Abstract
The MvaT and MvaU proteins belonging to the H-NS family were identified as DNA-binding proteins that interact with the regulatory region of the aotJQMOP-argR operon for arginine uptake and regulation. Recombinant MvaT and MvaU proteins were purified, and binding of these purified proteins to the aotJ regulatory region was demonstrated using electromobility shift assays. Polyclonal antibodies against purified MvaT and MvaU were prepared and employed in supershift assays to support these observations. Knockout mutations resulting in a single lesion in mvaT or mvaU, as well as knockout mutations resulting in double lesions, were constructed using biparental conjugation, and the absence of MvaT and MvaU in the resulting mutants was confirmed by immunoblot analysis. Using measurements of the β-galactosidase activities from aotJ::lacZ fusions in the mutants and the parental strain, it was found that MvaT and MvaU serve as repressors in control of aotJ expression. The effects of MvaT and MvaU on pyocyanin synthesis and CupA fimbrial expression in these mutants were also analyzed. Pyocyanin synthesis was induced in the single mutants but was completely abolished in the double mutant, suggesting that there is a complicated regulatory scheme in which MvaT and MvaU are essential elements. In comparison, MvaT had a more profound role than MvaU as a repressor of cupA expression; however, a combination of MvaT depletion and MvaU depletion had a strong synergistic effect on cupA. Moreover, prophage Pf4 integrated into the chromosome of Pseudomonas aeruginosa PAO1 was activated in an mvaT mvaU double mutant but not in a single mutant. These results were supported by purification and nucleotide sequencing of replicative-form DNA and by the release of phage particles in plaque assays. In summary, the mvaT mvaU double mutant was viable, and depletion of MvaT and MvaU had serious effects on a variety of physiological functions in P. aeruginosa.
The MvaT and MvaU proteins of Pseudomonas aeruginosa belong to the H-NS family of small DNA-binding proteins. MvaT was initially identified in P. mevalonii as a positive regulator for mevalonate catabolism (25). Subsequently, MvaT homologues have been identified in other pseudomonads based on structural and functional similarities; five homologues have been identified in P. putida, three homologues have been identified in P. fluorescens, four homologues have been identified in P. syringae, and two homologues have been identified in P. aeruginosa. In P. putida, the TurA protein represses the Pu promoter of the TOL plasmid in a temperature-dependent manner (24). In P. fluorescens, the MvaT and MvaV proteins regulate the expression of two biocontrol exoproducts, 2,4-diacetyl phloroglucinol and pyoluteorin (1). In P. aeruginosa, MvaT is involved in quorum-sensing responses and biofilm formation. Inactivation of mvaT resulted in increased production of PA-IL lectin and the toxic exoproduct pyocyanin, reduced biofilm formation, increased drug resistance, and reduced swarming motility (5, 33). In addition, the MvaT protein is involved in the phase-variable expression of the fimbrial cup genes involved in biofilm formation. Moreover, DNA microarray analysis has shown that more than 150 genes are influenced by the MvaT protein (27).
The MvaU protein shows high levels of similarity to MvaT and can perform some of the MvaT regulatory functions (28). Like other H-NS-related proteins, MvaT and MvaU possess two distinct domains: the N terminus for oligomerization and the C terminus for DNA-binding activity. The MvaT and MvaU proteins can interact through their N-terminal regions and form hetero- and homodimers (28). In general, mvaT mutation seems to have a much more profound effect than mvaU mutation. In a study using chromatin immunoprecipitation coupled with DNA microarrays, the potential MvaT and MvaU binding sites on the genome of P. aeruginosa were identified (3). In the same report, it was concluded that loss of both MvaT and MvaU from the cell cannot be tolerated.
In this study, we identified MvaT and MvaU as components of a nucleoprotein complex that controls the aotJQMOP-argR operon for arginine uptake and regulation (21, 22). Using mvaT and mvaU single- and double-mutant constructs, we also demonstrated the consequences of MvaT and MvaU depletion for prophage activation, pyocyanin synthesis, and fimbrial cupA gene expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains, plasmids, and constructs used in this study are listed in Table 1. Minimal medium P (12) containing carbon and nitrogen sources at concentrations of 20 mM was used for growth of P. aeruginosa. Luria-Bertani (LB) medium (17) was used for Escherichia coli and P. aeruginosa transformation, and the following supplements were added when they were required: 100 μg/ml ampicillin (E. coli), 100 μg/ml carbenicillin (P. aeruginosa), 100 μg/ml tetracycline (P. aeruginosa), 100 μg/ml gentamicin (P. aeruginosa), and 50 μg/ml 5-bromo-3-indolyl-β-d-galactopyranoside (X-Gal). For pyocyanin production, a defined minimal medium containing alanine and glycerol was used as described by Frank and Demoss (8).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) supE44 λ−thi-1 gyrA96 relA1 | Bethesda Research Laboratories |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 11 |
| BL21 | F−ompT lon hsdSB(rB− mB−) gal dcm | Novagen |
| P. aeruginosa strains | ||
| PAO1 | Wild type | 12 |
| PAO1-Sm | Spontaneous Smr mutant of PAO1 | 13 |
| PAO501 | argR::Gm | 23 |
| PAO4460 | rpoN::ΩKm | 20 |
| PAO-T | mvaT::Gmr mutant of PAO1 | This study |
| PAO-U | mvaU::Tetr mutant of PAO1 | This study |
| PAO-TU | mvaT::GmrmvaU::Tetr mutant of PAO1 | This study |
| CupA | cupA1::lacZ genomic reporter fusion of PAO1 | 28 |
| CupA-Sm | Spontaneous Smr mutant of CupA | This study |
| CupA-T | mvaT::Gmr mutant of CupA | This study |
| CupA-U | mvaU::Tetr mutant of CupA | This study |
| CupA-TU | mvaT::GmrmvaU::Tetr mutant of CupA | This study |
| Plasmids | ||
| pRTP2 | Apr Sms conjugation vector derived from pRTP1 with deletion of EcoRI site | 16 |
| pST500 | aotJ::lacZ translational fusion of pQF52 | 21 |
| pST500 M | pST500 with mutation of −10 site of promoter P2 | This study |
| pGEM-T Easy | Cloning vector | Promega |
| pBAD-HisA | Apr, protein expression vector by arabinose induction | Invitrogen |
| pHW1 | Apr, pBAD-HisA derivative for overexpression of MvaT | This study |
| pHW2 | Apr, pBAD-HisA derivative for overexpression of MvaU | This study |
| pUCP18 | E. coli-P. aeruginosa shuttle vector | 26 |
| pUCP-T | Complementation plasmid for mvaT gene, derived from pUCP18 | This study |
| pUCP-U | Complementation plasmid for mvaU gene, derived from pUCP18 | This study |
Purification of MvaT and MvaU from P. aeruginosa.
