AIF and cell death/survival
AIF is a bifunctional flavoprotein with a vital function in bioenergetics within mitochondria and a lethal function in cell death when it moves to the nucleus. During cortical development, AIF is required for neuronal cell survival (Cheung, et al., 2006). It is involved in normal mitochondrial respiration in neurons possibly by stabilizing mitochondrial complex I (Joza, et al., 2005) or maintaining mitochondrial structure (Cheung, et al., 2006). Loss of AIF in muscle leads to mitochondrial dysfunction, skeletal muscle atrophy and dilated cardiomyopathy (Joza, et al., 2005), but it protects cells from the consequences of obesity and diabetes (Pospisilik, et al., 2007). Forebrain-specific AIF null mice have defective cortical development and die by E17 (Cheung, et al., 2006). Harlequin mice, which have an 80% reduction in AIF expression, develop oxidative stress-mediated neurodegeneration (Klein, et al., 2002). Similarly, AIF null embryos die around embryonic day E12 (Cheung, et al., 2006). All these data suggest that AIF plays an important role in cell survival.
Apart from its cytoprotective role, AIF is also a key cell death effector that mediates PARP-1-dependent cytotoxicity in many different cell types (Ruchalski, et al., 2003, Yu, et al., 2002). Upon N-methyl-N′-nitro-N-nitrosoguanidine (MNNG)-induced DNA damage and PARP-1 activation in mouse embryonic fibroblasts, AIF translocates from the mitochondria to nucleus, resulting in large-scale DNA fragmentation and cell death (Yu, et al., 2002). AIF-deficient cortical neurons prepared from harlequin mice are more resistant to N-methyl-D-aspartic acid (NMDA) toxicity (Yu, et al., 2002). Moreover, PARP-1 chemical inhibitors, PARP-1 genetic ablation, AIF knockdown, or neutralizing anti-AIF antibodies prevents AIF translocation to the nucleus and inhibits alkylating DNA damage-mediated cell death in a variety of experimental paradigms (Lorenzo and Susin, 2007, Moubarak, et al., 2007, Xu, et al., 2006, Yu, et al., 2002), indicating a pivotal role of mitochondrial AIF release and nuclear translocation in parthanatos. This process is not blocked by the broad-spectrum caspase inhibitors, boc-aspartyl-fmk or Z-VAD-fmk in mouse embryonic fibroblasts (Yu, et al., 2002). In the presence of the pan-caspase inhibitor z-VAD-fmk, the release of AIF from mitochondria and translocation to the nucleus is also observed in HeLa cells and some other cell types (Gallego, et al., 2004, Susin, et al., 1999, Yu, et al., 2002). Therefore, AIF is the commitment point for parthanatos, which is independent of caspase activation.
However, in other forms of cell death where caspases are activated, AIF release from mitochondria has been observed (Arnoult, et al., 2002, Daugas, et al., 2000). Cell death stimuli, like tumor necrosis factor-α and CD95 triggers the sequential activation of caspase 9, apoptotic protease activating factor-1 (Apaf-1) and caspase 3. Caspases then activate endonucleases leading to DNA fragmentation. In this process, caspases are the key cell death effectors. The role of AIF is not clear even though it translocates. In camptothecin induced neuronal cell death when caspases are blocked, AIF mediates delayed cell death that is caspase-independent (Cregan, et al., 2002). Perhaps, AIF release from mitochondria helps consolidate the death process by providing an alternative mechanism of cell death that does not require caspase activation. Under situations where AIF translocates due to caspase activation, it may just represent epiphenomenon resulting from caspase activation and subsequent mitochondrial membrane depolarization. Thereby, AIF might not actually contribute to cell death under conditions of caspase activation.
3.2 Mitochondrial AIF release mechanisms
AIF release from the mitochondria and translocation to the nucleus is a central event in parthanatos. Interfering with AIF translocation to the nucleus could be a potential therapeutic target to prevent cell death. Several studies have shown that mechanisms for the release of AIF from mitochondria under different cell death stimuli or experimental conditions involve calpains or capthepsins. Recently, PAR polymer has been identified to serve as a cell death signal provoking AIF release from mitochondria (Andrabi, et al., 2006, Yu, et al., 2006).
3.2.1 PAR-mediated AIF release
3.2.1.1 PARP-1 activation, PAR generation and cell death
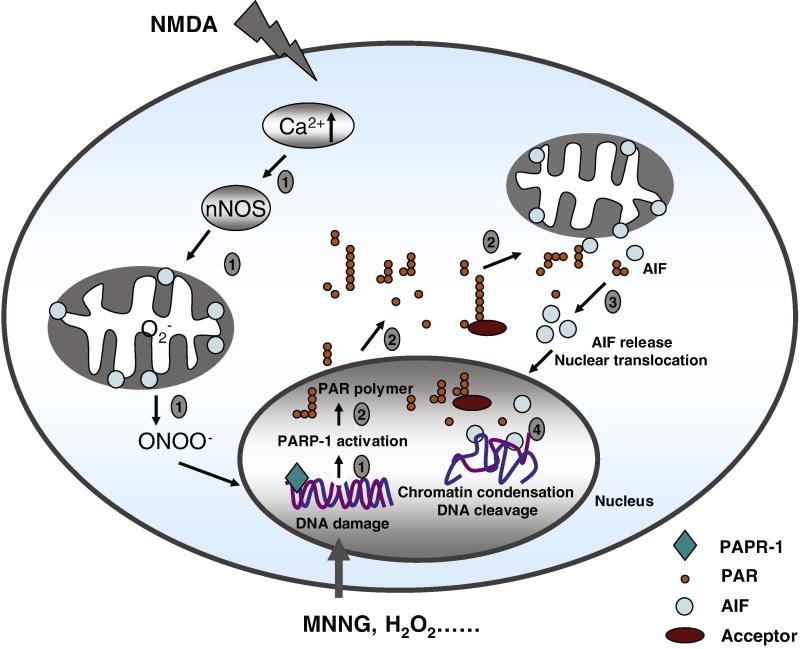
PARP-1 is the most extensively studied nuclear enzyme of the PARP superfamily which includes 17 putative PARP proteins based on protein domain homology and enzymatic functions (Dawson and Dawson, 2004, Schreiber, et al., 2006). PARP-1 consists of three major functional domains: 1) a 42-kDa N-terminal DNA-binding domain, containing two zinc-finger motifs and a nuclear localization sequence (NLS), which recognizes both double- and single-stranded DNA breaks; 2) a 16-kDa central automodification domain which is thought to be the target of self-poly-(ADP-ribosyl)ation and 3) a 55-kDa C-terminal catalytic domain containing both the NAD binding site and the catalytic domain which synthesizes PAR (Kameshita, et al., 1984). PARP-1 is potently activated by DNA strand nicks and breaks and facilitates DNA repair (Lautier, et al., 1993, Smulson, et al., 2000). Ischemic injury can also induce PARP-1 activation by massive release of the excitatory neurotransmitter glutamate. Activation of three subtypes of glutamatergic ionotropic receptors: N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainate receptors, leads to calcium influx, nitric oxide (NO) and reactive oxygen species (ROS) generation. Superoxide can combine with NO forming peroxynitrite (ONOO-). Excessive production of peroyxnitrite and other free radicals induces chromosomal DNA nicks and breaks, resulting in PARP-1 activation (Figure 3) (Dawson and Dawson, 2004). In addition, a variety of environmental stimuli, free radicals, hydrogen peroxide, hydroxyl radical, peroxynitrite, ionizing radiation and DNA-alkylating agent MNNG also triggers DNA strand nicks and breaks and subsequent PARP-1 activation.
