Abstract
As the heart ages, electrophysiological and biochemical changes can occur, and the ventricle in many cases loses distensibility, impairing diastolic function. How the proteomic signature of the aged ventricle is unique in comparison to young hearts is still under active investigation. We have undertaken a quantitative proteomics study of aging left ventricles (LVs) utilizing the isobaric Tagging for Relative and Absolute Quantification (iTRAQ) methodology. Differential protein expression was observed for 117 proteins including proteins involved in cell signaling, the immune response, structural proteins, and proteins mediating responses to oxidative stress. For many of these proteins, this is the first report of an association with the aged myocardium. Additionally, two proteins of unknown function were identified. This work serves as the basis for making future comparisons of the aged left ventricle proteome to that of left ventricles obtained from other models of disease and heart failure.
Keywords: aging, left ventricle, cardiac, MALDI-TOF, iTRAQ, proteomics
Introduction
Aging of the healthy heart is a complex process that is generally accompanied by mild cardiomyocyte hypertrophy, increased cellular senescence and replacement of cells with extracellular matrix. In aged myocardium, individual cardiomyocytes are subject to oxidative stress, alteration in the utilization of metabolic pathways, disruption of calcium homeostasis, and dysregulation of apoptotic functions. Physiologically, mild concentric hypertrophy in aged hearts is associated with changes in the myocardium including prolonged contraction, diminished contraction velocity, decreased β-adrenergic response, and increased myocardial stiffness1–3. Ultimately, impairment of heart function reduces the ability of the heart to respond to increased demand through the Frank Starling effect, decreasing the exercise capacity of aged individuals.
Cardiovascular proteomics has elucidated many pathways that contribute to hypertrophic processes and overall cardiac dysregulation, and offers the potential for clinical assessment of biomarkers for heart failure4–6. For example, studies of neonatal piglet right ventricles elucidated genetic heart defects at the protein level7, while an independent study assessed the cardioprotective effects of nitrites on the post-translational modification of proteins, particularly those involved in signaling, contraction, redox regulation and metabolism8. In studies of a heart failure model, proteomic assessment of mice subjected to transverse aortic constriction demonstrated differential protein expression of structural, signaling and redox proteins in comparison to healthy controls9. Through proteomics studies such as these, we begin to understand how heart failure develops mechanistically, and how transcriptome data literally translates to the protein level. However, heart function is complex, and it remains a challenge to unravel pathologic changes in protein expression from those that are compensatory, and acute changes from those that are more chronic and perhaps related indirectly to heart disease. Furthermore, proteomic studies of the heart on a global scale remain relatively infrequent. To address some of these concerns, we have chosen to elucidate the effect of the aging process on the heart using quantitative mass spectrometric method.
Aging is a risk factor for cardiovascular disease. It is not clear, however, whether the phenotype of the aging heart presents a signature similar in nature to that of chronic cardiovascular disease. More emphatically, the question arises whether any of the proteomic changes in the aged heart can be used to predict or dissect fundamental processes that underlie or contribute to the development of heart disease. Much of the recent effort to define the proteomic signature of aged ventricles has relied on microarray analysis to quantify differences in mRNA expression between young vs. aged tissue10–12. Microarray studies generate relevant data pertaining to gene expression. However, these studies do not report on quantitative changes in the expressed proteome or post-translational modifications that may influence cellular activity. Furthermore, levels of protein expression often do not correlate with levels of mRNA13,14. Relatively few proteomics studies have been performed on the aged heart. However, Dai, et al.15, identified 73 proteins that were differentially expressed in the aged mouse left ventricle using a 2-D gel approach,, mainly representing proteins involved in energy metabolism and proteins involved in the oxidative stress response. In a second 2D-gel study16, differential expression of 46 proteins, representing mostly metabolic or structural proteins, was observed in aged rat ventricles, accompanied by significant increases in protein nitration.
To achieve a greater depth of proteome coverage that reports on a broader range of protein classes, quantitative mass spectrometric measurement of differential protein expression in the aged rat left ventricle using the iTRAQ method17 was performed. This iTRAQ analysis allowed quantification of the relative expression of 1040 unique proteins, where a subset of 117 proteins demonstrated differential protein expression in young vs. aged hearts. Proteins with increased or decreased expression in aged myocardium represented a number of different protein functional classes, particularly those involved in immunological responses, cell signaling, energy metabolism, and oxidative stress responses. Only 27 of these proteins were found differentially expressed in either of the previous proteomics study reported by Dai, et al.15 or by Richardson, et al.16.
The first step to answering the question of whether aged heart shares a fundamental proteomic phenotype with that of diseased heart is to define the unique signature of the aged proteome. In this first report of iTRAQ analysis of the differential proteome of the aged left ventricle, we identified several candidate proteins that represented cellular networks including calcium signaling, the immune response, and the heart’s structural organization that were perturbed in aged heart. This work provides fundamental information necessary to make a direct comparison to differential protein expression in the failing heart.
Experimental Procedures
Reagents and experimental animal procurement
All procedures performed on the Fisher 344 rats used in this study were approved by the Institutional Animal Care and Use Committee, at the Medical University of South Carolina. Rats were purchased from the NIH Aging Rodent Colony18; six were 4 months-old (young) and six were 26 months-old (aged, and considered senescent.) The iTRAQ reagents and trypsin were acquired from Applied Biosystems (Framingham, MA). Unless otherwise noted, all reagents were purchased from Fisher Scientific (Waltham, MA).
Strategy
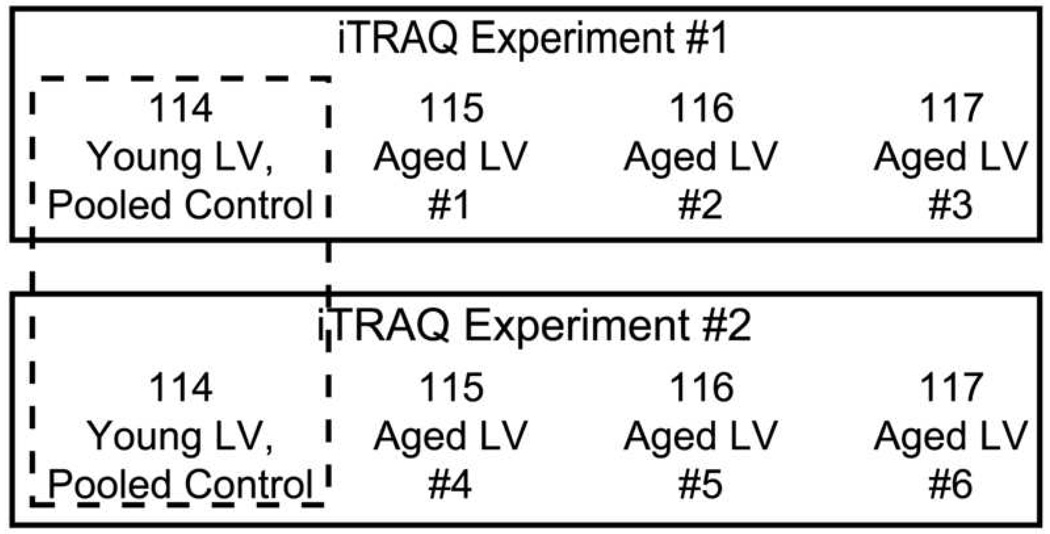
The four-plex iTRAQ procedure was used to assess protein levels in six aged rats by performing two iTRAQ experiments (Fig. 1). In both experiments, use of a pooled sample from young ventricles as the control (labeled with iTRAQ reagent 114) provided the baseline measurement of peptide levels for each 4-plex iTRAQ experiment, and allowed comparison of the two iTRAQ datasets to each other. The iTRAQ reagents 115, 116 and 117 were used to analyze the protein content of six individual aged ventricles over the course of the two separate iTRAQ experiments.
Figure 1.
Strategy for performing iTRAQ analysis of six aged left ventricles.
Preparation of crude tissue extracts
Six young male Fisher 344 rats (4 months-old) and 6 aged Fisher 344 rats (26 months-old) were anesthetized according to established procedure19. Each heart was removed and perfused with phosphate-buffered saline. The left ventricle was isolated. For each individual heart, 100 mg of the left ventricle was minced into 1 mm3 pieces, and treated with 500 µL of extraction buffer (20 mM HEPES pH 8.0, 1% CHAPS, and 1 M urea). The high concentration of CHAPS was sufficient to denature any endogenous proteases, minimizing the need for protease inhibitors, which could interfere with protein digestion during the iTRAQ protocol. Each sample was homogenized individually using a hand-held Polytron tissue homogenizer (Kinematic Inc.; Bohemia, NY). To disrupt cell membranes, preparations were sonicated over ice using a Branson (Danbury, CT) model 250 sonicator set at power setting 4 and operating at 100% duty cycle. Each sample was sonicated with three 2-second pulses with 3 seconds relaxation between pulses. This process was repeated every 15 minutes for 1 hour. Individual samples were centrifuged at 20,000 rpm for 30’ at 4 °C on an Optima TL ultracentrifuge (Beckman-Coulter, Fullerton, CA) to remove cell debris. The supernatant was aliquoted into 3 equal portions by volume, and total protein concentrations were determined using the bicinchoninic (BCA) protein determination concentration kit from Pierce (Rockford, Ill).
iTRAQ labeling and protein fractionation
Each aged rat ventricle preparation was assessed individually, where a 60 µg aliquot was placed in an individual eppendorf tube, and the volume of each was adjusted to 56 µL. The final protein concentration in each of the four samples was approximately 1.06 µg/µL. To create the control pool, an equal amount of protein (60 µg) from each of six young rats was combined in a total volume of 336 µL. Each sample was reduced, and alkylated according to the iTRAQ protocol17 (Applied Biosystems). To perform trypsin digestion, 50 µg of sequencing grade trypsin (Applied Biosystems) was resuspended in 50 µL of H20 and distributed equally between each of the four samples.