A culture of P. aeruginosa PAO4460 (rpoN::ΩKm) was grown in 6 liters of minimal medium P supplemented with 20 mM glutamate, 20 mM arginine, and 1 mM glutamine. The cells were harvested by centrifugation, washed once in 20 mM Tris-HCl (pH 7.5) with 1 mM EDTA, and resuspended in 50 ml (final volume) of the same buffer. Phenylmethylsulfonyl fluoride (1 mM) was added immediately before passage of the cell suspension through an Aminco French pressure cell at 16,000 lb/in2. The cell-free supernatant was collected by centrifugation at 48,000 × g for 20 min at 4°C and was then precipitated with 1% streptomycin sulfate and centrifuged under the same conditions. The resulting supernatant was applied to an ion-exchange column (HiLoad 26/10 Q-Sepharose HP; GE Healthcare) equilibrated with 20 mM Tris-HCl buffer (pH 7.5), and this was followed by elution with a linear 0 to 1 M KCl gradient. Fractions containing the target proteins with the desired DNA-binding activity eluted between 0.4 and 0.5 M KCl and were concentrated and desalted before injection into a heparin affinity column (GE Healthcare) equilibrated with 20 mM Tris-HCl buffer (pH 7.5). Fractions showing activity eluted between 0.56 and 0.72 M KCl in a linear 0 to 1 M KCl gradient. These fractions were concentrated and subjected to size exclusion chromatography (Superdex-200 HR 10/30; GE Healthcare) in 20 mM Tris-HCl (pH 7.5) with 0.5 M KCl.
Cloning of the mvaT and mvaU genes.
For protein overexpression, the mvaT and mvaU genes were PCR amplified from P. aeruginosa PAO1 chromosomal DNA using forward primers that contain a PciI restriction site and reverse primers that contain a BamHI restriction site, as follows: for mvaT, 5′-ACA TGT CCC TGA TCA ACG AAT ATC GC-3′ and 5′-GGA TCC TTA GCC GAG CAG GGT GGC CCA GCT-3′; and for mvaU, 5′-ACA TGT CCA AAC TTG CCG AGT TCC GC-3′ and 5′-GGA TCC TTA GCG TTG CAG CCA GGA TTG GAC-3′. The PCR products were purified and cloned into pGEM-T Easy (Promega), and positive clones were identified by sequence analysis. The genes were then cut from the pGEM-T Easy vector with PciI and BamHI and ligated into the pBAD-HisA expression vector digested with NcoI and BglII. The resulting plasmids used for overexpression of MvaT and MvaU were designated pHW1 and pHW2, respectively.
For complementation in P. aeruginosa, the intact mvaT and mvaU genes were amplified from PAO1 chromosomal DNA by using Pfu polymerase and the following primers: for mvaT, 5′-CAT GGA TCC AGC ACA GAC AAG GTA CCT GAC-3′ and 5′-ATC GGA TCC TTA GCC GAG CAG GGT GGC CCA-3′; and for mvaU, 5′-GGA TCC TTT AGA ACG GGA GTG AAA CGA ATG-3′ and 5′-GGA TCC TTA GCG TTG CAG CCA GGA TTC GAC-3′. The PCR products were digested with BamHI (underlined nucleotides) introduced by the primers and ligated into the pUCP18 vector digested with the same enzyme. The orientations and sequences of the insertions were confirmed by DNA sequence analysis. The resulting plasmids were designated pUCP-T and pUCP-U.
Purification of recombinant MvaT and MvaU.
Cultures of E. coli BL21 harboring pHW1 or pHW2 were grown in LB medium with ampicillin at 37°C. After induction with 0.2% arabinose for 4 h, the cells were harvested by centrifugation, washed once in 20 mM Tris-HCl (pH 7.5) with 1 mM EDTA, and stored at −80°C. For purification, the cell pellets were resuspended in the buffer mentioned above with 1 mM phenylmethylsulfonyl fluoride and broken by passage through an Aminco French pressure cell at 16,000 lb/in2 three times. The resulting crude extracts were treated on ice with DNase I for 1 h in the presence of 10 mM MgCl2. After centrifugation at 48,000 × g for 20 min at 4°C, the resulting pellets containing the overexpressed proteins were dissolved in a buffer containing 20 mM sodium phosphate (pH 7.0) and 1 mM EDTA and then injected into a HiPrep 16/10 SP FF column (GE Healthcare) and a HiPrep 16/10 heparin FF column (GE Healthcare). Fractions containing MvaT or MvaU were pooled and concentrated by filtration (Amicon Ultra-4 filter; Millipore), which was followed by separation with a size exclusion column (Superdex-200 HR 10/30; GE Healthcare) in 20 mM sodium phosphate (pH 7.0) with 1 M NaCl and 1 mM EDTA. The estimated molecular masses of purified MvaT and MvaU based on the elution volumes in gel filtration chromatography were 420 kDa (elution volume, 10.6 ml) and 90 kDa (13.9 ml), respectively.