Figure 3. Schematic model of PARP-1-, PAR polymer- and AIF-mediated death signal in parthanatos.
PARP-1 activation, PAR polymer formation, mitochondrial AIF release, AIF-mediated chromatin condensation and DNA fragmentation are four key steps in parthanatos. Step 1, DNA damage by NMDA or MNNG administration activates PARP-1. Excitotoxic activation of NMDA receptor can induce calcium influx, nNOS activation, NO production and reactive oxygen species generation. The reaction between NO and superoxide anion generates the potent oxidant preoxynitrite, which leads to DNA damage. Other agents like MNNG, free radicals, hydrogen peroxide, hydroxyl radical or ionizing radiation cause DNA damage, which activate PARP-1. Step 2, PARP-1 activation catalyzes PAR polymer formation, which translocates from the nucleus to the mitochondria. Step 3, PAR polymer mediates AIF release from mitochondria and translocation to nucleus. Step 4, AIF causes chromatin condensation and large DNA fragmentation through an unknown mechanism.
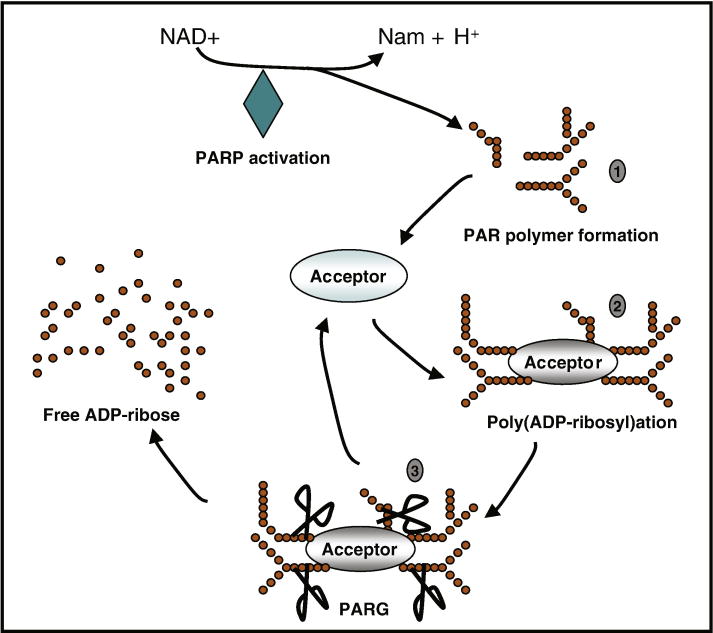
Upon activation, PARP-1 transforms nicotinamide adenine dinucleotide (NAD+) into long PAR polymers (Figure 2) and transfers them to a variety of nuclear proteins, including histones, DNA polymerases, topoisomerases, DNA ligase-2, transcription factors and PARP-1 itself (Lautier, et al., 1993, Smulson, et al., 2000). NAD+ is a cofactor for glycolysis and the tricarboxylic acid cycle, thus providing ATP for most cellular processes (Hageman and Stierum, 2001). Overactivation of PARP-1 results in NAD+ utilization and depletion of cellular NAD+ and ATP levels. It has been suggested that this might cause cell death, but subsequent studies indicate that energy depletion alone might not be sufficient to mediate PARP-1-dependent cell death. Data in an experimental stroke model showed that preservation of energy stores in PARP-1 knockout mice is not the mechanism underlying the reduction in infarct volume (Goto, et al., 2002). Moreover, studies in primary cell cultures indicate that energy depletion may not be a primary factor in PARP-1 mediated cell death (Fossati, et al., 2007, Moubarak, et al., 2007).
Figure 2. Regulation of PAR polymer generation and degradation.
Severe DNA damage causes excessive activation of PARPs, especially PARP-1. PARPs use nicotinamide adenine dinucleotide (NAD+) as substrate to generate PAR polymer and nicotinamide (Nam) (step 1). Numerous nuclear proteins are acceptors of PAR polymer. PARPs catalyze the addition of PAR onto itself and other acceptor proteins, resulting in poly(ADP-ribosyl)ation. This modification might affect the functions of acceptor proteins (step 2). PARG can quickly catalyze the hydrolysis of PAR polymer into free ADP-ribose (step 3).
In the PARP superfamily, PARP-1 is responsible for > 90% PAR polymer generation (D’Amours, et al., 1999, Dawson and Dawson, 2004). The basal levels of PAR are very low. However, excessive activation of PARP-1 leads to 10–500-fold increase in PAR polymer formation (D’Amours, et al., 1999). PAR polymers could reach lengths of hundreds of ADP-ribose units depending on the extent and type of DNA damage (Hassa, et al., 2006). Poly (ADP-ribosyl)ation is a post-translational modification that is involved in a wide range of physiological and pathophysiological processes (Hassa, et al., 2006). PARP-1-mediated poly (ADP-ribosyl)ation modifies the charge distribution of acceptor proteins and increases the hindrance of proteins by the addition of a bulky and complex negatively charged structure (Hong, et al., 2004). This modification can result in inhibition of the physiological functions of acceptor proteins. The exact function of this modification may also vary among different acceptor proteins. Severe poly (ADP-ribosyl)ation of key cellular proteins might lead to cell death. Recently, it was shown that PARP-1-mediated p53-poly(ADP-ribosyl)ation blocked the interaction between p53 and the nuclear export receptor Crm1, resulting in nuclear accumulation of p53 (Kanai, et al., 2007). Although complex functions of poly (ADP-ribosyl)ation are involved in intracellular signaling, transcriptional regulation, DNA repair pathways and maintenance of genomic stability, telomere dynamics, cell differentiation and proliferation and cell death (Chiarugi, 2002, Hassa, et al., 2006), the physiological function of the poly (ADP-ribosyl)ation of individual acceptor proteins is still under investigation.