Each iTRAQ experiment (4 samples) was then subjected to the iTRAQ labeling protocol. After 14 hours of digestion, iTRAQ labeling was performed on the control pool by applying the iTRAQ tagging reagent 114, while each of the three aged ventricle preparations were treated individually with iTRAQ labeling reagents 115, 116, and 117, respectively. The labeling proceeded for 1 hour, at which time the reactions were quenched with 10 µL of 1M Tris pH 8.0. After labeling, all four iTRAQ-labeled samples were combined.
Initial fractionation of proteolytic peptides was performed using strong cation exchange (SCX) chromatography over a polysulfoethyl A column (200 × 2.1 mm; PolyLC) at 250 µL/min. using a Waters (Milford, MA) model 600 HPLC pump. Absorbance at 214 nm was monitored, and fractions were collected every 5 minutes. Peptides were chromatographed using the following gradient program. First, peptides were allowed to bind to the column for 20 minutes at 90% low salt buffer A (10 mM KH2PO4 in 25% ACN; pH 2.7–3.0) and 10% high salt buffer B (10 mM KH2PO4 and 0.5 M KCl in 25% ACN; pH 2.7–3.0). Over 40 minutes, the concentration of high-salt buffer B was increased linearly to 50%. At this stage, peptides were eluted isocratically for 40 minutes, followed by a 10 minute gradient to 100% buffer B. Individual SCX fractions were lyophilized to dryness.
Peptides were further fractionated by performing reversed-phase (RP) chromatography on each SCX fraction using an Ultimate/Famos LC system (Dionex; Sunnyvale, CA) running Chromeleon v. 6.8 software. Peptides were first desalted using an LC-Packings C-18 precolumn cartridge (Pepmap 100, 5 mm x 300 um ID). RP-HPLC was performed using a Microtech C18 column (i.d. 100 um x 15 cm; 300A pore size). HPLC solvent A was composed of 2% ACN and 0.1% TFA In brief, a linear gradient from 12% of HPLC solvent B (85% ACN, 10% isopropanol and 0.1% TFA) to 95% solvent B was used to elute peptides. Absorbance at 214 nm was used to monitor peptide elution. A Probot spotting device (Dionex), connected in-line with the HPLC system, was used to spot droplet fractions onto an Optitof LC/MALDI target (Applied Biosystems), where the eluate from the C18 column was mixed with a MALDI matrix solution at a ratio of 1:2 v/v. The MALDI matrix was prepared by dissolving α-cyano-4-hydroxy-cinnamic acid (Bruker Daltonics) at a concentration of 8 mg/mL in a solution of 70% ACN and 0.1% TFA, which also contained also 0.15 mg/mL of ammonium citrate.
Spectra were acquired on a 4800 MALDI TOF-TOF mass spectrometer (Applied Biosystems). MS spectra were collected over m/z 800–3500 to identify parent ions, and the top 10 most abundant ions were selected for MS/MS analysis using a collision energy of 1 keV. Data were processed using GPS Explorer v. 3.6. Peptide data were searched against the rat Refseq database (3/17/07), using a precursor error tolerance of 200 ppm and a MS/MS error tolerance of 0.5 Da. Two missed trypsin cleavages were allowed. S-methylmethanethiosulfonate-labeling of cysteine, iTRAQ labeling of lysine and iTRAQ labeling of each peptide N-terminus were set as fixed modifications. Methionine oxidation and iTRAQ labeling of tyrosine were allowed as variable modifications. Tandem MS summary reports, including best peptide sequence, protein name, spot index, accession number, start sequence position, modifications, and normalized iTRAQ reagent cluster areas were generated using GPS Explorer.
Occasionally, the GPS Explorer software, employing a MASCOT search, would identify a protein from Rattus norvegicus as a “predicted” protein. For each “predicted” protein, a BLAST search20 was performed against either the Refseq protein database or the nonredundant protein database using the sequence identified during the original MASCOT search. In the event that an “extant” or confirmed protein from Rattus norvegicus (non-theoretical) was identified in the mascot search, care was taken to confirm that the tryptic peptide sequences observed in these iTRAQ experiments were also present in the “extant” protein. In the case where tryptic peptides identified through the MASCOT search matched the confirmed protein, the GI number of the confirmed protein is reported.
Quantification of relative protein expression
We employed a customized software package, iQuantitator, to infer the magnitude of change in protein expression. The software infers treatment-dependent changes in expression using Bayesian statistical methods. Inference is based on a previously published statistical model21 and uses Markov-Chain Monte Carlo methods22 (Gibbs Sampling). The software was developed as a collection of R scripts23 with the Gibbs sampler implemented in the C programming language. Basically, this approach was used to generate means, medians, and 95% credible intervals (upper and lower) for each treatment-dependent change in protein expression by using peptide-level data for each component peptide, and integrating data across the two experiments. For proteins whose iTRAQ ratios were down-regulated in aged rats, the extent of down-regulation was considered further if the null value of 1 was above the upper limit of the credible interval. Conversely, for proteins whose iTRAQ ratios were up-regulated in aged rats, the extent of up-regulation was considered further if the lower limit of the credible interval had a value greater than 1. The width of these credible intervals depends on the data available for a given protein. Since the number of peptides observed and the number of spectra used to quantify the change in expression for a given protein are taken into consideration, it is possible to detect small but significant changes in up- or down-regulation when many peptides are available. A detailed description of the statistical parameters employed by this program will be presented elsewhere.
Data from a total of 71,296 spectra from two iTRAQ experiments were considered using this customized software, with 668 spectra eliminated due to the presence of disallowed modifications (iTRAQ-labeled tyrosines) and 5 due to missing data from one or more iTRAQ reporter ions. Of the total, 12,306 spectra were used to identify 4085 peptides (1040 unique proteins). For each protein, and each peptide associated with a given protein, the mean, median, and 95% credible intervals were computed for each of the protein- and peptide-level treatment effects.
Network analysis
To model the signaling networks being affected by aging in the rat, the accession number of each protein found up- or down-regulated in the aging study was subjected to network analysis using the Ingenuity Pathways Analysis (IPA) v.5 software suite (Ingenuity, Inc). The IPA software identified the top three major networks affected as 1) carbohydrate metabolism, cell signaling and energy production; 2) cell morphology, connective tissue disorders and inflammation; and 3) cell assembly and organization.
Western blotting
Western blotting for calnexin, prohibitin and the voltage-dependent anion channel 1 was performed to confirm the results of iTRAQ analysis. A portion of the ventricles from young and aged rats used for iTRAQ analysis were solubilized in RIPA buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% Sodium deoxycholate, 0.1% SDS with Roche protease inhibitors) and the amount of soluble protein present was quantified by BCA analysis as per manufacturer’s instructions (Pierce). Equal amounts of protein from each young and aged ventricle were separated by SDS-PAGE on 3–8% or 4–12% gradient gels under reducing conditions and analyzed by Western blot as described24. Primary antibodies used were anti-calnexin (Calbiochem; San Diego, CA), anti-prohibitin (Abcam; Cambridge, MA), and anti-VDAC1 (Fisher Scientific).
Quantification was performed by histogram analysis using Photoshop Imaging Software (Adobe). Basically, blot images were inverted, and the mean intensity (arbitrary units) of and number of pixels spanned by each individual protein band were quantified. The average absolute intensity and the standard deviation were determined. The protein ratio (aged vs. young) was determined using these averaged values. The two-tailed, paired T-test, was used to generate p-values.
Results
Morphometric Analysis
Determination of the wet weight of the left ventricle demonstrated the anticipated large increase in the mass of the heart over the lifespan of the animal (Table 1). However, measurement of ventricle mass, normalized to either the body weight (BW) or tibia length (TL) of the aged and young rats (Table 1), allowed quantification of the extent of hypertrophy of the ventricles as a function of aging in consideration of the overall animal’s weight. While the ratio of right ventricle mass to tibia length (RV/TL) decreased slightly in the aged rat, the ratio of left ventricle mass to tibia length (LV/TL) of aged rats was slightly elevated from that of young rats. The small differences in these values reflect minimal hypertrophy of the aged left ventricle, which is consistent with other reports, but represents a trend towards hypertrophy in the aged left ventricle.
Table 1.
Morphometric analysis of young and aged rats.
| Age (months) | 4 | 26 | p-value |
|---|---|---|---|
| n (number of animals) | 6 | 6 | |
| Body weight (BW, g) | 322 +/− 5.4 | 397 +/− 29 | 0.0022 |
| LV weight, wet (VW, mg) | 577 +/− 46 | 727 +/− 71 | 0.17 |
| RV/BW (mg/g) | 0.5 +/− 0.03 | 0.40 +/− 0.05 | 0.362 |
| LV/BW (mg/g) | 1.73 +/− 0.10 | 1.75 +/− 0.19 | 0.897 |
| RV/TL (mg/mm) | 3.55 +/− 0.24 | 3.41 +/− 0.41 | 0.43 |
| LV/TL (mg/mm) | 12.0 +/− 0.1 | 13.8 +/− 1.5 | 0.079 |
| TL (mm) | 46.5 +/− 0.8 | 50.3 +/− 1.9 | 0.0070 |
iTRAQ analysis
A total of 71,296 MS/MS spectra were acquired over two iTRAQ runs. Of these, 12,306 spectra were utilized to assign 4085 unique peptides, representing 1040 unique proteins. To estimate the false positive rate for protein identification, the Swiss-Prot sequences within the rat Refseq database were randomized using an in-house Perl script. Only proteins that were present at a C.I. value of 95% or greater and were identified by at least two peptides were considered. A MASCOT search of the original iTRAQ data was made against the shuffled database, and 11 proteins were identified using 22 peptides. This correlates to a false-positive rate of 1.1%.