Production of MvaT and MvaU antibodies and immunoblot analysis.
Polyclonal antibodies were raised against the MvaT and MvaU proteins by subcutaneous injection of the proteins into two rabbits (one rabbit for each protein) seven times, using a total of 7 mg purified MvaT or MvaU. Sera were collected 3 weeks after the final injection and stored at −20°C. For immunoblot analysis, protein samples were separated by electrophoresis and then electroblotted onto a polyvinylidene difluoride membrane and analyzed by using standard protocols with the MvaT and MvaU antibodies (1:1,500 dilution) and goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugates (1:3,000 dilution; Bio-Rad).
Electromobility shift assay (EMSA).
The pST500 plasmid was used as a template for PCR amplification of probe F501 (Fig. 1) with the following primers: 5′-TTG GCA ATT TAT GCT TGA GAT TTG-3′ and 5′-GTG CGA GCT TCT TCA TCT GGA AC-3′. The 316-bp amplified fragment was purified using a QIAquick gel extraction kit (Qiagen) and 5′ end labeled with [γ-32P]ATP (111 TBq/mmol; Perkin Elmer) in a reaction mixture (20 μl) containing 5 ng of the purified DNA fragment. The reaction was stopped by heat inactivation at 65°C for 10 min. The probe was purified by passage through a G-50 Sephadex column (Roche) to remove unincorporated [γ-32P]ATP.
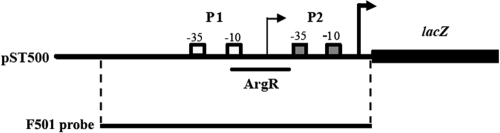
FIG. 1.
Schematic diagram of the aotJ regulatory region of pST500 and the F501 probe. The −10 and −35 regions of promoters P1 and P2 of the aotJQMOP-argR operon on pST500 are labeled. The arrowheads indicate the transcriptional start sites of these two promoters, and the ArgR binding site of ArgR is also indicated. Probe F501 is the DNA fragment used for demonstration of DNA-protein interactions in this study.
The EMSA was conducted using a reaction mixture (20 μl) containing 50 mM Tris-HCl (pH 7.0), 50 mM KCl, 1 mM EDTA, 10% (vol/vol) glycerol, 100 μg/ml acetylated bovine serum albumin (omitted in EMSA with crude extracts), and specific amounts of probe and protein samples. The reaction mixtures were equilibrated for 20 min at 25°C and loaded onto a 4% gel running in 1× Tris-acetate-EDTA. After drying, the gel was exposed to a phosphorimager plate (Fuji) and analyzed with a Fuji bioimaging analyzer.
To prepare crude extracts for EMSA, the cell cultures were grown in minimal medium P with supplements until mid-log phase. Cells were harvested by centrifugation at 7,000 × g for 10 min and washed once in 20 mM potassium phosphate (pH 7.0) containing 1 mM EDTA. The washed pellets were suspended in the same buffer, and the cells were ruptured by passage through a French pressure cell at 16,000 lb/in2. Crude cell extracts were obtained by centrifugation at 27,000 × g for 20 min to remove the cell debris. Protein concentrations were determined by the Bradford method with bovine serum albumin as the standard. The radioactively labeled DNA probe (2.08 × 10−9 M) was allowed to interact with protein extracts (4 μg) or purified ArgR (6 × 10−9 M). For EMSA with purified MvaT or MvaU, the 32P-labeled DNA probe (9.0 × 10−12 M) was allowed to interact with these proteins at the concentrations indicated below.
For EMSA with MvaT and MvaU antibodies, a PAO1 crude cell extract was prepared using a bacterial culture grown in minimal medium P supplemented with glutamate and arginine. The reaction mixture contained crude protein extract (2 μg) and the 32P-labeled DNA probe (9.4 × 10−10 M) in the buffer described above along with antibodies (10-fold serial dilutions from the stock preparations).
Construction of mvaT and mvaU mutants.