PARP-1 has emerged as a key death mediator in a number of experimental models including diabetes, inflammation, stroke, trauma, cerebral hypoxia/ischemia, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine (MPTP) toxicity, and also in various diseases (Cosi and Marien, 1999, Culmsee, et al., 2005, Eliasson, et al., 1997, Endres, et al., 1997, Mandir, et al., 2000, Mandir, et al., 1999, Plesnila, et al., 2004, Wang, et al., 2004, Zhang, et al., 1994). It also plays a prominent role in NMDA excitotoxicity (Mandir, et al., 2000, Zhang, et al., 1994). The PARP-1 specific inhibitor, 3,4-dihydro-5-(4-(1-piperidinyl)butoxyl)-1(2H)-isoquinolinone (DPQ), rescues NMDA-induced neuronal death (Yu, et al., 2002). Moreover, PARP-1 KO mice are remarkably resistant to the excitotoxic effects of glutamate and NMDA both in vitro and in vivo (Pieper, et al., 1999, Yu, et al., 2002). PARP-1-induced cell death involves PAR polymer formation and requires AIF nuclear translocation (Andrabi, et al., 2006, Yu, et al., 2006, Yu, et al., 2002).
3.2.1.2 PAR-induced AIF translocation from mitochondria to nucleus
In response to NMDA treatment in cortical neuronal cultures, PARP-1 activation generates PAR polymer in a time-dependent manner (Andrabi, et al., 2006, Yu, et al., 2006, Yu, et al., 2002). 15 min after NMDA treatment, PAR polymer formation is mainly present in the nuclear fraction and was also noted in the cytosolic fraction. 30 min after NMDA application, PAR polymer is also observed in mitochondria fraction (Yu, et al., 2006), where it was colocalized with an integral mitochondrial protein cytochrome oxidase 1 (Yu, et al., 2006). 1 h after NMDA administration, the levels of PAR reached ~80 nM causing ~60% neuronal death (Andrabi, et al., 2006). These observations suggest that PAR polymer, which is mainly produced in the nucleus, can translocate to cytosol and mitochondria, where it may play a role in mitochondrial AIF release (Figure 3).
PAR polymer participates directly in cell death signaling (Andrabi, et al., 2006). Transporting in vitro synthesized and purified PAR polymer into living cells using a lipid-based delivery system (Andrabi, et al., 2006), PAR polymer is directly toxic to cells. It caused cell death in a PAR-size-dependent and dose-dependent manner. This event was not prevented by the pan-caspase inhibitor zVAD-fmk, but abrogated by pretreatment of the PAR polymer with the PAR degrading enzymes poly (ADP-ribose) glycohydrolase (PARG) or phosphodiesterase 1.
PAR-mediated cell death is AIF-dependent. Cortical neurons from Harlequin mice are resistant to both PAR and NMDA toxicity (Yu, et al., 2006). Upon exposure to PAR-containing nuclear supernatant, AIF is released from isolated mitochondria. Pretreatment of nuclear supernatant with proteinase K failed to prevent mitochondrial AIF release, indicating that a nonproteinaceous factor might be crucial for mitochondria AIF release, consistent with the involvement of PAR. Further evidence showed that PAR polymer injected into living cells induces AIF release from mitochondria and translocation to nucleus (Yu, et al., 2006). Neutralizing antibodies to PAR or catabolism of PAR by over-expression of PARG prevented AIF translocation to the nucleus and cell death (Andrabi, et al., 2006, Yu, et al., 2006). These results reveal that PAR polymer is an AIF-releasing factor, which plays important roles in PARP-1-dependent cell death. Recently, Poirier and coworkers showed that AIF is a PAR polymer-binding protein determined by LC-MS/MS analysis (Gagne, et al., 2008). Presumably, in parthanatos, free PAR polymer or poly (ADP-ribosy)lated acceptor protein, produced by the excessive activation of PARP-1, translocates to cytosol and mitochondria, where it interacts with mitochondrial AIF and triggers its release and nuclear translocation (Figure 3). Some other factors might also be involved in this process. Although it is clear that PAR polymer plays an important role in mitochondrial AIF release, the mechanism by which PAR polymer causes AIF release is not known. It is important to understand the detailed molecular mechanism of PARP-1-dependent neuronal death, which might lead to identification of new target molecules to treat patients with neurologic diseases.
3.2.1.3 PARG: regulation of PAR levels and cell death
Upon PARP-1 activation induced by DNA damage, the levels of poly (ADP-ribosyl)ation rapidly increase. This modification is dynamically regulated by PARG (Kameshita, et al., 1984, Whitacre, et al., 1995). PARG is the catabolic enzyme responsible for the catabolism of PAR polymer. It rapidly hydrolyzes the ribose-ribose bonds of both linear and branched portions of PAR bound to acceptor proteins (Davidovic, et al., 2001). PARG may play a prominent role in the DNA damage response and repair by removing PAR from modified proteins.
PARG cDNA, which was initially isolated from bovine tissue, encodes a protein with a predicted molecular weight of 110 kDa (Lin, et al., 1997). Recently, the PARG gene and protein functions were further characterized in other species (Ame, et al., 1999, Meyer-Ficca, et al., 2004, Winstall, et al., 1999). Human PARG consists of 18 exons and 17 introns. It has a regulatory N-terminal domain and a catalytic center domain (Bonicalzi, et al., 2005). The full-length 110 kDa (PARG110) is mainly localized to the nucleus. Several alternative spliced PARG isoforms were also identified (Bonicalzi, et al., 2005, Haince, et al., 2006). PARG103 and PARG99 are cytoplasmic spliced PARG isoforms that lack of exon 1 or exon 1 and 2, respectively; PARG60 is another spliced isoform that appears to be mainly localized to the mitochondria and cytosol (Meyer, et al., 2007). Caspase-3 cleavage of PARG releases two enzymatically active C-terminal fragments PARG85 and PARG74, which are primarily localized to the cytosol (Bonicalzi, et al., 2003). All these isoforms are able to catalyze PAR turnover in different cellular compartments. However, the significance of the different subcellular location of these isoforms still remains largely unknown.