Of the 1040 proteins identified in this study, 256 were shared between the two complete iTRAQ data sets, which is proportionally consistent with other reports commenting on the technical variance between iTRAQ datasets25,26. Of these, statistically significant differential expression was found for 117 proteins that had a confidence interval (%CI) above 95% and were identified by at least two peptides. Of these, 57 were down-regulated in the aging left ventricle, while the remaining 60 were up-regulated.
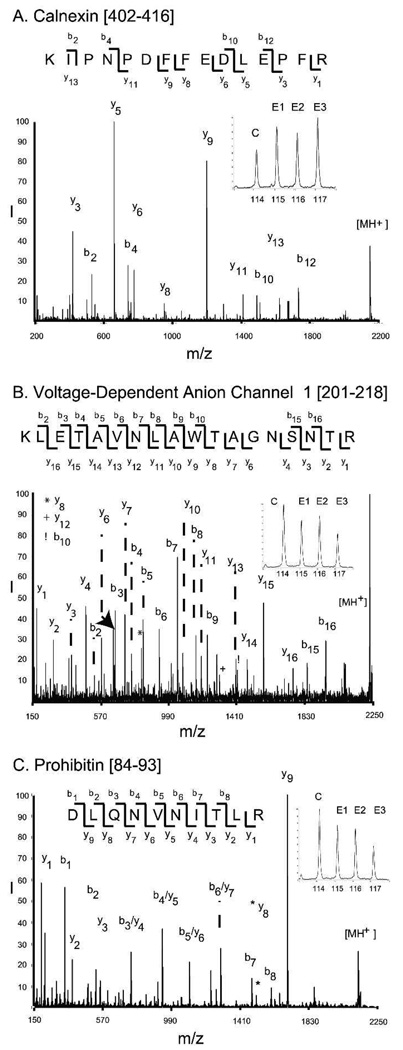
We identified 23 signaling proteins (Table 2), which comprised the second-most abundant class of proteins found differentially expressed in this aged rat study. Of these, γ-synuclein, a centrosome-associated protein, was the most highly up-regulated (iTRAQ ratio 1.44). Although γ-synuclein is found at high concentrations in the brain, it is also robustly expressed in the heart27,28. Currently, the role of synuclein in the heart is poorly understood, but it may play a role in cell proliferation of non-myocyte cell types or sympathetic innervation of the heart. Calnexin (iTRAQ ratio 1.29) was also found to be up-regulated in this study, and the tandem mass spectrum for the calnexin peptide [402–416] is presented in Fig. 2A.
Table 2.
Differential expression of signaling proteins.
| Protein Identity | Accession Numbera |
iTRAQ Ratio |
Credible Interval |
# Pep.d |
Seq. Cover.e |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | γ-Synuclein | 13928954 | 1.44 | 1.13 | 1.80 | 2 | 19 |
| 2 | Nuclease-sensitive element binding protein 1 | 92373398 | 1.43 | 1.20 | 1.71 | 5 | 27 |
| 3 | Caveolin 1 α-isoform | 19526761 | 1.31 | 1.07 | 1.60 | 3 | 20 |
| 4 | Calnexin | 25282419 | 1.29 | 1.03 | 1.60 | 3 | 9 |
| 5 | 14 kDa phosphohistidine phosphatasef | 109467870 | 1.28 | 1.11 | 1.49 | 5 | 38 |
| 6 | Brain abundant, membrane attached signal protein 1 | 11560135 | 1.26 | 1.07 | 1.49 | 7 | 48 |
| 7 | Calreticulin | 11693172 | 1.25 | 1.13 | 1.38 | 17 | 52 |
| 8 | Predicted: similar to Myristoylated alanine-rich C-kinase substrate | 109510177 | 1.25 | 1.08 | 1.46 | 6 | 42 |
| 9 | Sarcalumenin | 157818487 | 1.25 | 1.07 | 1.47 | 7 | 18 |
| 10 | EH-domain containing 2 | 67846074 | 1.23 | 1.00 | 1.51 | 4 | 12 |
| 11 | Coiled-coil helix-coiled-coil-helix domain containing 3f | 157817027 | 1.21 | 1.05 | 1.40 | 9 | 26 |
| 12 | Milk Fat globule-EGF factor 8 isoform 1 | 93277126 | 1.21 | 1.01 | 1.43 | 6 | 18 |
| 13 | Calmodulin 1 | 14010863 | 1.20 | 1.02 | 1.42 | 9 | 49 |
| 14 | Predicted: similar to AHNAK nucleoprotein isoform 1 isoform 5 | 109463547 | 1.19 | 1.12 | 1.27 | 45 | 27 |
| 15 | EF hand domain containing 2 | 72255531 | 1.18 | 1.01 | 1.37 | 6 | 23 |
| 16 | Calumenin homolog (Hypothetical protein LOC360380) | 76559925 | 1.15 | 1.01 | 1.30 | 9 | 39 |
| 17 | Laminin receptor 1 | 8393693 | 0.870 | 0.761 | 0.992 | 9 | 49 |
| 18 | Phosphatidylethanolamine binding Protein | 8393910 | 0.861 | 0.766 | 0.972 | 10 | 72 |
| 19 | Predicted: similar to Ras-GTPase-protein activating binding protein 1 | 109488113 | 0.805 | 0.662 | 0.971 | 5 | 17 |
| 20 | Voltage-dependent anion channel 1 | 13786200 | 0.803 | 0.707 | 0.908 | 9 | 44 |
| 21 | Prohibitin | 13937353 | 0.778 | 0.663 | 0.892 | 7 | 33 |
| 22 | Expressed in non-metastatic cells 2 | 55926145 | 0.778 | 0.697 | 0.868 | 10 | 74 |
| 23 | N-Myc downstream regulated gene 2 | 19424278 | 0.744 | 0.617 | 0.816 | 7 | 33 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
A predicted protein whose observed peptides matched an existing protein in the Refseq database.
Figure 2. Mass spectra of three representative proteins.
The MS/MS spectra and the iTRAQ regions (inset) are presented for three representative peptides. Fig. 2A: calnexin[402–416], Fig. 2B: the voltage-dependent anion channel 1 [201–208], and Fig. 2C: Prohibitin[84–93].
Calreticulin (iTRAQ ratio 1.25) and calmodulin 1 (iTRAQ ratio 1.20), are calcium-related signaling proteins found up-regulated in aged myocardium. Interestingly, the AHNAK nucleoprotein (iTRAQ ratio 1.19) and the calumenin homolog (hypothetical protein LOC360380, iTRAQ ratio 1.15), two proteins that regulate the activity of calcium channels29,30, were found to be up-regulated. Down-regulation of the voltage dependent anion channel 1 (iTRAQ ratio 0.803; VDAC1) and prohibitin (iTRAQ ratio 0.778) was observed by iTRAQ analysis. As well, a representative tandem mass spectrum is provided for one peptide of VDAC 1 (Fig. 2B) and one of prohibitin (Fig. 2C). The N-myc downstream regulated gene 2 (NRDG2, iTRAQ ratio 0.744) exhibited the greatest degree of down-regulation of signaling proteins identified in the aged rat. NRDG2, a signaling protein that is a tumor suppressor and a stress response protein, promotes T-cell proliferation and can be phosphorylated by Akt31. Overall, of the 23 signaling proteins observed at either increased or decreased levels in the aging rat, only a very few have been implicated in aging.
In this iTRAQ study, metabolic proteins represented the largest class of proteins observed differentially regulated during aging, identifying 32 metabolic proteins (Table 3 & Table 4). Plasma glutamate carboxypeptidase (iTRAQ ratio 1.28) and D-dopachrome tautomerase (iTRAQ ratio 1.26) were found to have the greatest level of up-regulation. In contrast, the peroxisomal form of the enoyl CoA hydratase demonstrated the greatest decrease in expression in aged hearts (iTRAQ ratio 0.557).
Table 3.
Differential expression of metabolic proteins.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq. Cover.e |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | Plasma glutamate carboxypeptidase | 13928880 | 1.28 | 1.08 | 1.50 | 6 | 20 |
| 2 | D-dopachrome tautomerase | 13162287 | 1.26 | 1.10 | 1.45 | 7 | 76 |
| 3 | ADP-ribosylation factor 3 | 18266716 | 1.26 | 1.01 | 1.59 | 4 | 26 |
| 4 | Triosephosphate isomerase 1 | 117935064 | 1.23 | 1.15 | 1.32 | 18 | 84 |
| 5 | Apolipoprotein E | 20301954 | 1.21 | 1.07 | 1.37 | 11 | 33 |
| 6 | Glycogenin 1 | 13591969 | 1.19 | 1.02 | 1.40 | 6 | 22 |
| 7 | Glyceraldehyde-3- phosphate dehydrogenasef | 109484558 | 1.18 | 1.04 | 1.34 | 9 | 38 |
| 8 | Peptidylprolyl isomerase A | 8394009 | 1.17 | 1.05 | 1.29 | 11 | 55 |
| 9 | Glyoxylase 1 | 46485429 | 1.15 | 1.02 | 1.29 | 8 | 47 |
| 10 | Apolipoprotein A-I | 6978515 | 1.04 | 1.02 | 1.26 | 14 | 58 |
| 11 | Fatty acid binding protein 4 | 16758094 | 1.12 | 1.01 | 1.23 | 7 | 43 |
| 12 | Aldehyde reductase 1 | 6978491 | 1.11 | 1.02 | 1.21 | 17 | 56 |
| 13 | Malate dehydrogenase, mitochondrial | 42476181 | 1.08 | 1.02 | 1.13 | 22 | 69 |
| 14 | Aldolase A | 6978487 | 0.836 | 0.755 | 0.927 | 15 | 43 |
| 15 | Succinate dehydrogenase complex, subunit A, flavoprotein | 18426858 | 0.834 | 0.721 | 0.965 | 10 | 22 |
| 16 | Thiosulfate sulfurtransferase | 57528682 | 0.791 | 0.639 | 0.977 | 3 | 17 |
| 17 | Glyoxalase domain-containing 4 (Hypothetical protein LOC363644) | 62079189 | 0.744 | 0.619 | 0.890 | 5 | 18 |
| 18 | Dihydrolipoamide S-succinyl transferase | 55742725 | 0.743 | 0.658 | 0.840 | 10 | 28 |
| 19 | Phosphoglycerate kinase 1 | 40254752 | 0.712 | 0.579 | 0.869 | 4 | 55 |
| 20 | Hydroxysteroid (17-β) dehydrogenase 10 | 13994225 | 0.708 | 0.592 | 0.851 | 6 | 24 |
| 21 | Fumarate hydratase 1 | 8393358 | 0.698 | 0.613 | 0.794 | 9 | 27 |
| 22 | Glutamate oxoloacetate transaminase 2 | 6980972 | 0.687 | 0.572 | 0.823 | 5 | 17 |
| 23 | 3-Hydroxyisobutyrate dehydrogenase | 83977457 | 0.670 | 0.521 | 0.863 | 3 | 15 |
| 24 | Pyruvate dehydrogenase E1 component α-subunit, somatic form, mitochondriaf | 124430510 | 0.631 | 0.537 | 0.741 | 8 | 20 |
| 25 | Pyruvate dehydrogenase (lipoamide) β | 56090293 | 0.582 | 0.517 | 0.652 | 10 | 33 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
A predicted protein whose observed peptides matched an existing protein in the Refseq database.