To knock out mvaT or mvaU, the flanking regions were PCR amplified with the following primer pairs: for the upstream arm of mvaT, 5′-CAT AAG CTT CGC GGG CGA TCG GGG CGA AAG-3′ and 5′-ATC GAA TTC TTG TCT GTG CTG AGT GGC GGT-3′; for the downstream arm of mvaT, 5′-ATC GAA TTC ACG AAG AAC GCC AGC CCA GTG-3′ and 5′-CAT GGA TCC GAT GTC CGC GCC ACC ATT GCC-3′; for the upstream arm of mvaU, 5′-AAG CTT CAA GGC GAT CTT CAA GCC GAT CTA-3′ and 5′-GAA TTC TTC GTT TCA CTC CCG TTC TAA AAA-3′; and for the downstream arm of mvaU, 5′-GAA TTC AGC CGG TTT TCC CGA CGG CAT CCT-3′ and 5′-GGA TCC CCG CCA TTG TCG CAT TCG CGC AGC-3′. The two flanking arms for each gene with the configurations 5′-HindIII-left arm-EcoRI-3′ and 5′-EcoRI-right arm-BamHI-3′ were ligated and cloned into the HindIII and BamHI sites of pRTP2. After insertion of a gentamicin resistance cassette (for mvaT) or a tetracycline resistance cassette (for mvaU) into the EcoRI site, the final recombinant plasmids were introduced into E. coli SM10 to serve as the donors in biparental conjugation. To construct mvaT or mvaU single-knockout mutants, PAO1-Sm or CupA-Sm was used as the recipient strain. After incubation at 37°C for 6 h, transconjugants were selected on plates containing gentamicin and streptomycin for the mvaT mutant or tetracycline and streptomycin for the mvaU mutant. To construct an mvaT mvaU double-knockout mutant, the mvaU single-knockout mutant was used as the recipient strain for further knockout of mvaT. After incubation at 37°C overnight, transconjugants were selected on plates supplemented with gentamicin, tetracycline, and streptomycin. The authenticity of the mutants was confirmed by PCR and Western blotting.
Construction of pST500M.
Plasmid pST500 was reported previously to be an aotJ::lacZ fusion covering the intact aotJ regulatory region of two tandem promoters, P1 and P2 (21). Plasmid pST500M was derived from pST500 by site-directed mutagenesis to degenerate the −10 sequence of the P2 promoter (Fig. 1). The mutation was generated by using a site-directed mutagenesis kit (Stratagene) with the following primers (the mutated sites are underlined): 5′-TTC GGC CTG TCG CCG ACG CGC CGC TCA CCC CAG-3′ and 5′-CTG GGG TGA GCG GCG CGT CGG CGA CAG GCC GAA-3′.
Pyocyanin quantitation assay.
Cells from fresh plates were inoculated into glycerol-alanine medium to obtain an initial optical density at 600 nm of 0.04 and then grown for 25 h. For pyocyanin extraction, 4 ml of the cell culture was mixed with 3 ml of chloroform with vortexing for 30 s. After centrifugation at 6,000 × g for 5 min, 2 ml of the chloroform layer containing a blue-green pigment was retrieved, and pyocyanin was extracted into 2 ml of 0.2 M HCl as a pink pigment. After appropriate dilution, the absorbance at 520 nm was determined, and the concentration of pyocyanin was determined using an extinction coefficient of 2,460 M−1 cm−1.
Phage plaque assays.
To prepare P. aeruginosa PAO1 cells as plating bacteria, the cell pellet from an overnight culture grown in LB medium was washed once with sterile 10 mM MgSO4 and suspended in the same buffer to obtain an adjusted optical density at 600 nm of around 1.0. For plaque assays, phages released into the spent medium of the cells grown overnight in LB medium were collected after centrifugation at 4,500 × g for 10 min. An aliquot of the supernatant (600 μl) was mixed with 90 μl of the plating bacterial suspension and incubated at 37°C for 30 min to allow attachment of phage particles to the cells. To plate the mixture, 3 ml of melted 0.7% top agar was added, vortexed briefly, and poured immediately onto a LB medium plate. After the agar solidified, the plate was incubated at 37°C overnight for plaque formation.
cupA gene expression.
Strain CupA was derived from PAO1 by insertion of a lacZ reporter gene right after the cupA1 gene, and the expression of lacZ was under the control of the cupA promoter (28). Further knockout of mvaT and/or mvaU was then conducted to obtain a set of CupA serial mutants. To check cupA gene expression, cells from fresh overnight cultures were inoculated and grown until the log phase in LB medium at 37°C with vigorous shaking. The β-galactosidase activity was then measured with ortho-nitrophenyl-β-d-galactopyranoside as the substrate.
RESULTS
Presence of an ArgR-independent nucleoprotein complex in the aotJ regulatory region.
The ArgR protein serves as an arginine-responsive transcriptional regulator in control of arginine and glutamate metabolism (22). Expression of the aotJQMOP-argR operon is subject to autoactivation by ArgR in response to the presence of l-arginine. In an attempt to search for transcriptional regulators other than ArgR that may participate in control of argR expression, we performed EMSA with crude extracts prepared from the argR mutant and its parental strain, PAO1, using the 316-bp aotJ regulatory region as the probe. The results of the EMSA revealed the presence of a DNA-protein complex independent of ArgR (C1 complex) (Fig. 2A).
FIG. 2.
EMSA with crude extracts or purified MvaT or MvaU. The reaction conditions and signal detection method used are described in Materials and Methods. (A) EMSA with crude cell extracts of P. aeruginosa PAO1 and mutant derivatives. The radioactively labeled DNA probe (2.08 × 10−9 M) was allowed to interact with protein extracts (4 μg) or purified ArgR (6 × 10−9 M). Control, probe only; PAO1, wild-type strain PAO1; PAO501, argR::Gm mutant; PAO4460, rpoN::ΩKm mutant; ArgR, control with purified ArgR. (B) Supershift by MvaT and MvaU antibodies. The reaction mixtures contained protein extracts (2 μg) of PAO1, 32P-labeled DNA probe (9.4 × 10−10 M), and antibodies diluted as indicated. C, control (DNA probe only); P, protein extract with no antibodies added; 1, 10−1, 10−2, 10−3, reaction mixtures with undiluted and 10-, 100-, and 1,000-fold-diluted MvaT or MvaU antibody, respectively. (C and D) EMSA performed with purified MvaT or MvaU at the concentrations indicated. The concentration of the 32P-labeled DNA probe used was 9.0 × 10−12 M.