PARG is a key enzyme required for the hydrolysis of PAR in mammals. However, the physiological functions of PARG and the consequences of PAR hydrolysis are a mystery. To date, some studies suggest a role for PARG in cell death. Knocking down PARG110 and PARG60 by small interfering RNA (siRNA) protected against H2O2-induced cell death (Blenn, et al., 2006). PARG110-deficient mice created by deletion of exon 2 and exon 3 are viable and fertile (Cortes, et al., 2004) and are resistant renal ischemia/reperfusion injury (Patel, et al., 2005). Interestingly, these mice still have PARG activity in all organs and cells tested. Although deletion of PARG110 decreased PARG activity in cytosolic and nuclear fractions, it dramatically increased PARG activity in mitochondrial fractions by 331% (Cortes, et al., 2004). In line with these observations, PARP-1 automodification was reduced and no apparent accumulation of PAR was observed in PARG110-deficient embryonic fibroblasts following H2O2 treatment (Cortes, et al., 2004). Thus, these mice cannot be used to explore the role of PARG deficiency as they inappropriately express PARG in the mitochondria. True, PARG null mice are embryonic lethal (Koh, et al., 2004). These mice are devoid of any PARG activity. The lethality occurred at approximately day E3.5, likely due to the inability to hydrolyze PAR polymer (Koh, et al., 2004). PARG null embryonic trophoblast stem cells derived from periimplantation PARG null embryos grew only in the presence of the PARP inhibitor benzamide and they were hypersensitive to DNA damaging agents. This is in agreement with the notion that the regulation of PAR levels is critical for cell survival and the failure to degrade PAR results in deleterious consequences to the cell. In addition, other studies show that PARG over expression, which degrades PAR polymer, prevents PARP-1-dependent mitochondrial AIF release and cell death (Yu, et al., 2006). Thus, experimental evidence points to PARG playing a protective role in the cellular response to cell stress. The biologic importance of PARG in poly (ADP-ribosy)lation suggests that PARG may be a potential target to prevent neuronal cell death by regulating the level of PAR.
3.2.2 Calpain mediates mitochondrial AIF release
Recently, some studies indicate that the release of AIF from mitochondria requires cleavage by calpains (Cao, et al., 2007, Moubarak, et al., 2007, Polster, et al., 2005). Calpain is a calcium-dependent intracellular cysteine protease. Its activity is tightly controlled by calcium. Binding of Ca2+ and phospholipids to calpain might induce conformational changes, forming a functional catalytic site (Suzuki, et al., 2004). So far, calpain I (μ-calpain) and calpain II (m-calpain) are the two most important and most studied isoforms in the brain (Bevers and Neumar, 2008). Micromolar or millimolar Ca2+ concentration is sufficient to activate μ- and m-calpain, respectively (Goll, et al., 2003). Both calpains share a common small regulatory subunit protein of 28 kDa encoded by the capn4 gene. This regulatory subunit protein is critical for calpain function, as genetic deletion of the capn4 gene completely blocks calpain catalytic activity (Tan, et al., 2006). Calpain is thought to be primarily localized in the cytoplasm, associating with the membranes of the endoplasmic reticulum and Golgi apparatus (Hood, et al., 2003). Recently, Garcia et al. showed that calpain I is present in isolated mitochondria (Garcia, et al., 2005). Cao et al. further showed that clapain I localizes in the mitochondrial intermembrane space (Cao, et al., 2007).
Calpain plays an important role in postischemic neuronal cell death via proteolysis of a wide variety of substrates (Suzuki, et al., 2004). To date, no specific amino acid sequence has been found to be uniquely recognized by calpains. For protein substrates, tertiary structure rather than primary amino acid sequences are likely to direct cleavage. For small-peptide substrates, the amino acid specificity involves small hydrophobic amino acids (e.g. leucine, valine or isoleucine) at the P2 position, and large hydrophobic amino acids (e.g., phenylalanine or tryptophan) at the P1 position (Cuerrier, et al., 2005).
Polster et al. showed that calpain I was able to cleave AIF in vitro and induced AIF release from isolated mitochondria (Polster, et al., 2005). However, isolated mitochondria might be different from the mitochondria in the intact cell system. It still remains unknown whether calpain I efficiently cleaves AIF in intact cells. Recently, Cao et al. showed that calpain I activity was required for AIF translocation following ischemic neuronal injury (Cao, et al., 2007). AIF was cleaved from the 62 kDa to the 57 kDa form following ischemic injury (Figure 1B) and translocated from the mitochondria to the nucleus in a calpain-dependent manner. Over expression of endogenous calpain inhibitor, calpastatin, inhibited AIF translocation in CA1 neurons after global ischemia, indicating a role for calpain in the release of AIF and subsequent neuronal injury after stroke (Cao, et al., 2007). Susin and coworkers showed that sequential activation of PARP-1, calpains and Bax may play a role in MNNG-induced necrotic programmed cell death (Moubarak, et al., 2007). In that study, MEFs were cultured in medium without glucose for 48 h before MNNG treatment. Both MNNG-induced necrotic programmed cell death and ischemic forms of neuronal injury are thought to utilize the glycolytic pathway, thus they may utilize similar pathways to release AIF. Therefore, under specific experimental conditions such as glycolysis, calpain may play a role in mitochondrial AIF cleavage and release.
Unlike necrotic programmed cell death, parthanatos does not require the absence of glucose. Different signaling events appear to be involved in necrotic programmed cell death and parthanatos. Parthanatos involves PAR as a cell death signal (Andrabi, et al., 2006) and AIF release requires PAR (Yu, et al., 2006). However, it is unknown whether calpain-mediated release of AIF is a parallel pathway independent of PAR, or whether calpain plays a role in PAR-mediated AIF release during parthanatos. Further work in experimental neuronal systems is required to identify the role of calpain in mitochondrial AIF release in parthanatos.
3.3 AIF causes DNA fragmentation and chromatin condensation
Upon PARP-1 activation, AIF is released from mitochondria and translocates to the nucleus. This event seems to be controlled by heat-shock protein 70 (HSP70), as it blocks the AIF-induced chromatin condensation of purified nuclei in a cell-free system (Ravagnan, et al., 2001). The mechanism underlying this HSP70 antagonism of AIF may involve physical interactions of HSP70 with AIF at amino acids 150–228 (Gurbuxani, et al., 2003, Ravagnan, et al., 2001). The ATPase domain of HSP70 is critical for sequestering AIF in the cytosol (Ruchalski, et al., 2006), thereby inhibiting AIF nuclear translocation (Gurbuxani, et al., 2003). A deletion mutant (Δ150–268) of AIF that does not bind HSP70, facilitated the nuclear import of AIF and increased AIF-mediated chromatin condensation (Gurbuxani, et al., 2003). Current evidence indicates that HSP70 can inhibit cell death by neutralizing and interacting with AIF.