Table 4.
Differential expression of proteins involved in fatty acid metabolism.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq. Cover.e |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | Acyl-coenzyme A dehydrogenase, long chain | 6978431 | 1.07 | 1.01 | 1.13 | 29 | 55 |
| 2 | Acyl-coenzyme A dehydrogenase, short chain | 11968090 | 0.813 | 0.751 | 0.879 | 16 | 52 |
| 3 | Acetyl-coenzyme A acetyltransferase 1 | 8392836 | 0.779 | 0.665 | 0.910 | 5 | 20 |
| 4 | Acetyl-coenzyme A synthetase 2-likef | 157818027 | 0.729 | 0.629 | 0.841 | 8 | 17 |
| 5 | Enoyl coenzyme A hydratase, short chain, 1, mitochondrial | 17530977 | 0.684 | 0.571 | 0.820 | 5 | 26 |
| 6 | L-3-Hydroxyacyl-coenzyme A Dehydrogenase | 17105336 | 0.675 | 0.572 | 0.799 | 5 | 20 |
| 7 | Enoyl coenzyme A hydratase 1, peroxisomal | 12018256 | 0.557 | 0.477 | 0.653 | 7 | 33 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
A predicted protein whose observed peptides matched an existing protein in the Refseq database.
Although five glycolytic enzymes were observed in this iTRAQ experiment, only 3 were differentially expressed in the aged heart. Phophoglycerate kinase, which produces 2 ATP molecules per cycle, was found at diminished levels (iTRAQ ratio 0.712) in aged hearts while triosephosphate isomerase (iTRAQ ratio 1.23) and glyceraldehyde-3-phosphate dehydrogenase (iTRAQ ratio 1.18) were found at enhanced levels. Hexokinase and enolase were also observed, however, their iTRAQ ratios did not differ significantly from 1.0. These data are consistent with the increase in glycolysis found in surgical models of hypertrophy and chronically failing hearts32,33.
The concentration of succinate dehydrogenase (iTRAQ ratio 0.834) and fumarate hydratase (iTRAQ ratio 0.698), decreased in the aged rat; however, mitochondrial malate dehydrogenase (iTRAQ ratio 1.08) was observed at slightly up-regulated levels. Aconitase, isocitrate dehydrogenase and malate dehydrogenase were also detected, but their expression ratios did not change in the aged rat.
The iTRAQ data indicated that three out of four enzymes of fatty acid β-oxidation were found at decreased levels in aged ventricles (Table 4): the short-chain acyl-CoA dehydrogenase (iTRAQ ratio 0.813), the mitochondrial enoyl CoA hydratase (iTRAQ ratio 0.684) and L-3-hydroxyacyl-CoA dehydrogenase (iTRAQ ratio 0.675). Although acetyl-CoA acyltransferase 2 (β-ketothiolase) was identified and showed a trend towards down-regulation, these data did not meet statistical criteria for a down-regulated response.
Additional proteins involved in fatty acid metabolism were shown to be present at depressed levels in aged LV. The ketothiolase responsible for catabolizing isoleucine into acetyl- and proprionyl-CoAs, acetyl-CoA acyltransferase 1 (iTRAQ ratio 0.779), was found at decreased levels. Pyruvate dehydrogenase activity affects entry of citrate into the Krebs cycle, formation of Acyl-CoA fatty acids, and also the synthesis of cholesterol, and in aged rats the α- and β- subunits were found at decreased levels (iTRAQ ratios of 0.631 and 0.582, respectively). Overall, these data demonstrate a general depression in the function of fatty acid-related pathways in the aged LV.
Nineteen proteins participating in either electron transfer and/or ATP production were found differentially expressed in the aging rat LV (Table 5). The results were as follows: the ATPase inhibitor factor 1 (iTRAQ ratio 1.27) was found at enhanced levels. The electron-transfer-flavoprotein β-polypeptide was found down-regulated (iTRAQ ratio 0.775), as were two polypeptides from ubiquinol-cytochrome c reductase complex: Rieske iron-sulfur polypeptide 1 (iTRAQ ratio 0.796), and core protein II (iTRAQ ratio 0.765). The somatic form of cytochrome C (iTRAQ ratio 0.894) as well as cytochrome c-1 (iTRAQ ratio 0.799) were found at decreased levels in aged LV.
Table 5.
Differential expression of proteins involved in energy metabolism.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq. Cover. |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | NADH dehydrogenase 1β7 | 157823197 | 1.60 | 1.36 | 1.89 | 4 | 37 |
| 2 | NADH dehydrogenase 1β8 | 157822261 | 1.40 | 1.21 | 1.62 | 5 | 38 |
| 3 | ATPase inhibitory factor 1 | 77917528 | 1.27 | 1.03 | 1.55 | 3 | 15 |
| 4 | NADH dehydrogenase 1β9f | 1879370 | 1.19 | 1.00 | 1.42 | 5 | 45 |
| 5 | Predicted: similar to NADH dehydrogenase 1α13 | 109481899 | 1.17 | 1.03 | 1.34 | 8 | 53 |
| 6 | ATP synthase, H+ transporting, mitochondrial F0 complex, F6 | 16758388 | 1.10 | 1.00 | 1.20 | 11 | 56 |
| 7 | Predicted: similar to Cytochrome c, somaticg | 109490661 | 0.894 | 0.820 | 0.975 | 9 | 62 |
| 8 | NADH dehydrogenase 1α5 | 6981260 | 0.869 | 0.785 | 0.963 | 10 | 73 |
| 9 | NADH dehydrogenase flavoprotein 3- like | 12018282 | 0.848 | 0.743 | 0.972 | 11 | 37 |
| 10 | NADH dehydrogenase flavoprotein 2 | 51092268 | 0.806 | 0.721 | 0.902 | 10 | 46 |
| 11 | ATP synthase, H+ transporting, mitochondrial F1 complex, α1 | 40538742 | 0.805 | 0.753 | 0.860 | 25 | 49 |
| 12 | Predicted: similar to cytochrome c-1 | 109482353 | 0.799 | 0.698 | 0.917 | 8 | 43 |
| 13 | Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 57114330 | 0.796 | 0.716 | 0.885 | 11 | 48 |
| 14 | ATP synthase, H+ transporting, mitochondrial F1 complex, β | 54792127 | 0.784 | 0.748 | 0.785 | 24 | 60 |
| 15 | Electron-transfer-flavoprotein, β | 51948412 | 0.775 | 0.685 | 0.878 | 8 | 35 |
| 16 | Ubiquinol-cytochrome c reductase core protein II | 55741544 | 0.765 | 0.679 | 0.864 | 10 | 33 |
| 17 | Creatine kinase, muscle | 6978661 | 0.761 | 0.701 | 0.828 | 19 | 53 |
| 18 | NADH dehydrogenase Fe-S protein 2 | 58865384 | 0.712 | 0.557 | 0.909 | 3 | 9 |
| 19 | Creatine kinase, ubiquitous, mitochond. | 60678254 | 0.602 | 0.486 | 0.742 | 2 | 6 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
A predicted protein whose observed peptides matched an existing protein in the Refseq database.
The analysis of the effect of age on NADH dehydrogenase 1 was more complex, where the β7 (iTRAQ ratio 1.60), β8 (iTRAQ ratio 1.40), β9 (iTRAQ ratio 1.19) and α13 (iTRAQ ratio 1.17) subunits were found up-regulated. However, the α5 polypeptide (iTRAQ ratio 0.869) was found at diminished levels. Similarly, while the F6 subunit of the F0 ATP synthase complex was found at elevated levels (iTRAQ ratio 1.10), the α (iTRAQ ratio 0.805) and β (iTRAQ ratio 0.784) subunits of the F1 ATP synthase complex were found at decreased levels. The iTRAQ data from aging rat hearts also demonstrate an energy deficiency similar to that of an energy-stressed hypertrophic heart32,33, where decreased levels of the mitochondrial (iTRAQ ratio 0.761) and muscle isoforms of creatine (iTRAQ ratio 0.602) kinase were observed.