Identification of MvaT and MvaU as proteins bound to the aotJ regulatory region.
To identify the DNA-binding protein(s) for the C1 nucleoprotein complex, a series of column chromatography experiments was performed as described in Materials and Methods to purify the protein(s) of interest from the crude extract of the PAO4460 (rpoN) mutant, which resulted in more intense formation of the C1 complex than experiments with PAO1 (Fig. 2A, lanes 2 and 4). After three column purification steps, the DNA-binding activity was in a set of protein fractions that consistently contained two polypeptides with molecular masses in the range from 14 to 16 kDa (Fig. 3A). An N-terminal sequence analysis of the first 10 amino acid residues of these two polypeptides revealed that the two proteins are encoded by two genes annotated as mvaT and mvaU on the P. aeruginosa genome website (www.pseudomonas.com). The mvaT and mvaU genes encode two homologous DNA-binding proteins belonging to the H-NS family, and the calculated molecular masses of these proteins are 14.2 kDa for MvaT and 13.4 kDa for MvaU.
FIG. 3.
Purification of MvaT and MvaU. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of fractions from size exclusion chromatography for purification of MvaT and MvaU from PAO4460 (rpoN). Lane M, protein markers; lanes 1 to 8, elution fractions1 to 8, respectively. (B and C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis purification profiles for recombinant MvaT and MvaU, respectively. Lane M, protein markers; lane CUI, crude extract of uninduced culture; lane CI, crude extract of induced culture; lane S, sample from a HiPrep 16/10 SP FF column; lane H, sample from a HiPrep 16/10 heparin FF column fraction; lane GF, sample from a gel filtration column.
Purification of recombinant MvaT and MvaU.
To demonstrate binding of MvaT and MvaU to the aotJ regulatory region, recombinant plasmids pHW1 and pHW2 were constructed by employing the pBAD system and introduced into E. coli Top10 for expression of MvaT and MvaU using arabinose induction. However, it was observed that there was significant proteolytic degradation of overexpressed MvaT and MvaU in Top10, mostly due to the presence of the LonA protease (7). Therefore, E. coli BL21, a lonA mutant strain, was employed as the host to prevent degradation. As shown in Fig. 3B and 3C, both the MvaT and MvaU proteins were successfully overexpressed and purified by column chromatography as described in Materials and Methods. The N-terminal sequences of the first 10 amino acids of the purified MvaT and MvaU were identical to the sequences annotated by the Pseudomonas genome project, with the first Met residue removed in both cases.
Interactions of purified MvaT and MvaU with the aotJ regulatory region.
The purified MvaT and MvaU proteins were tested to determine their interactions with the aotJ regulatory region. Probe F501 (Fig. 1) was 32P labeled and added to reaction mixtures with increasing concentrations of either MvaT or MvaU. As shown in Fig. 2C and 2D, multiple bands of nucleoprotein complexes were obtained for the reaction mixtures with low concentrations of MvaT or MvaU, and a complete shift of F501 to a low-mobility complex was observed with higher concentrations of the proteins. These patterns of multinucleoprotein complex formation were similar to what has been reported previously for H-NS of E. coli (6, 36).
Supershift assays with anti-MvaT and anti-MvaU antibodies.
To further demonstrate the involvement of MvaT and MvaU in C1 complex formation, different concentrations of anti-MvaT or anti-MvaU antibody were incubated with a crude cell extract of PAO1 to titrate these two proteins specifically. As shown in Fig. 2B, the C1 complex disappeared when a high concentration of antibodies was used in the reaction mixture, but the ArgR-DNA complex (C2 complex) was retained. These results support the hypothesis that MvaT and MvaU participate in the formation of the C1 complex.
Activation of the aotJQPOP-argR operon in an mvaT mvaU double mutant.
To analyze potential effects of mvaT and/or mvaU through the C1 complex on expression of the aotJQMOP-argR operon, mutants with a single lesion in mvaT or mvaU, as well as mutants with mvaT mvaU double lesions, were constructed, and the MvaT and MvaU expression patterns in these mutants and parental strain PAO1 were analyzed by immunoblotting (Fig. 4), which confirmed the successful construction of the mutants.
FIG. 4.

Immunoblot analysis of MvaT and MvaU. Crude extracts were prepared from PAO1, an mvaT mutant (PAO-T), an mvaU mutant (PAO-U), and mvaT mvaU double mutants (PAO-TU1 and PAO-TU2). Anti-MvaU antibodies were diluted 1,500-fold for detection of both MvaU and MvaT (using cross-reactions) using the standard protocol with chromogenic substrates. The control was nonspecific binding of anti-MvaU antibodies to an unknown peptide that served as an internal control for crude extracts loaded onto each lane.
Plasmid pST500 carrying the entire regulatory region of aotJ promoters was introduced into these strains to test the potential effects of mvaT and mvaU on expression of the aotJQMOP-argR operon. Compared to the activity in parental strain PAO1, the aotJ promoter activity in the mvaU mutant was similar, but the aotJ promoter activity in the mvaT mutant was slightly increased. However, significant three- to fourfold induction was detected in the mvaT mvaU double mutants (Table 2).
TABLE 2.