Recombinant AIF causes purified nuclei to undergo peripheral condensation of chromatin and loss of DNA (Susin, et al., 1999). However, the mechanism by which AIF induces chromatin condensation and DNA fragmentation still remains a mystery. Ye et al. showed that recombinant human AIF interacted with DNA. But the strong positive electrostatic potential at the AIF surface did not seem to be important for AIF-DNA interaction (Ye, et al., 2002). DNA-binding defective mutants of AIF failed to induce cell death (Ye, et al., 2002), suggesting that the interaction between AIF and DNA is required for AIF to induce chromatin condensation and DNA fragmentation. However, mouse AIF, unlike its human counterpart, does not have obvious DNA-binding structural motifs. This observation suggests that mechanisms other than AIF-DNA interaction might be responsible for AIF-induced DNA and chromatin reconfiguration.
AIF itself does not display any intrinsic endonuclease properties (Mate, et al., 2002, Ye, et al., 2002). Although it cannot be excluded that AIF has hitherto uncharacterized cryptic nuclease activity, AIF is likely to recruit other protein factors with a nuclease properties that are involved in DNA fragmentation. The interaction of AIF with DNA may increase the susceptibility of DNA to latent endogenous nucleases. The identity of the AIF-associated endonuclease is not known. In Caenorhabditis elegans, the AIF homolog wah-1 associates and cooperates with CPS-6, an endonuclease G (EndoG) homolog, to promote DNA degradation (Wang, et al., 2002). However, EndoG in mammals appears to play little if any role in DNA fragmentation. Moreover, EndoG null mice are viable and develop to adulthood without showing any obvious abnormalities (David, et al., 2006, Irvine, et al., 2005), indicating that EndoG is not essential for the early embryogenesis and cell death in mammals. Future studies are required to identify the AIF-associated endonuclease.
4. AIF, parthanatos and neurologic diseases
As a caspase-independent cell death effector, AIF plays an important role in ischemia and stroke. Stroke is the third cause of death and disability in the United States, just behind diseases of the heart and cancer (Koh, et al., 2005). Neuronal damage and cell death following stroke and ischemia is a primary cause of subsequent morbidity and mortality (Koh, et al., 2005, Koh, et al., 2005). Over the past decade, a large body of evidence demonstrates that activation of PARP-1 significantly increases during brain ischemia and plays a pivotal role in ischemia-induced neuronal injury (Chiarugi, 2005). PARP-1-deficient neurons are protected against cell death caused by NMDA treatment and oxygen glucose deprivation in vitro. Moreover, PARP-1 knockout mice are resistant to neuronal injury in vivo, following middle cerebral artery occlusion (Eliasson, et al., 1997). AIF is a key factor that mediates PARP-1-dependent neuronal death (Yu, et al., 2002). The biologic importance of PARP-1 and AIF in focal brain ischemia was further elucidated by different studies (Hong, et al., 2004, Komjati, et al., 2004, Plesnila, et al., 2004).
The causal involvement of AIF in neurodegenerative diseases was first supported by the observation that cortical neurons prepared from harlequin mice were resistant to glutamate excitotoxicity (Cheung, et al., 2005). It was also suggested that loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) following MPTP treatment was triggered by PARP-1-mediated AIF nuclear translocation (Wang, et al., 2003). PARP-1 knockout mice are dramatically protected against the loss of dopaminergic neurons in the SNpc following MPTP-induced neurotoxicity (Mandir, et al., 1999). Further investigation is required to elucidate the pivotal role of PARP-1 and AIF in different neurologic disorders.
5. Therapeutic targets
PARP-1 activation, PAR polymer formation, mitochondrial AIF release and nuclear translocation, AIF-mediated chromatin condensation/DNA fragmentation are four key steps in parthanatos (Figure 3), which has been implicated to play a pivotal role in multiple neurologic diseases. Therefore, PARP-1, PAR polymer and AIF could be potential targets for therapy of neurologic disorders.
PARP inhibitors can block PARP-1 activation, thereby regulating PAR levels. Several inhibitors, like 1, 5-dihydroxyisoquinoline (DHIQ) and DPQ, have been shown to completely block NMDA- and MNNG-induced toxicity (Yu, et al., 2002), reduce post-traumatic spinal cord injury (Genovese, et al., 2005) and may have protective benefit in chronic neurologic models (Mandir, et al., 1999). These investigations provide the basis for potential clinical applications of PARP inhibitors. Recently, imidazoquinolinone, imidazopyridine and isoquinolindione derivatives have been identified as novel and potent PARP inhibitors (Eltze, et al., 2008). The protective role of these inhibitors in neurologic diseases still needs to be elucidated.
PAR polymer functions as a cell death signal (Andrabi, et al., 2006). PARG has been reported to directly regulate the levels of PAR polymer. Overexpression of PARG in mice significantly reduced infarct volumes after focal ischemia (Andrabi, et al., 2006). In contrast, reduction of PARG expression in mice increased infarct volumes after focal ischemia. This suggests that interference with PAR polymer signaling may offer innovative therapeutic approaches for the treatment of cellular injury following neurologic diseases.
AIF is a key mediator in parthanatos. Upon PARP-1 activation, AIF releases from mitochondria and translocates to nucleus, where it causes chromatin condensation and DNA fragmentation (Figure 3). So far, no AIF inhibitor has been identified to prevent these two events. But HSP70 might be applied to prevent mitochondrial AIF translocation to nucleus. The mechanism of AIF inhibition by HSP70 involves cytosolic retention of AIF. Further evidence indicates that endogenous HSP70 protein levels are sufficiently elevated to modulate the lethal action of AIF. Knocking down endogenous HSP70 sensitizes to the lethal effect of AIF (Ravagnan, et al., 2001). Understanding the mechanisms how AIF releases from mitochondria and how it induces cell death will help to identify a novel therapeutic approach to protect the brain from different neurologic diseases.