Oxidative stress was implicated by the list of differentially expressed proteins (Table 6). Oxidative stress often results in changes in the distribution of protein folding chaperones; therefore these proteins were also included in Table 6. Of those proteins associated with chaperone activity and protein folding, two were found at enhanced iTRAQ levels in aged rats: αB-crystallin (iTRAQ ratio 1.31), and heat shock 27 protein 1 (iTRAQ ratio 1.13). Increased expression of these two protein chaperones is consistent with a similar increase in expression found in aging skeletal muscle34. In contrast, mitochondrial stress protein 70 (iTRAQ ratio 0.762), and heat shock protein 8 (iTRAQ ratio 0.596) were found at reduced levels. Further evaluation is necessary to understand the functional significance of the increase in αB-crystallin and heat shock protein 27 with the concomitant decrease in stress protein 70 and heat shock protein 8.
Table 6.
Differential expression of antioxidant proteins or chaperones.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq. Cover. |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | αB-Crystallin | 16905067 | 1.31 | 1.19 | 1.43 | 9 | 59 |
| 2 | Glutathione-S-transferase π isoform | 25453420 | 1.25 | 1.12 | 1.39 | 8 | 52 |
| 3 | Complement component 1 Q subunit binding protein | 48675371 | 1.21 | 1.07 | 1.36 | 7 | 36 |
| 4 | Heat shock 27 protein 1 | 94400790 | 1.13 | 1.02 | 1.26 | 9 | 60 |
| 5 | Glutathione peroxidase 1 | 30520381 | 1.12 | 1.01 | 1.25 | 9 | 51 |
| 6 | DJ-1 protein | 16924002 | 0.831 | 0.692 | 0.998 | 4 | 38 |
| 7 | Superoxide dismutase 2 (SOD2) | 8394331 | 0.813 | 0.734 | 0.900 | 9 | 42 |
| 8 | Glutathione-S-transferase μ type 3 | 13592152 | 0.802 | 0.714 | 0.902 | 13 | 61 |
| 9 | Glutathione-S-transferase Ω1 | 56090550 | 0.770 | 0.601 | 0.987 | 2 | 10 |
| 10 | Stress-70 protein, mitochondrial precursor | 116242506 | 0.762 | 0.67 | 0.867 | 11 | 20 |
| 11 | Peroxiredoxin 5 precursor | 16758404 | 0.756 | 0.659 | 0.869 | 7 | 40 |
| 12 | Peroxiredoxin 3 | 11968132 | 0.729 | 0.629 | 0.847 | 5 | 21 |
| 13 | Heat shock protein 8 | 13242237 | 0.596 | 0.540 | 0.657 | 18 | 37 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
Three additional proteins that participate in responses to oxidative stress were found up-regulated including: glutathione S-transferase π isoform (iTRAQ ratio 1.25), the Q subunit of complement component 1 (iTRAQ ratio 1.21), and glutathione peroxidase (iTRAQ ratio 1.12). Five stress proteins found at reduced levels included the DJ-1 protein (iTRAQ ratio 0.831), superoxide dismutase 2 (SOD2, iTRAQ ratio 0.813), glutathione transferase μ type 3 (iTRAQ ratio 0.802), glutathione-S-transferase Ω1 (iTRAQ ratio 0.770), peroxiredoxin 5 (iTRAQ ratio 0.756), and peroxiredoxin 3 (iTRAQ ratio 0.729).
Eighteen structural proteins were found differentially expressed (Table 7). Myosin heavy chain 7 (iTRAQ ratio 2.07) was found up-regulated to relatively high levels. Microtubule-associated protein 1A (iTRAQ ratio 1.27) and microtubule-associated protein 4 (iTRAQ ratio 1.21) were found at enhanced levels. Cysteine-rich protein 3, which is also known as the Muscle LIM protein that acts as a molecular biosensor that promotes myogenic differentiation35, was found at elevated levels (iTRAQ ratio 1.22). The “four and a half LIM domains 1” protein (FHL1, iTRAQ ratio 1.33) was present at elevated levels in the aging rat, a finding in agreement with a previous study on levels of FHL1 mRNA in β-adrenergic receptor-induced cardiomyopathy36. The heart-specific protein FHL2, which regulates myofibrillogenesis and connects the sarcomere to metabolic enzymes37, was found at a diminished ratio of 0.644 in aged ventricular tissue.
Table 7.
Differential expression of structural proteins.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq. Cover. |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | Myosin heavy chain, polypeptide 7, β | 8393807 | 2.07 | 1.91 | 2.24 | 22 | 14 |
| 2 | Predicted: similar to Band 4.1-like protein 2 | 109460528 | 1.44 | 1.14 | 1.82 | 2 | 3 |
| 3 | Four and a half LIM domains 1 isoform 1 | 76781465 | 1.33 | 1.19 | 1.50 | 10 | 41 |
| 4 | α-spectrin 2 | 31543764 | 1.33 | 1.24 | 1.43 | 52 | 27 |
| 5 | Microtubule-associated protein 1 A | 13591886 | 1.27 | 1.02 | 1.59 | 4 | 27 |
| 6 | Predicted: similar to tensin | 109487302 | 1.24 | 1.10 | 1.38 | 14 | 12 |
| 7 | Cysteine-rich protein 3 | 16924004 | 1.22 | 1.10 | 1.36 | 10 | 65 |
| 8 | Microtubule-associated protein 4 | 66730360 | 1.21 | 1.03 | 1.41 | 9 | 14 |
| 9 | Myosin heavy chain, polypeptide 6, α | 8393804 | 1.19 | 1.13 | 1.25 | 74 | 38 |
| 10 | Spectrin β2 | 61557085 | 1.18 | 1.05 | 1.33 | 19 | 11 |
| 11 | Desmin | 11968118 | 0.912 | 0.850 | 0.976 | 25 | 62 |
| 12 | Troponin T2, cardiac | 6981666 | 0.906 | 0.847 | 0.970 | 20 | 48 |
| 13 | Tropomyosin 1, α isoform a | 78000188 | 0.903 | 0.856 | 0.952 | 35 | 68 |
| 14 | Troponin 1, type 3 | 8394469 | 0.896 | 0.820 | 0.979 | 15 | 65 |
| 15 | Tropomodulin 1 | 48675841 | 0.837 | 0.720 | 0.973 | 7 | 22 |
| 16 | Myosin, light polypeptide 3 | 6981240 | 0.835 | 0.790 | 0.885 | 20 | 85 |
| 17 | Predicted: similar to actinin α2 | 109506033 | 0.689 | 0.608 | 0.782 | 14 | 21 |
| 18 | Four and a half LIM domains 2 | 13928940 | 0.644 | 0.569 | 0.724 | 12 | 51 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
Five immune-related proteins were detected, all at enhanced levels (Table 8). Orosomucoid 1 (α1-acid glycoprotein; iTRAQ ratio 1.48) is a T-cell mitogen that can be absorbed by T-cells38, and is a hallmark of an acute phase response. Hemopexin (iTRAQ ratio 1.39), modulates lymphocyte activity by controlling their heme/iron levels, and also stimulates protein kinase C activity39. An actin-sequestering protein that promotes lymphocyte proliferation, thymosin β4 (iTRAQ ratio 1.38) is synthesized in many rat organs including the heart and brain40 and is particularly abundant within macrophages. Thymosin β4 has also been implicated in cardiac repair mechanisms41. Annexin 5 (iTRAQ ratio 1.40) and complement factor H (iTRAQ ratio 1.30) were also observed.
Table 8.
Differential expression of immune proteins.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq Cover. |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | Orosomucoid 1 | 16757980 | 1.48 | 1.18 | 1.86 | 3 | 13 |
| 2 | Annexin 5 | 6978505 | 1.40 | 1.31 | 1.50 | 21 | 76 |
| 3 | Hemopexin | 16758014 | 1.39 | 1.29 | 1.50 | 22 | 44 |
| 4 | Thymosin, β4 | 13592119 | 1.38 | 1.16 | 1.34 | 5 | 45 |
| 5 | Complement component factor H | 77861917 | 1.30 | 1.10 | 1.55 | 8 | 10 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
An additional 7 proteins that did not readily fit into any of the aforementioned groups were observed differentially expressed in aged ventricles compared to young ones (Table 9). Notably, we observed a decrease in the level of prolyl-4-hydroxylase β-polypeptide (iTRAQ ratio 0.783), the enzyme that is responsible for generating hydroxyproline within collagen and is a widely recognized marker for collagen synthesis42. The function of the ES1 protein (iTRAQ ratio 0.852) remains unclear, and a protein identified only as an open reading frame, hypothetical protein LOC362946 (iTRAQ ratio 0.763) has no known function.
Table 9.
Miscellaneous proteins demonstrating differential protein expression.
| Protein Identity | Accession Number |
iTRAQ Ratio |
Credible Interval |
# Pep. |
Seq. Cover. |
||
|---|---|---|---|---|---|---|---|
| Lower | Upper | (%) | |||||
| 1 | Transferrin | 61556986 | 1.44 | 1.34 | 1.53 | 37 | 50 |
| 2 | Hemoglobin α1 | 6981010 | 1.24 | 1.12 | 1.33 | 10 | 70 |
| 3 | Hemoglobin β-chain complex | 17985949 | 1.10 | 1.03 | 1.16 | 16 | 95 |
| 4 | Serine protease inhibitor α1 | 17985949 | 1.10 | 1.03 | 1.16 | 16 | 95 |
| 5 | ES1 protein | 51948422 | 0.852 | 0.756 | 0.964 | 9 | 47 |
| 6 | Prolyl-4-hydroxylase β polypeptide | 6981324 | 0.783 | 0.639 | 0.956 | 5 | 12 |
| 7 | Hypothetical protein LOC362946 | 56090401 | 0.763 | 0.603 | 0.960 | 2 | 14 |
The gi accession number obtained from the NCBI database
Cr. Int. 1 denotes the Credible interval #1
Cr. Int 2 denotes the Credible interval #2
# Pep. indicates the number of peptides observed for each protein
Seq Cover. Reports the sequence coverage reported on by the iTRAQ data, in percent.