Effect of mvaT mvaU knockout mutation on aotJ promoter activity
| Strain | Genotype | pST500
|
pST500M
|
||
|---|---|---|---|---|---|
| Sp act (nmol/min/mg)a | Change (fold)b | Sp act (nmol/min/mg)a | Change (fold)b | ||
| PAO1 | Wild type | 8,389 | 1.0 | 1,223 | 1.0 |
| PAO-T | mvaT::Gm | 7,588 | 0.9 | 1,143 | 0.9 |
| PAO-U | mvaU::Tc | 11,791 | 1.4 | 1,524 | 1.2 |
| PAO-TU1 | mvaT::Gm mvaU::Tc | 25,694 | 3.1 | 12,360 | 10.1 |
| PAO-TU2 | mvaT::Gm mvaU::Tc | 30,142 | 3.6 | 12,211 | 10.0 |
Specific activity with ortho-nitrophenyl-β-galactoside as the substrate.
Change compared to wild-type strain PAO1.
As characterized in a previous report (21), the aotJ promoter activity in pST500 is due to two promoters, the ArgR-repressible P1 promoter and the ArgR-inducible P2 promoter, and the ArgR binding site covers the −10 region of P1 and the −35 region of P2 (Fig. 1). To further analyze the effects of mvaT and mvaU on the aotJ promoter, plasmid pST500M was constructed; this plasmid was derived from pST500 and lacked a functional P2 promoter due to destruction of the −10 region of P2 by site-directed mutagenesis (change from TAGAAT to CGACGC). The loss of induction by arginine in pST500M was confirmed using strain PAO1 (data not shown). As shown in Table 2, a single lesion in mvaT or mvaU did not cause a significant change in the aotJ promoter activities; however, the promoter activities were increased 10-fold in the mvaT mvaU double mutants. These results indicated that MvaT and MvaU may exert a promoter-silencing effect on expression of the aotJQMOP-argR operon in wild-type strain PAO1.
Prophage activation in an mvaT mvaU double mutant.
During construction of an mvaT mvaU double mutant, we observed that colonies of the desired mutant grew much slower than colonies of mvaT or mvaU single mutants. Fortuitously, an extrachromosomal DNA species of unknown identity was detected specifically in samples of the mvaT mvaU double mutant but not in samples of parental strain PAO1 or the single mutants (Fig. 5A). When subjected to EcoRI digestion (Fig. 5B), this DNA species displayed a distinct restriction pattern. Cloning and sequencing of most of the EcoRI-digested fragments revealed that this DNA species covers a contiguous region on the chromosome corresponding to the PA0715-PA0729.1 locus, which was reported to be the integrated genome of filamentous phage Pf4 (31). Therefore, we considered this DNA species the replicative form (RF) of prophage Pf4, which was activated when both mvaT and mvaU were deleted. The spent media of the mvaT mvaU double mutants were also tested for the presence of phage particles by using plaque assays, and the results were positive (data not shown). Surprisingly, once prophage Pf4 was activated in the double mutant, it could not be suppressed by mvaT or mvaU carried on plasmids, as shown by the persistence of RF DNA production in the resulting recombinant strains (data not shown).
FIG. 5.
Activation of prophage Pf4 in mvaT mvaU double mutants. (A and B) Agarose gel electrophoresis of purified RF DNA from the cells before and after EcoRI digestion, respectively. Overnight cell cultures were used for extraction of RF DNA with a Qiagen miniprep kit using the manufacturer's instructions. The RF DNA was separated on a 1% agarose gel. The samples loaded in each well were equivalent to 0.7 ml of overnight cell cultures. The sizes of the DNA markers were (from bottom to top) 200 bp, 300 bp, 400 bp, 500 bp, 1.0 kb, 1.5 kb, 2.0 kb, 3.0 kb, 4.0 kb, 6.0 kb, 8.0 kb, 10.0 kb, and 12.0 kb.
Elimination of pyocyanin synthesis without MvaT and MvaU.
It has been reported that synthesis of pyocyanin, a water-soluble cytotoxic pigment, can be enhanced in an mvaT mutant (4, 5) as a result of increased concentrations of Pseudomonas quinolone signal (PQS) in quorum sensing. To test the effect of mvaT and mvaU on pyocyanin synthesis, pyocyanin was extracted from cultures of mutant strains and parental strain PAO1 grown in alanine-glycerol medium as described in Materials and Methods and then was quantified. All of the strains except the double mutant were dark blue (Fig. 6A). The blue pigment containing pyocyanin was extracted with chloroform and then back-extracted in an acidic solution, resulting in a pink pigment (Fig. 6B, top layer). Compared to PAO1, the mvaT and mvaU single mutants generated twofold more pyocyanin, consistent with what has been reported previously (5). In contrast, pyocyanin synthesis did not occur in the double mutant, even after incubation for a long time (Fig. 6), and this deficiency could be complemented by mvaT or mvaU carried on plasmids (Table 3). Similar to the pattern of pyocyanin production, compounds that were retained in the organic phase and fluoresced blue under UV light (excitation wavelength, 254 nm) were found in the single mutants and the parental strain but not in the double mutant (Fig. 6C).
FIG. 6.

Pyocyanin extraction from strain PAO1 and mutants of this strain. (A) Cell cultures grown for 72 h. (B) Back-extraction of pyocyanin in an acidic solution under nature light. (C) Samples in panel B under UV light.
TABLE 3.
Quantitation of pyocyanin in P. aeruginosa PAO1 and mutant derivatives of this strain
| Strain | Genotype | Pyocyanin concn (μg/ml) with:
|
||
|---|---|---|---|---|
| No plasmid | pUCP-T | pUCP-U | ||
| PAO1 | Wild type | 29 | 30 | 33 |
| PAO-T | mvaT::Gm | 71 | 50 | 68 |
| PAO-U | mvaU::Tc | 71 | 74 | 58 |
| PAO-TU1 | mvaT::Gm mvaU::Tc | NDa | 33 | 42 |
| PAO-TU2 | mvaT::Gm mvaU::Tc | ND | 73 | 48 |
ND, not detectable.