6. Concluding remarks
Excessive activation of PARP-1 leads to cell death through a mechanism designated parthanatos. The molecular mechanism for parthanatos involves the release of AIF from mitochondria and translocation to the nucleus. PAR polymer generated by PARP-1 activation plays a pivotal role in this deadly crosstalk between the nucleus and mitochondria. PAR polymer itself functions as a cell death signal that translocates from nucleus to mitochondria to mediate AIF release from mitochondria. The exact mechanisms how PAR polymer causes AIF nuclear translocation and how AIF induces chromatin condensation and large DNA fragmentation still remain to be elucidated. Understanding the molecular mechanism of parthanatos, including PARP-1 activation, PAR polymer formation, AIF nuclear translocation and lethal effect of AIF in nucleus, might lead to more options to develop new therapeutic targets for neurologic diseases that are associated with cell death caused by mitochondrial dysfunction and nuclear DNA damage.
Acknowledgments
This work was supported by grants from the NIH (NS39148), the American Heart Association Postdoctoral Fellowship Award to YW. T.M.D. is the Leonard and Madlyn Abramson Professor of Neurodegenerative Disease at Johns Hopkins University.
Abbreviation
- AIF
apoptosis-inducing factor
- Cyt c
cytochrome c
- EndoG
endonuclease G
- HSP70
heat-shock protein 70
- MLS
mitochondrial localization sequence
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- MPTP
1-methyl-4- phenyl-1,2,5,6-tetrahydropyridine
- NAD+
nicotinamide adenine dinucleotide
- NMDA
N-methyl-D-aspartic acid
- NLS
nuclear localization sequence
- NO
nitric oxide
- PAR
poly(ADP-ribose)
- PARG
poly (ADP-ribose) glycohydrolase
- PARP-1
poly (ADP-ribose) polymerase-1
- ROS
reactive oxygen species
- PD1
phosphodiesterase 1
- SNpc
substantia nigra pars compacta
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ame JC, Apia F, Jacobson EL, Jacobson MK. Assignment of the poly(ADP-ribose) glycohydrolase gene (PARG) to human chromosome 10q11.23 and mouse chromosome 14B by in situ hybridization. Cytogenet Cell Genet. 1999;85:269–270. doi: 10.1159/000015310. [DOI] [PubMed] [Google Scholar]
- 2.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnoult D, Parone P, Martinou JC, Antonsson B, Estaquier J, Ameisen JC. Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. J Cell Biol. 2002;159:923–929. doi: 10.1083/jcb.200207071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevers MB, Neumar RW. Mechanistic role of calpains in postischemic neurodegeneration. J Cereb Blood Flow Metab. 2008;28:655–673. doi: 10.1038/sj.jcbfm.9600595. [DOI] [PubMed] [Google Scholar]
- 5.Blenn C, Althaus FR, Malanga M. Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. Biochem J. 2006;396:419–429. doi: 10.1042/BJ20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 2005;62:739–750. doi: 10.1007/s00018-004-4505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonicalzi ME, Vodenicharov M, Coulombe M, Gagne JP, Poirier GG. Alteration of poly(ADP-ribose) glycohydrolase nucleocytoplasmic shuttling characteristics upon cleavage by apoptotic proteases. Biol Cell. 2003;95:635–644. doi: 10.1016/j.biolcel.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Boujrad H, Gubkina O, Robert N, Krantic S, Susin SA. AIF-mediated programmed necrosis: a highly regulated way to die. Cell Cycle. 2007;6:2612–2619. doi: 10.4161/cc.6.21.4842. [DOI] [PubMed] [Google Scholar]
- 9.Cao G, Xing J, Xiao X, Liou AK, Gao Y, Yin XM, Clark RS, Graham SH, Chen J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J Neurosci. 2007;27:9278–9293. doi: 10.1523/JNEUROSCI.2826-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, MacLaurin JG, Rippstein P, Park DS, Shore GC, McBride HM, Penninger JM, Slack RS. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. Embo J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung EC, Melanson-Drapeau L, Cregan SP, Vanderluit JL, Ferguson KL, McIntosh WC, Park DS, Bennett SA, Slack RS. Apoptosis-inducing factor is a key factor in neuronal cell death propagated by BAX-dependent and BAX-independent mechanisms. J Neurosci. 2005;25:1324–1334. doi: 10.1523/JNEUROSCI.4261-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiarugi A. Inhibitors of poly(ADP-ribose) polymerase-1 suppress transcriptional activation in lymphocytes and ameliorate autoimmune encephalomyelitis in rats. Br J Pharmacol. 2002;137:761–770. doi: 10.1038/sj.bjp.0704934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiarugi A. Poly(ADP-ribosyl)ation and stroke. Pharmacol Res. 2005;52:15–24. doi: 10.1016/j.phrs.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosi C, Marien M. Implication of poly (ADP-ribose) polymerase (PARP) in neurodegeneration and brain energy metabolism. Decreases in mouse brain NAD+ and ATP caused by MPTP are prevented by the PARP inhibitor benzamide. Ann N Y Acad Sci. 1999;890:227–239. doi: 10.1111/j.1749-6632.1999.tb07998.x. [DOI] [PubMed] [Google Scholar]
- 16.Cregan SP, Fortin A, MacLaurin JG, Callaghan SM, Cecconi F, Yu SW, Dawson TM, Dawson VL, Park DS, Kroemer G, Slack RS. Apoptosis-inducing factor is involved in the regulation of caspase-independent neuronal cell death. J Cell Biol. 2002;158:507–517. doi: 10.1083/jcb.200202130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuerrier D, Moldoveanu T, Davies PL. Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach: the importance of primed side interactions. J Biol Chem. 2005;280:40632–40641. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- 18.Culmsee C, Zhu C, Landshamer S, Becattini B, Wagner E, Pellecchia M, Blomgren K, Plesnila N. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and Bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25:10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342 (Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 20.Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prevost MC, Leber B, Andrews D, Penninger J, Kroemer G. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. Faseb J. 2000;14:729–739. [PubMed] [Google Scholar]