Western blot quantitation
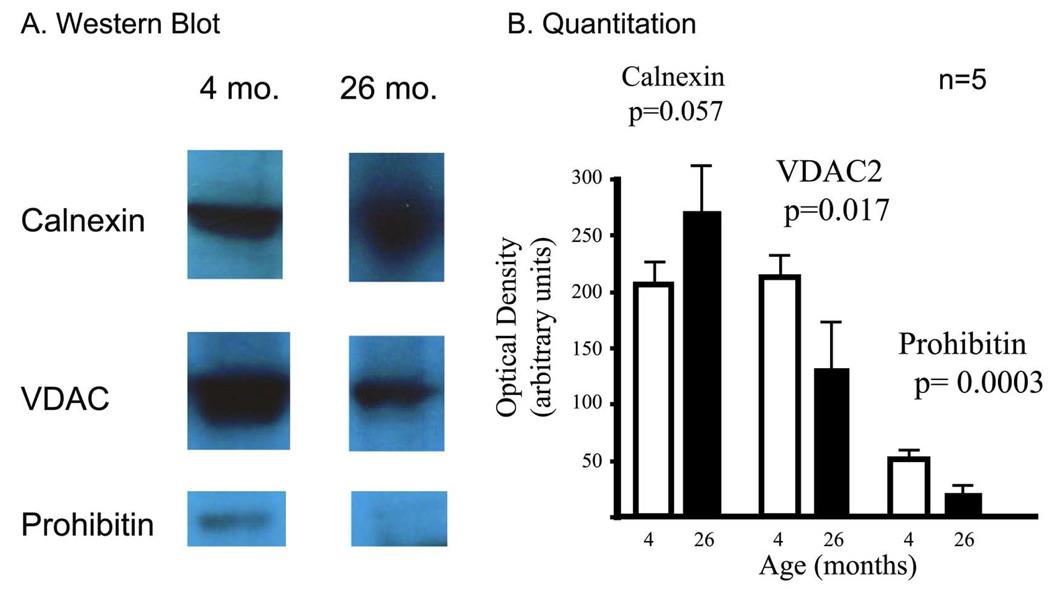
Using Western blots, the up- or down-regulation of three representative proteins (calnexin, the voltage dependent anion channel 2, and prohibitin) was assessed. The mean of 5 young and 5 aged rats, as well as the standard deviation and p-values were determined. It was not possible to assay the sixth homogenate from each group because iTRAQ analysis required consumption of these entire samples. These data are summarized in histogram format (Fig. 3). In all cases the trend towards up- or down-regulation for these three proteins confirmed the directionality revealed by the iTRAQ ratio. It is noteworthy that calnexin bands were markedly more diffuse in the 26-month old rats than in the 4-month old rats. Indeed, in the case of calnexin where the trend towards up-regulation was observed (blot ratio aged/young = 1.34), it was more difficult to conclude that the two data sets (young vs. old) were statistically different (p value = 0.57). However, for VDAC1 (blot ratio 0.62) and prohibitin (blot ratio 0.39) the results were more easily interpreted with p-value scores of 0.017 and 0.0003, respectively.
Figure 3. Western blotting, and quantification of western blot staining intensity.
Western blotting was performed on samples from young (4 month old) and aged tissue (26 months old). Fig. 3A: Representative blots are shown for calnexin[402–416], the voltage-dependent anion channel[201–208], and prohibitin[84–93]. Fig. 3B: A histogram demonstrating quantitation data is reported, using average intensity values for five of the six aged ventricles compared to five of the six young ventricles. A students’ T-Test was used to determine p-values.
Pathway analysis
As is the case for most proteomics studies, low abundance proteins may not be detected during iTRAQ analysis. Ingenuity Pathway Analysis software was used to generate network diagrams to elucidate signaling pathways in the left ventricle impacted by the aging process based on the iTRAQ results. The advantage that pathway analysis offers is that data-based results can be viewed in their cellular context and can be used to infer changes in proteins that were not detected. The top three networks that were reported on by proteins observed in our study were networks describing: 1) carbohydrate metabolism, cell signaling and energy production; 2) cell morphology, connective tissue disorders and inflammation, and 3) cellular assembly and organization.
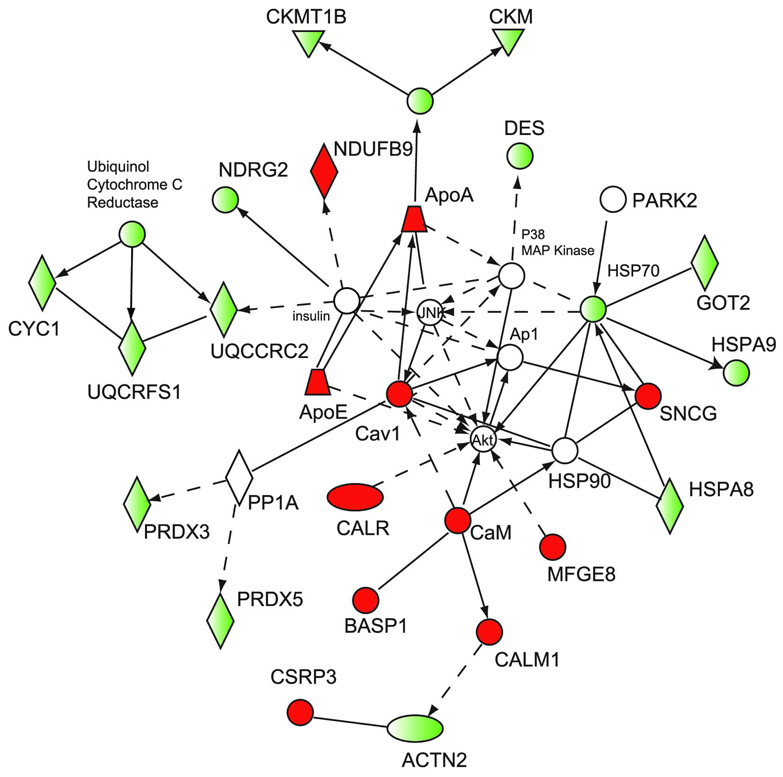
Of 27 proteins observed in the iTRAQ experiment, and assigned to the IPA network describing cell signaling and energy production, and 11 were found up-regulated in the iTRAQ experiment. In contrast, 16 were found down-regulated (Fig. 4). By inference, this network suggests that Akt is up-regulated in the aging ventricle, even though it was not observed directly. Akt, also known as protein kinase B, participates in anti-apoptotic signaling and cellular proliferation, and has been implicated in signaling events that direct hypertrophic cardiac remodeling. Of 16 proteins assigned to the network describing cellular organization, 12 were found up-regulated. Only one, superoxide dismutase 2, was found at decreased levels. Three additional proteins of this group were identified in our iTRAQ study but were not differentially expressed in the aged rat ventricle. In addition, analysis of the cellular assembly and organization network allowed us to infer that JNK, PKC and F-actin should be found at up-regulated levels in the aged left ventricle. This hypothesis will be evaluated in future work.
Figure 4. Proteins described by the network: cell signaling and energy production.
Network analysis of differentially expressed proteins was performed using the Ingenuity Pathways Analysis software. The first network describing proteins affecting cellular organization and morphology is presented. Proteins are represented as nodes. Nodes in red represent up-regulated proteins, while nodes in green represent down-regulated proteins. Proteins represented by white nodes were not observed. Solid lines indicate direct interactions or regulation, while dashed lines indicate indirect effects mediated by additional proteins.
The following abbreviations were used: Akt, protein kinase B; Ap1, adapter protein 1; ApoA, Apolipoprotein A; ApoE, Apolipoprotein E; BASP1, brain-attached signaling peptide 1; CalR, Calreticulin; Cav1, caveolin 1; CKM, creatine kinase (muscle); CKMT1B, creatine kinase (mitochondrial); CYC1, cytochrome c1; DES, desmin; GOT, glutamate oxaloacetate transaminase 2; HSPA8, Stress 70 protein; HSPA9, Heat shock protein 9; HSP70, heat shock protein 70; HSP90, heat shock protein 90; MFGE8, milf-fat globular-EGF factor 8; NDRG2, N-myc downstream regulated gene 2;; NDUF B9, NADH dehydrogenase (ubiquinol) β9; PARK2, parkin 2; PRDX3, perioxiredoxin 3; PRDX5, peroxiredoxin 5; PP1A, protein phosphatase 1A; UQCRFS1, uniquinol cytochrome C reductase Fe-S protein.
Discussion
The goal of this study was to broaden our understanding of the aging process in the heart by using the iTRAQ quantitative proteomics method to investigate differential protein expression between young and old rats. Even in the absence of cardiovascular disease there are changes in LV mass, volume, and geometry which occur during advancing age and can be important predictors of functional and clinical outcome. As a result of aging alone, the mass of the left ventricle can increase up to 15–20% during a person’s lifetime.
We chose to create a pooled control sample representing young ventricles, which offered two main advantages. First, the same pooled control sample representing young ventricles was used in both iTRAQ runs, allowing direct comparison of the data between runs. Second, use of a pooled control sample permitted investigation of 6 individual aged ventricles between the two iTRAQ runs, as opposed to individual assessment of young ventricles, which would have limited the number of aged ventricles investigated in one iTRAQ study. Additional issues concerning the distribution of means and variance in pooled proteomics samples have been described elsewhere43,44.