Differential effects of MvaT and MvaU on cupA expression.
Another genetic trait that demonstrated the physiological functions of MvaT and MvaU was the expression of cupA genes for fimbrial biogenesis. It has been reported that cupA genes are induced by a lesion in mvaT due to phase variation of unknown mechanisms (28). The potential effects of mvaT and/or mvaU on cupA gene expression were analyzed by measuring the β-galactosidase activities of a series of strains derived from strain CupA, a well-characterized reporter strain which has a lacZ gene inserted right after the cupA1 gene (28). As shown in Table 4, a lesion in mvaT increased the cupA gene expression 18-fold, which could be completely suppressed by plasmid pUCP-T and partially suppressed by pUCP-U. Double lesions in mvaT and mvaU increased the expression 197-fold, and introduction of pUCP-T had a much stronger complementation effect than introduction of pUCP-U. However, the level of cupA expression in the mvaU single mutant was only slightly higher (less than twofold higher) than the level of cupA expression in the parental strain.
TABLE 4.
Effect of mvaT mvaU knockout mutation on cupA gene expression
| Strain | Genotype |
cupA gene expression (nmol/min/mg) witha:
|
||
|---|---|---|---|---|
| No plasmid | pUCP-T | pUCP-U | ||
| CupA | cupA1::lacZ, wild type | 62 | 49 | 48 |
| CupA-T | mvaT::Gm | 1,098 | 83 | 268 |
| CupA-U | mvaU::Tc | 103 | 52 | 82 |
| CupA-TU1 | mvaT::Gm mvaU::Tc | 12,048 | 993 | 4,928 |
| CupA-TU2 | mvaT::Gm mvaU::Tc | 14,998 | 1,148 | 4,692 |
cupA gene expression was measured by quantitation of β-galactosidase activity, and the values are the averages of two measurements with standard deviations less than 5%.
DISCUSSION
mvaT mvaU double mutants are viable.
MvaT and MvaU are global transcriptional regulators belonging to the H-NS family. Involvement of these two homologous proteins in several aspects of bacterial physiology in P. aeruginosa has been reported previously (3, 5, 27, 28, 33, 34). However, most of the previous studies involved genetic analysis of mutants devoid of either mvaT or mvaU but not genetic analysis of mvaT mvaU double mutants. Moreover, using an elegant genetic design for conditional depletion of both the MvaT and MvaU proteins, it was concluded in a recent report (3) that survival of P. aeruginosa requires the presence of either mvaT or mvaU and that an mvaT mvaU double mutation is lethal. In contrast, the double mutants were constructed using conventional gene knockout approaches and were confirmed by immunoblotting in our study. We observed that colonies of the double mutants grew to a visible size much slower than colonies of the single mutants in the last step of mutant construction. The slow growth of the mvaT mvaU double mutants may be related to prophage activation in these mutants.
MvaT and MvaU as transcriptional silencers.
Activation of prophage Pf4 in a double mutant was indicated by detection of RF DNA inside the cells and by release of phage particles into the spent medium (Fig. 5). The phenomenon of prophage activation by depletion of MvaT and MvaU was very intriguing, as it was suggested previously that these two global regulators silence transcription of foreign DNA elements in P. aeruginosa (3). Previously, mature biofilms of P. aeruginosa were found to release the filamentous phage Pf4 (35), which may be linked to developmental events during biofilm formation (32). Prophage activation in biofilms has been reported to be regulated by quorum sensing (29) and may play a pivotal role in modulation of colony morphology in P. aeruginosa (2, 14, 31). Therefore, based on our finding that there was prophage activation in the mvaT mvaU double mutants, it is reasonable to speculate that the intracellular MvaT and MvaU might be depleted by unknown mechanisms in response to environmental stress and as a result prophage production and other events that are otherwise silenced by these two proteins are activated.
Like H-NS from enteric bacteria, MvaT and MvaU were shown to bind preferentially to AT-rich regions of the target genes, and these two proteins coordinate in regulation of the same set of target genes in P. aeruginosa (3). Using chromatin immunoprecipitation coupled with DNA microarrays, the MvaT and MvaU binding sites in genes of prophage Pf4, CupA fimbrial formation, and biosynthesis of pyocyanin (derived from phenazine) were identified, among other things. Although the regulatory region of the aotJQMOP-argR operon for arginine uptake and regulation was missing in the published list, the effect of MvaT and MvaU on the aotJ promoter activity was clearly demonstrated using biochemical and genetic analyses in this study (Fig. 2 and Table 2).
Pyocyanin synthesis.
Although deletion of mvaT or mvaU alone increased pyocyanin synthesis in the mutants, we observed that no pyocyanin was detected in the growth medium of the mvaT mvaU double mutants (Table 3 and Fig. 6). Pyocyanin is a blue redox pigment produced in the stationary phase as a secondary metabolite. Enzymes involved in pyocyanin biosynthesis are encoded by the phzH, phzM, and phzS genes and the two phzABCDEFG operons (10, 18). Pyocyanin biosynthesis is an integral part of the quorum-sensing network (9) and is subject to control by many transcriptional regulators, including the lasR, rhlR, and pqsR regulators (15). It has been reported that failure to produce PQS resulted in total elimination of pyocyanin synthesis (19). In our analysis of quasi-mvaT mvaU mutants (unpublished data), the absence of pyocyanin synthesis was found to be correlated with an undetectable level of PQS (30). Therefore, it is tempting to speculate that pyocyanin synthesis may be subject to two tiers of regulation by MvaT and MvaU, one involving PQS with MvaT and MvaU that is essential for the synthesis and the other involving MvaT- and MvaU-dependent repression of phz genes. Further experiments are required to elucidate the mechanisms by which MvaT and MvaU regulate pyocyanin and/or PQS production.