- 21.David KK, Sasaki M, Yu SW, Dawson TM, Dawson VL. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13:1147–1155. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- 22.Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- 23.Dawson VL, Dawson TM. Deadly conversations: nuclear-mitochondrial cross-talk. J Bioenerg Biomembr. 2004;36:287–294. doi: 10.1023/B:JOBB.0000041755.22613.8d. [DOI] [PubMed] [Google Scholar]
- 24.Delettre C, Yuste VJ, Moubarak RS, Bras M, Lesbordes-Brion JC, Petres S, Bellalou J, Susin SA. AIFsh, a novel apoptosis-inducing factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J Biol Chem. 2006;281:6413–6427. doi: 10.1074/jbc.M509884200. [DOI] [PubMed] [Google Scholar]
- 25.Delettre C, Yuste VJ, Moubarak RS, Bras M, Robert N, Susin SA. Identification and characterization of AIFsh2, a mitochondrial apoptosis-inducing factor (AIF) isoform with NADH oxidase activity. J Biol Chem. 2006;281:18507–18518. doi: 10.1074/jbc.M601751200. [DOI] [PubMed] [Google Scholar]
- 26.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson TM, Snyder SH, Dawson VL. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 28.Eltze T, Boer R, Wagner T, Weinbrenner S, McDonald MC, Thiemermann C, Burkle A, Klein T. Imidazoquinolinone, Imidazopyridine, and Isoquinolindione Derivatives as Novel and Potent Inhibitors of the Poly(ADP-ribose) Polymerase (PARP): A Comparison with Standard PARP Inhibitors. Mol Pharmacol. 2008;74:1587–1598. doi: 10.1124/mol.108.048751. [DOI] [PubMed] [Google Scholar]
- 29.Endres M, Wang ZQ, Namura S, Waeber C, Moskowitz MA. Ischemic brain injury is mediated by the activation of poly(ADP-ribose)polymerase. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Fossati S, Cipriani G, Moroni F, Chiarugi A. Neither energy collapse nor transcription underlie in vitro neurotoxicity of poly(ADP-ribose) polymerase hyper-activation. Neurochem Int. 2007;50:203–210. doi: 10.1016/j.neuint.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallego MA, Joseph B, Hemstrom TH, Tamiji S, Mortier L, Kroemer G, Formstecher P, Zhivotovsky B, Marchetti P. Apoptosis-inducing factor determines the chemoresistance of non-small-cell lung carcinomas. Oncogene. 2004;23:6282–6291. doi: 10.1038/sj.onc.1207835. [DOI] [PubMed] [Google Scholar]
- 33.Garcia M, Bondada V, Geddes JW. Mitochondrial localization of mu-calpain. Biochem Biophys Res Commun. 2005;338:1241–1247. doi: 10.1016/j.bbrc.2005.10.081. [DOI] [PubMed] [Google Scholar]
- 34.Genovese T, Mazzon E, Muia C, Patel NS, Threadgill MD, Bramanti P, De Sarro A, Thiemermann C, Cuzzocrea S. Inhibitors of poly(ADP-ribose) polymerase modulate signal transduction pathways and secondary damage in experimental spinal cord trauma. J Pharmacol Exp Ther. 2005;312:449–457. doi: 10.1124/jpet.104.076711. [DOI] [PubMed] [Google Scholar]
- 35.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 36.Goto S, Xue R, Sugo N, Sawada M, Blizzard KK, Poitras MF, Johns DC, Dawson TM, Dawson VL, Crain BJ, Traystman RJ, Mori S, Hurn PD. Poly(ADP-ribose) polymerase impairs early and long-term experimental stroke recovery. Stroke. 2002;33:1101–1106. doi: 10.1161/01.str.0000014203.65693.1e. [DOI] [PubMed] [Google Scholar]
- 37.Gurbuxani S, Schmitt E, Cande C, Parcellier A, Hammann A, Daugas E, Kouranti I, Spahr C, Pance A, Kroemer G, Garrido C. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene. 2003;22:6669–6678. doi: 10.1038/sj.onc.1206794. [DOI] [PubMed] [Google Scholar]
- 38.Hageman GJ, Stierum RH. Niacin, poly(ADP-ribose) polymerase-1 and genomic stability. Mutat Res. 2001;475:45–56. doi: 10.1016/s0027-5107(01)00078-1. [DOI] [PubMed] [Google Scholar]
- 39.Haince JF, Ouellet ME, McDonald D, Hendzel MJ, Poirier GG. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim Biophys Acta. 2006;1763:226–237. doi: 10.1016/j.bbamcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Harraz MM, Dawson TM, Dawson VL. Advances in neuronal cell death 2007. Stroke. 2008;39:286–288. doi: 10.1161/STROKEAHA.107.511857. [DOI] [PubMed] [Google Scholar]
- 41.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25:259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Hood JL, Logan BB, Sinai AP, Brooks WH, Roszman TL. Association of the calpain/calpastatin network with subcellular organelles. Biochem Biophys Res Commun. 2003;310:1200–1212. doi: 10.1016/j.bbrc.2003.09.142. [DOI] [PubMed] [Google Scholar]
- 44.Irvine RA, Adachi N, Shibata DK, Cassell GD, Yu K, Karanjawala ZE, Hsieh CL, Lieber MR. Generation and characterization of endonuclease G null mice. Mol Cell Biol. 2005;25:294–302. doi: 10.1128/MCB.25.1.294-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joza N, Oudit GY, Brown D, Benit P, Kassiri Z, Vahsen N, Benoit L, Patel MM, Nowikovsky K, Vassault A, Backx PH, Wada T, Kroemer G, Rustin P, Penninger JM. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25:10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kameshita I, Matsuda Z, Taniguchi T, Shizuta Y. Poly (ADP-Ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J Biol Chem. 1984;259:4770–4776. [PubMed] [Google Scholar]
- 47.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 48.Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman SL. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 49.Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res. 2005;52:5–14. doi: 10.1016/j.phrs.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 50.Koh DW, Dawson TM, Dawson VL. Poly(ADP-ribosyl)ation regulation of life and death in the nervous system. Cell Mol Life Sci. 2005;62:760–768. doi: 10.1007/s00018-004-4508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komjati K, Mabley JG, Virag L, Southan GJ, Salzman AL, Szabo C. Poly(ADP-ribose) polymerase inhibition protect neurons and the white matter and regulates the translocation of apoptosis-inducing factor in stroke. Int J Mol Med. 2004;13:373–382. [PubMed] [Google Scholar]
- 53.Krantic S, Mechawar N, Reix S, Quirion R. Apoptosis-inducing factor: a matter of neuron life and death. Prog Neurobiol. 2007;81:179–196. doi: 10.1016/j.pneurobio.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Kroemer G, El-Deiry WS, Golstein P, Peter ME, Vaux D, Vandenabeele P, Zhivotovsky B, Blagosklonny MV, Malorni W, Knight RA, Piacentini M, Nagata S, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2005;12(Suppl 2):1463–1467. doi: 10.1038/sj.cdd.4401724. [DOI] [PubMed] [Google Scholar]