In all, we identified 117 proteins that were observed in each of two complete iTRAQ runs that examined differential protein expression in six aged rat left ventricles compared to a control pool of six young rats. Metabolic proteins represented the largest class of proteins differentially expressed in the iTRAQ study. In agreement with numerous other studies of aged ventricles32,33, the iTRAQ data also demonstrate impairments in fatty acid oxidation and cellular respiration, and it is noteworthy that four of the five enzymes participating in fatty acid oxidation were quantified in this study. The production of ATP by mitochondria is known to be compromised in aged hearts. Preston, et al.45, demonstrated changes in the transcriptome of mitochondrial proteins in aged hearts involved in fatty acid oxidation consistent with reduced oxygen utilization and ATP synthesis. Similarly, in hypertrophy, the heart increasingly relies on glycolysis for generating energy molecules, and this trend is reflected in the data by the down-regulation of two enzymes of the Krebs cycle, succinate dehydrogenase and fumarate hydratase. Decreased fatty acid oxidation is also an important consequence of the hypertrophic response, particularly in failing hearts; in the healthy myocardium fatty acid metabolism normally accounts for 60% of the heart’s energy requirements2.
In addition, creatine kinase, an enzyme necessary for the storage of energy in the form of phosphocreatine, was found depressed in the aged rat ventricles. Under normal physiological conditions ATP is converted to ADP; however, in the energy-stressed hypertrophic heart, for example, an energy deficiency develops, and, consequently, creatine kinase is actively repressed in order to conserve ATP32. In sum, the iTRAQ data paint a global picture of changes in energy metabolism in the aged ventricle that are similar in some regards to those associated with hypertrophic ventricles.
One protein signature of aged myocardium is an isoform switch in myosin heavy chain from α to β forms46. This switch is postulated to result from changes in gene expression of the two forms of myosin heavy chain and contribute to alterations in contractility characteristic of aged vs. young myocardium. The β isoform of myosin heavy chain displayed the highest differential in protein expression in aged ventricles increasing 2.07-fold over that of young tissue. Other proteins that might contribute to contractility that were differentially expressed in the aged heart include troponins, tropomyosin and actin α2.
A reduced capacity to respond to β2-adrenergic stimulation is another hallmark of aged ventricles. An uncoupling of β-adrenergic receptor to adenylate cyclase in aged myocytes has been proposed to limit cyclic adenine monophosphate (cAMP)-dependent phosphorylation of essential proteins that influence protein function and enhance contractility. iTRAQ analysis revealed a decrease in FHL2, a protein also decreased in failing myocardium47. Whether reductions in FHL2 in aged myocytes influences adenylate cyclase cellular localization and contributes to decreased β-adrenergic stimulation is an interesting prospect requiring further study.
Several circulatory proteins were observed, despite perfusion of the rat hearts prior to dissection. MALDI is an exquisitely sensitive technique, and it may be the case that even relatively low levels of remaining proteins (i.e. hemoglobin) are being detected. Alternatively, perhaps the circulating proteins are interacting with extracellular matrix. Although it would be ideal to remove these circulatory and/or blood proteins with a depletion column, it is yet unclear whether the depletion column methods would adversely affect quantitative analysis of the extracts.
Relationship to the literature on aging ventricle proteomics
In the literature there are a relatively limited number of proteomics studies that address aging of cardiac ventricles. Using a 2D PAGE approach, Dai, et al.14, reported age-related differential expression of 73 proteins within the left ventricle of 23-month old male mice, the majority of which were metabolic proteins. In the iTRAQ study reported here, 17 of these 73 proteins were found differentially expressed. Compared to the present study, the directionality of differential protein expression (i.e. up- or down-regulated) was the same for 10 proteins observed in the 2D PAGE study: aldolase A, creatine kinase, desmin, the DJ-1 protein, electron transferring flavoprotein, malate dehydrogenase, peroxisomal enoyl-CoA hydratase, phosphoglycerate kinase, prohibitin, SOD2, and VDAC1. However, in the present study increased concentrations of apolipoprotein A1, long-chain acetyl-CoA dehydrogenase, G3P dehydrogenase were observed, in contrast to the results of Dai, et al.14. Finally, 11 proteins that were differentially expressed in the 2D-PAGE study of mice were not found differentially expressed in the iTRAQ data (aconitase 2, adenylate kinase 1, cytochrome c oxidase Va, superoxide dismutase 1, the mitochondrial GrpE-like 1, lactate dehydrogenase B, myoglobin, 3-oxoacid dehydrogenase, peroxiredoxin 1 and the voltage-dependent anion-selective channel 2). Further investigation will be necessary to establish whether methodological differences between these two studies or innate differences between the aged ventricles from mice and rats are responsible for these contrasting results.
There are numerous potential factors accounting for the differences between this report and the work of Dai, et al.14 Perhaps the most compelling is that the 23-month old mice employed in the aforementioned 2D PAGE study were pre-senescent. In contrast, the rats employed in the iTRAQ work reported here are senescent, where the literature reports marked physiological changes in rats of this age in terms of energy balance and physiology. The 2D PAGE study also reported multiple protein spots for each of the following proteins: fumarate hydratase, L-3-hydroxyacyl-CoA dehydrogenase, peroxiredoxin 3, and triose phosphate isomerase, where individual gel spots demonstrated varying degrees and directionality of differential protein expression. The presence of multiple 2D gel spots for each protein may reflect post-translational modification of these four proteins. Post-translational modifications were not assessed in the present study, therefore the iTRAQ data reflect overall changes in the expression of these two proteins. It is also noteworthy that the 2D PAGE work analyzed both soluble and insoluble extracts prepared from the left ventricle, while the iTRAQ protocol employed in the present study used 1M urea and 1% CHAPS as detergents to solubilize ventricular tissue.
In independent work, Richardson, et al.15, used 2D PAGE to identify 46 differentially expressed proteins in aging rats, of which 7 of these were identified in the present iTRAQ study. The iTRAQ analysis confirmed up-regulation of heat shock protein 27 and transferrin, and both studies demonstrated down-regulation of creatine kinase in aged rats. In contrast to the Richardson, et al.15 study, however, desmin, myosin light chain 3, phosphatidylethanolamine binding protein, pyruvate dehydrogenase (lipoamide) β and tropomyosin were observed to be down-regulated in the aged rat iTRAQ study reported here. Again, the reasons for these discrepancies are not clear, but the ages of the rats and some of the methodological differences could be factors.
Concluding remarks
At present, a direct quantitative proteomic comparison of failing hearts to aged hearts has not been made, so it is not yet clear if age-related left ventricular hypertrophy possesses its own unique protein signature distinct from that of more chronic, pathological forms of LVH. The 117 proteins found differentially expressed in the aged left ventricle should be examined further in mechanistic studies of aging and heart failure models to determine whether they are uniquely affected by aging, or are impacted equally by heart failure.
We are currently investigating additional methods for isolating cellular organelles and compartments in order to enrich proteins of interest in ventricular extracts. It is common knowledge that, when mass spectrometry is employed in these proteomics experiments on total cell extracts, that high-abundance proteins are preferentially identified with greater confidence, at the expense of those present at lower abundance. This is readily observed in this experiment, where we see many circulating and metabolic proteins, at the expense of extracellular matrix proteins-related (2 observed). Enhanced enrichment strategies will allow investigation of lower-abundance proteins that may provide greater insight into the aged ventricle proteome.
In sum, a relatively large number of differentially expressed proteins representing seven functional classes were identified by using the iTRAQ approach, establishing a baseline for the proteomic signature of AR-LVH as determined by iTRAQ analysis. These observations pave the way for direct comparison of differential protein expression in the aged ventricle to ventricles exposed to extreme stress or injury, such as models of active heart failure.
Supplementary Material
ACKNOWLEDGMENT
We thank Tyler Rentz for his technical assistance on this project. We also thank Drs. Craig Beeson and Daniel Knapp for many insightful discussions and Dr. Lashanda Waller for proofreading this manuscript. The authors acknowledge the following sources of support: NHLBI contract (NIH NO1-HV-28281, KLS) and an IRACDA scholarship NIH K12GMO81265 to JEG.
SUPPORTING INFORMATION AVAILABLE:
The supplementary file presents the printout generated by the iQuantitator statistical software suite. This material is available free of charge on the Internet at http://pubs.acs.org.