Oligomeric composition of MvaT and MvaU.
Like H-NS and StpA of E. coli (7), MvaT and MvaU inside the cells are likely to form homomeric and heteromeric complexes. When purified from P. aeruginosa, fractions containing the MvaT and MvaU proteins from the size exclusion column were widespread (Fig. 3A), and most of these proteins in their native forms eluted as molecules with molecular masses of at least 90 kDa or more. Since the calculated molecular mass of MvaT or MvaU is around 13 to 14 kDa, the homomeric and/or heteromeric forms of MvaT and MvaU are apparently composed of more than two subunits when they are purified in vitro. In addition, multinucleoprotein complexes were detected in the EMSA with the aotJ probe and purified MvaT and MvaU from recombinant strains of E. coli, supporting the hypothesis that homomeric MvaT and MvaU are present in different oligomeric conformations.
Acknowledgments
We thank Simon L. Dove of Harvard Medical School for providing the CupA strain.
This project was supported by National Science Foundation grant 0415608.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Baehler, E., P. de Werra, L. Y. Wick, M. Pechy-Tarr, S. Mathys, M. Maurhofer, and C. Keel. 2006. Two novel MvaT-like global regulators control exoproduct formation and biocontrol activity in root-associated Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 19:313-329. [DOI] [PubMed] [Google Scholar]
- 2.Brockhurst, M. A., A. Buckling, and P. B. Rainey. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. Biol. Sci. 272:1385-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castang, S., H. R. McManus, K. H. Turner, and S. L. Dove. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. USA 105:18947-18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diggle, S. P., K. Winzer, S. R. Chhabra, K. E. Worrall, M. Camara, and P. Williams. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50:29-43. [DOI] [PubMed] [Google Scholar]
- 5.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dole, S., V. Nagarajavel, and K. Schnetz. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52:589-600. [DOI] [PubMed] [Google Scholar]
- 7.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 8.Frank, L. H., and R. D. Demoss. 1959. On the biosynthesis of pyocyanine. J. Bacteriol. 77:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas, D., B. W. Holloway, A. Schambock, and T. Leisinger. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol. Gen. Genet. 154:7-22. [DOI] [PubMed] [Google Scholar]
- 13.Itoh, Y. 1997. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J. Bacteriol. 179:7280-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo, M. Y., M. K. Yang, W. P. Chen, and T. T. Kuo. 2000. High-frequency interconversion of turbid and clear plaque strains of bacteriophage f1 and associated host cell death. Can. J. Microbiol. 46:841-847. [PubMed] [Google Scholar]
- 15.Lau, G. W., D. J. Hassett, H. Ran, and F. Kong. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599-606. [DOI] [PubMed] [Google Scholar]
- 16.Li, C., and C. D. Lu. 2009. Unconventional integration of the bla gene from plasmid pIT2 during ISlacZ/hah transposon mutagenesis in Pseudomonas aeruginosa PAO1. Curr. Microbiol. 58:472-477. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishijyo, T., D. Haas, and Y. Itoh. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 40:917-931. [DOI] [PubMed] [Google Scholar]
- 21.Nishijyo, T., S. M. Park, C. D. Lu, Y. Itoh, and A. T. Abdelal. 1998. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 180:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 179:5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rescalli, E., S. Saini, C. Bartocci, L. Rychlewski, V. De Lorenzo, and G. Bertoni. 2004. Novel physiological modulation of the Pu promoter of TOL plasmid: negative regulatory role of the TurA protein of Pseudomonas putida in the response to suboptimal growth temperatures. J. Biol. Chem. 279:7777-7784. [DOI] [PubMed] [Google Scholar]
- 25.Rosenthal, R. S., and V. W. Rodwell. 1998. Purification and characterization of the heteromeric transcriptional activator MvaT of the Pseudomonas mevalonii mvaAB operon. Protein Sci. 7:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 27.Vallet, I., S. P. Diggle, R. E. Stacey, M. Camara, I. Ventre, S. Lory, A. Lazdunski, P. Williams, and A. Filloux. 2004. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186:2880-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallet-Gely, I., K. E. Donovan, R. Fang, J. K. Joung, and S. L. Dove. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 102:11082-11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wally, H. 2006. Identification and characterization of MvaT and MvaU global regulators in arginine catabolism and quorum sensing of Pseudomonas aeruginosa. Ph.D. thesis. Georgia State University, Atlanta.
- 31.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:8066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westfall, L. W., N. L. Carty, N. Layland, P. Kuan, J. A. Colmer-Hamood, and A. N. Hamood. 2006. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 255:247-254. [DOI] [PubMed] [Google Scholar]
- 34.Westfall, L. W., A. M. Luna, M. San Francisco, S. P. Diggle, K. E. Worrall, P. Williams, M. Camara, and A. N. Hamood. 2004. The Pseudomonas aeruginosa global regulator MvaT specifically binds to the ptxS upstream region and enhances ptxS expression. Microbiology 150:3797-3806. [DOI] [PubMed] [Google Scholar]
- 35.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 36.Wojtuszewski, K., M. E. Hawkins, J. L. Cole, and I. Mukerji. 2001. HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry 40:2588-2598. [DOI] [PubMed] [Google Scholar]