- 55.Lautier D, Lagueux J, Thibodeau J, Menard L, Poirier GG. Molecular and biochemical features of poly (ADP-ribose) metabolism. Mol Cell Biochem. 1993;122:171–193. doi: 10.1007/BF01076101. [DOI] [PubMed] [Google Scholar]
- 56.Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 57.Loeffler M, Daugas E, Susin SA, Zamzami N, Metivier D, Nieminen AL, Brothers G, Penninger JM, Kroemer G. Dominant cell death induction by extramitochondrially targeted apoptosis-inducing factor. Faseb J. 2001;15:758–767. doi: 10.1096/fj.00-0388com. [DOI] [PubMed] [Google Scholar]
- 58.Lorenzo HK, Susin SA. Therapeutic potential of AIF-mediated caspase-independent programmed cell death. Drug Resist Updat. 2007;10:235–255. doi: 10.1016/j.drup.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Mandir AS, Poitras MF, Berliner AR, Herring WJ, Guastella DB, Feldman A, Poirier GG, Wang ZQ, Dawson TM, Dawson VL. NMDA but not non-NMDA excitotoxicity is mediated by Poly(ADP-ribose) polymerase. J Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandir AS, Przedborski S, Jackson-Lewis V, Wang ZQ, Simbulan-Rosenthal CM, Smulson ME, Hoffman BE, Guastella DB, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc Natl Acad Sci U S A. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mate MJ, Ortiz-Lombardia M, Boitel B, Haouz A, Tello D, Susin SA, Penninger J, Kroemer G, Alzari PM. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struct Biol. 2002;9:442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 62.Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res. 2007;313:2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 64.Miramar MD, Costantini P, Ravagnan L, Saraiva LM, Haouzi D, Brothers G, Penninger JM, Peleato ML, Kroemer G, Susin SA. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 65.Modjtahedi N, Giordanetto F, Madeo F, Kroemer G. Apoptosis-inducing factor: vital and lethal. Trends Cell Biol. 2006;16:264–272. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 66.Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Menissier-de Murcia J, Susin SA. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol Cell Biol. 2007;27:4844–4862. doi: 10.1128/MCB.02141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. Embo J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, Wang ZQ, Thiemermann C. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolase are protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 2005;16:712–719. doi: 10.1681/ASN.2004080677. [DOI] [PubMed] [Google Scholar]
- 69.Pieper AA, Verma A, Zhang J, Snyder SH. Poly (ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol Sci. 1999;20:171–181. doi: 10.1016/s0165-6147(99)01292-4. [DOI] [PubMed] [Google Scholar]
- 70.Plesnila N, Zhu C, Culmsee C, Groger M, Moskowitz MA, Blomgren K. Nuclear translocation of apoptosis-inducing factor after focal cerebral ischemia. J Cereb Blood Flow Metab. 2004;24:458–466. doi: 10.1097/00004647-200404000-00011. [DOI] [PubMed] [Google Scholar]
- 71.Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J Biol Chem. 2005;280:6447–6454. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- 72.Pospisilik JA, Knauf C, Joza N, Benit P, Orthofer M, Cani PD, Ebersberger I, Nakashima T, Sarao R, Neely G, Esterbauer H, Kozlov A, Kahn CR, Kroemer G, Rustin P, Burcelin R, Penninger JM. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 73.Ravagnan L, Gurbuxani S, Susin SA, Maisse C, Daugas E, Zamzami N, Mak T, Jaattela M, Penninger JM, Garrido C, Kroemer G. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat Cell Biol. 2001;3:839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- 74.Ruchalski K, Mao H, Li Z, Wang Z, Gillers S, Wang Y, Mosser DD, Gabai V, Schwartz JH, Borkan SC. Distinct hsp70 domains mediate apoptosis-inducing factor release and nuclear accumulation. J Biol Chem. 2006;281:7873–7880. doi: 10.1074/jbc.M513728200. [DOI] [PubMed] [Google Scholar]
- 75.Ruchalski K, Mao H, Singh SK, Wang Y, Mosser DD, Li F, Schwartz JH, Borkan SC. HSP72 inhibits apoptosis-inducing factor release in ATP-depleted renal epithelial cells. Am J Physiol Cell Physiol. 2003;285:C1483–1493. doi: 10.1152/ajpcell.00049.2003. [DOI] [PubMed] [Google Scholar]
- 76.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 77.Smulson ME, Simbulan-Rosenthal CM, Boulares AH, Yakovlev A, Stoica B, Iyer S, Luo R, Haddad B, Wang ZQ, Pang T, Jung M, Dritschilo A, Rosenthal DS. Roles of poly(ADP-ribosyl)ation and PARP in apoptosis, DNA repair, genomic stability and functions of p53 and E2F-1. Adv Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- 78.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53(Suppl 1):S12–18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- 80.Tan Y, Dourdin N, Wu C, De Veyra T, Elce JS, Greer PA. Conditional disruption of ubiquitous calpains in the mouse. Genesis. 2006;44:297–303. doi: 10.1002/dvg.20216. [DOI] [PubMed] [Google Scholar]
- 81.Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G. AIF deficiency compromises oxidative phosphorylation. Embo J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, Shimoji M, Yu SW, Dawson TM, Dawson VL. Apoptosis inducing factor and PARP-mediated injury in the MPTP mouse model of Parkinson’s disease. Ann N Y Acad Sci. 2003;991:132–139. doi: 10.1111/j.1749-6632.2003.tb07471.x. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 85.Whitacre CM, Hashimoto H, Tsai ML, Chatterjee S, Berger SJ, Berger NA. Involvement of NAD-poly(ADP-ribose) metabolism in p53 regulation and its consequences. Cancer Res. 1995;55:3697–3701. [PubMed] [Google Scholar]
- 86.Winstall E, Affar EB, Shah R, Bourassa S, Scovassi AI, Poirier GG. Poly(ADP-ribose) glycohydrolase is present and active in mammalian cells as a 110-kDa protein. Exp Cell Res. 1999;246:395–398. doi: 10.1006/excr.1998.4321. [DOI] [PubMed] [Google Scholar]
- 87.Xu Y, Huang S, Liu ZG, Han J. Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. J Biol Chem. 2006;281:8788–8795. doi: 10.1074/jbc.M508135200. [DOI] [PubMed] [Google Scholar]
- 88.Ye H, Cande C, Stephanou NC, Jiang S, Gurbuxani S, Larochette N, Daugas E, Garrido C, Kroemer G, Wu H. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat Struct Biol. 2002;9:680–684. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- 89.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]