Abbreviations
- 2D PAGE
2-dimensional polyacrylamide gel electrophoresis
- ACN
acetonitrile
- AR-LVH
age-related left ventricular hypertrophy
- BCA
bicinchoninic acid
- BW
body weight
- CHAPS
3-[(3-cholamidopropyl)-dimethylamino]-1-propanesulfonate
- EDTA
ethylene diamine tetraacetic acid
- HCl
hydrochloric acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HPLC
high performance liquid chromatography
- IPA
ingenuity pathways analysis
- iTRAQ
isobaric Tagging for Absolute and Relative Quantification
- IPA
ingenuity pathways analysis
- KCl
potassium chloride
- KH2PO4
potassium phosphate monobasic
- K2HPO4
potassium phosphate dibasic
- LC-MALDI
liquid chromatography Matrix Associated Laser Desorption Ionization Mass Spectrometry
- LV
left ventricle
- RP
reversed-phase
- RV
right ventricle
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- RIPA
radio-immunoprecipitation assay
- SCX
strong cation exchange
- SDS
sodium dodecyl sulfate
- TFA
trifluoroacetic acid
- TL
tibia length
- TOF
time of flight
- VDAC
voltage-dependent anion channel
REFERENCES
- 1.Walker EM, Jr, Nillas MS, Mangiarua EI, Cansino S, Morrison RG, Perdue RR, Triest WE, Wright GL, Studeny M, Wehner P, Rice KM, Blough ER. Age-associated changes in hearts of male Fischer 344/Brown Norway F1 rats. Ann Clin Lab Sci. 2006;36(4):427–438. [PubMed] [Google Scholar]
- 2.Bernhard D, Laufer G. The aging cardiomyocyte: a mini-review. Gerontology. 2008;54(1):24–31. doi: 10.1159/000113503. [DOI] [PubMed] [Google Scholar]
- 3.Lakatta EG. Cardiovascular aging in health sets the stage for cardiovascular disease. Heart, Lung, and Circ. 2002;11:76–91. doi: 10.1046/j.1444-2892.2002.00126.x. [DOI] [PubMed] [Google Scholar]
- 4.Schoenhoff PS, Fu Q, van Eyk JE. Cardiovascular proteomics: implications for clinical applications. Clin. Lab. Med. 2009;29(1):87–99. doi: 10.1016/j.cll.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ping P. Getting to the heart of proteomics. N. Engl. J. Med. 2009;360(5):532–534. doi: 10.1056/NEJMcibr0808487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didangelos A, Simper D, Monaco C, Mayr M. Proteomics of acute coronary syndromes. Curr. Atheroscler. Rep. 2009;11(3):188–185. doi: 10.1007/s11883-009-0030-x. [DOI] [PubMed] [Google Scholar]
- 7.Sheikh AM, Barret C, Villamizar N, et al. J. Right ventricular hypertrophy with early dysfunction: a proteomics study in a neonatal model. Thoracic. Cardiovasc. Surg. 2009;137(5):1146–1153. doi: 10.1016/j.jtcvs.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Perlman DH, Bauer SM, Ashrafian H, et al. Mechanistic insights into nitrite-induced cardioprotection using an integrated metabolomic/proteomic approach. Circ. Res. 2009;104(6):796–804. doi: 10.1161/CIRCRESAHA.108.187005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsey ML, Goshorn DK, Comte-Walters S, et al. A multidimensional proteomic approach to identify hypertrophy-associated proteins. Proteomics. 2006;6(7):2225–2235. doi: 10.1002/pmic.200500013. [DOI] [PubMed] [Google Scholar]
- 10.Dwyer JP, Ritchie ME, Smyth GK, Harrap SB, Delbridge L, Domenighetti AA. Di Nicolantonio, R. Myocardial gene expression associated with genetic cardiac hypertrophy in the absence of hypertension. Hypertens Res. 2008;31(5):941–955. doi: 10.1291/hypres.31.941. [DOI] [PubMed] [Google Scholar]
- 11.Rysä J, Leskinen H, Ilves M, Ruskoaho H. Distinct upregulation of extracellular matrix genes in transition from hypertrophy to hypertensive heart failure. Hypertension. 2005;45(5):927–933. doi: 10.1161/01.HYP.0000161873.27088.4c. [DOI] [PubMed] [Google Scholar]
- 12.Boluyt MO, Bing OH. Matrix gene expression and decompensated heart failure: the aged SHR model. Cardiovasc Res. 2000;46(2):239–249. doi: 10.1016/s0008-6363(00)00043-2. [DOI] [PubMed] [Google Scholar]
- 13.Anderson L, Sielhamer J. A comparison of selected mRNA and protein abundances in the human liver. Electrophoresis. 1997;18:553–557. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 14.Unwin RD, Smith DL, Blinco D, Wilson CL, et al. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood. 2006;107(12):4687–4694. doi: 10.1182/blood-2005-12-4995. [DOI] [PubMed] [Google Scholar]
- 15.Dai Q, Escobar GP, Hakala KW, Lambert JM, Weintraub ST, Lindsey ML. The left ventricle proteome differentiates middle-aged old left ventricles in mice. J. Proteome Res. 2008;7(2):756–765. doi: 10.1021/pr700685e. [DOI] [PubMed] [Google Scholar]
- 16.Richardson MR, Lai X, Mason SB, Miller SJ, Witzmann FA. Differential protein expression during aging in ventricular myocardium of Fischer 344 x Brown Norway hybrid rats. Exp. Gerontol. 2008;43(10):909–919. doi: 10.1016/j.exger.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54(11):492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- 19.Bradshaw AD, Baicu CF, Rentz TJ, Van Laer AO, Boggs Lacy JM, Zile MR. Circulation. 2009;119(2):269–280. doi: 10.1161/CIRCULATIONAHA.108.773424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Hill EG, Schwacke JH, Comte-Walters S, Slate EH, Oberg AL, Eckel-Passow JE, Therneau TM, Schey KL. A statistical model for iTRAQ data analysis. J. Proteome Res. 2008;7(8):3091–3101. doi: 10.1021/pr070520u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlin BP, Louis TA. Boca Raton: Chapman & Hall/CRC; 2000. Bayes and Empirical Bayes methods for data analysis. [Google Scholar]
- 23.Team RDC. Computer Software. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- 24.Rentz TJ, PF, Bornstein P, Sage EH, Bradshaw AD. SPARC regulates processing of procollagen I and collagen fibrillogenesis in dermal fibroblasts. J Biol Chem. 2007;282(30):22062–22071. doi: 10.1074/jbc.M700167200. [DOI] [PubMed] [Google Scholar]
- 25.Oberg AL, Mahoney JE, Eckel-Passow CJ, et al. Statistical Analysis of Relative Labeled Mass Spectrometry Data from Complex Samples Using ANOVA. J. Proteome Res. 2008;7(1):225–233. doi: 10.1021/pr700734f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 27.Surguchov A, Palazzo RE, Surgucheva I. Gamma synuclein: subcellular localization in neuronal and non-neuronal cells and effect on signal transduction. Cell Motil. Cytoskeleton. 2001;49(4):218–228. doi: 10.1002/cm.1035. [DOI] [PubMed] [Google Scholar]
- 28.Ltic S, PM, Mladenovic A, Raicevic N, Ruzdijic S, Rakic L, Kanazir S. Alpha-synuclein is expressed in different tissues during human fetal development. J. Mol. Neurosci. 2004;22(3):199–204. doi: 10.1385/jmn:22:3:199. [DOI] [PubMed] [Google Scholar]
- 29.Komuro A, Masuda Y, Kobayashi K, Babbitt R, Gunel M, Flavell RA, Marchesi VT. The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc. Natl. Acad. Sci USA. 2004;101(12):4053–4058. doi: 10.1073/pnas.0308619101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahoo SK, Kim DH. Calumenin interacts with SERCA2 in rat cardiac sarcoplasmic reticulum. Mol. Cells. 2008;26(3):265–269. [PubMed] [Google Scholar]
- 31.Burchfield JG, Lennard AJ, Narasimhan S, Hughes WE, Wasinger VC, Corthals GL, Okuda T, et al. Akt mediates insulin-stimulated phosphorylation of NRDG2: evidence for cross-talk with protein kinase C theta. J. Biol. Chem. 2004;279(18):18623–18632. doi: 10.1074/jbc.M401504200. [DOI] [PubMed] [Google Scholar]
- 32.Taha M, Lopaschuk GD. Alterations in energy metabolism in cardiomyopathies. Ann. Med. 2007;39(8):594–607. doi: 10.1080/07853890701618305. [DOI] [PubMed] [Google Scholar]
- 33.Smith CS, Bottomley PA, Schulman SP, Gerstenblith G, Weiss RG. Altered creatine kinase adenosine triphosphate kinetics in failing hypertrophied human myocardium. Circulation. 2006;114(11):1151–1158. doi: 10.1161/CIRCULATIONAHA.106.613646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung L, Ng YC. Age-related alterations in expression of apoptosis regulatory proteins and heat shock proteins in rat skeletal muscle. Biochim Biophys Acta. 2005;1762(1):103–109. doi: 10.1016/j.bbadis.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79(2):221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 36.Gaussin V, Tomlinson JE, Depre C, et al. Common genomic response in different mouse models of β-adrenergic-induced cardiomyopathy. Circulation. 2003;108(21):2926–2933. doi: 10.1161/01.CIR.0000101922.18151.7B. [DOI] [PubMed] [Google Scholar]
- 37.Kong Y, Shelton JM, Rothermel B, et al. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to β-adrenergic stimulation. Circulation. 2001;103(22):2731–2738. doi: 10.1161/01.cir.103.22.2731. [DOI] [PubMed] [Google Scholar]
- 38.Dirienzo W, Stefanini GF, Miribel L, et al. Alpha 1-acid glycoprotein (alpha 1-AGP) on the membrane of human lymphocytes: possible involvement in cellular activation. Immunol. Lett. 1987;15(2):167–170. doi: 10.1016/0165-2478(87)90049-6. [DOI] [PubMed] [Google Scholar]
- 39.Smith A, Eskew JD, Borza CM, Pendrak M, Hunt RC. Role of heme-hemopexin in human T-lymphocyte proliferation. Exp. Cell. Res. 1997;232(2):246–254. doi: 10.1006/excr.1997.3526. [DOI] [PubMed] [Google Scholar]
- 40.Hannappel E, Xu GJ, Morgan J, Hempstead J, Horecker BL. Thymosin &beta4: a ubiquitous peptide in rat and mouse tissues. Proc. Nat. Acad. Sci., USA. 1982;79(7):2172–2175. doi: 10.1073/pnas.79.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432(7016):466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- 42.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22(1):15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 43.Karp NA, Lilley KS. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics. 2009;9(2):388–397. doi: 10.1002/pmic.200800485. [DOI] [PubMed] [Google Scholar]
- 44.Gan CS, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J. Proteome Res. 2007;6(2):821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 45.Preston CC, Oberlin AS, Holmuhamedov EL, et al. Aging-induced alterations in gene transcripts and functional activity of mitochondrial oxidative phosphorylation complexes in the heart. Mech. Aging Dev. 2008;129(6):304–312. doi: 10.1016/j.mad.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long X, Boluyt MO, O’Neill L, et al. Myocardial retinoid X receptor, thyroid hormone receptor, and myosin heavy chain gene expression in the rat during adult aging. J Gerontol. A Biol. Sci. Med. Sci. 1999;54(1):B23–B27. doi: 10.1093/gerona/54.1.b23. [DOI] [PubMed] [Google Scholar]
- 47.Bovil E, Westaby S, Crisp A, Jacobs S, Shaw T. Reduction of four-and-a-half LIM-protein 2 expression occurs in human left ventricular failure and leads to altered localization reduced activity of metabolic enzymes. J Thorac. Cardiovasc. Surg. 2009;137(4):853–861. doi: 10.1016/j.jtcvs.2008.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.