Abstract
Pathogens lacking the enzymatic pathways for de novo purine biosynthesis are required to salvage purines and pyrimidines from the host environment for synthesis of DNA and RNA. Two key enzymes in purine salvage pathways are IMP dehydrogenase (GuaB) and GMP synthase (GuaA), encoded by the guaB and guaA genes, respectively. While these genes are typically found on the chromosome in most bacterial pathogens, the guaAB operon of Borrelia burgdorferi is present on plasmid cp26, which also harbors a number of genes critical for B. burgdorferi viability. Using molecular genetics and an experimental model of the tick-mouse infection cycle, we demonstrate that the enzymatic activities encoded by the guaAB operon are essential for B. burgdorferi mouse infectivity and provide a growth advantage to spirochetes in the tick. These data indicate that the GuaA and GuaB proteins are critical for the survival of B. burgdorferi in the infection cycle and highlight a potential difference in the requirements for purine salvage in the disparate mammalian and tick environments.
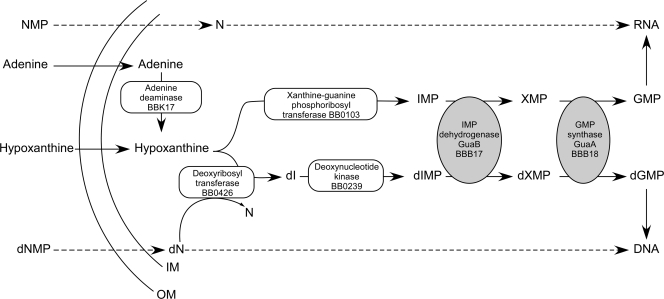
Purine metabolism is critical for the growth and virulence in mammals of many bacterial pathogens (11, 26, 29, 33, 51). Borrelia burgdorferi, the infectious agent of Lyme borreliosis, lacks the genes encoding the enzymes required for de novo nucleotide synthesis (8, 12) and therefore must rely on salvage of purines and pyrimidines from its hosts for nucleic acid biosynthesis (21, 35). Furthermore, B. burgdorferi lacks the genes encoding key enzymes required for a classic purine salvage pathway, including hpt (hypoxanthine-guanine phosphoribosyltransferase), purA (adenylosuccinate synthase), purB (adenylosuccinate lyase), and the locus encoding a ribonucleotide reductase (4, 8, 12, 35, 66). Despite the absence of a ribonucleotide reductase, an enzyme critical for the generation of deoxynucleotides through enzymatic reduction of ribonucleotides (32), a novel purine salvage pathway that involves salvage of deoxynucleosides from the host and interconversion of purine bases to deoxynucleosides by BB0426, a deoxyribosyl transferase, has recently been demonstrated for B. burgdorferi (23) (Fig. 1).
FIG. 1.
Pivotal role of the GuaAB proteins in the purine salvage pathway of B. burgdorferi. A novel pathway for purine salvage has recently been elucidated for B. burgdorferi (23). Extracellular adenine and hypoxanthine are salvaged by B. burgdorferi from mammalian and tick host environments (61). Following transport, adenine can be converted to hypoxanthine by adenine deaminase (BBK17) (21). This pathway proposes two possible fates for hypoxanthine, as follows. (i) Hypoxanthine is converted to IMP by a putative xanthine-guanine phosphoribosyl transferase (BB0103), IMP is converted to XMP by IMPDH (GuaB or BBB17), and XMP is converted to GMP by GMP synthase (GuaA or BBB18), resulting in guanine nucleotides for RNA synthesis. (ii) Direct transport of deoxynucleosides appears to provide a source of deoxyribose for interconversion of hypoxanthine to deoxyinosine by a deoxyribosyl transferase (BB0426) (23). dIMP is generated by a putative deoxynucleotide kinase (BB0239). GuaB converts dIMP to dXMP, and GuaA converts dXMP to dGMP, providing guanine deoxynucleotides for DNA synthesis (23). Salvage of free guanine nucleosides and guanine deoxynucleosides, when they are available in the host environment, may allow B. burgdorferi to circumvent the GuaAB requirement for GMP and dGMP biosynthesis. The dashed arrows indicate dephosphorylation of nucleotide monophosphate or deoxynucleotide monophosphate prior to transport by the spirochete and rephosphorylation of nucleoside and deoxynucleoside to nucleotide triphosphate and deoxynucleotide triphosphate, respectively, for RNA and DNA synthesis. NMP, nucleotide monophosphate; N, nucleoside; dN, deoxynucleoside; dNMP, deoxynucleotide monophosphate; OM, outer membrane; IM, inner membrane.
In its infection cycle, B. burgdorferi passages between two disparate environments with potentially distinct purine availabilities, the tick vector and a mammalian host. Hypoxanthine is the most abundant purine in mammalian blood (17), and it is available for salvage by B. burgdorferi during the blood meal of an infected tick and during the spirochete's transient presence in the mammalian bloodstream. Despite the absence of the hpt gene, we and others have shown that B. burgdorferi is able to transport and incorporate low levels of hypoxanthine (23, 35). During mammalian infection B. burgdorferi resides in various tissues, including the skin, heart, bladder, and joints. Adenine has been shown to be ubiquitous in mammalian tissues (61) and therefore is available for salvage by B. burgdorferi. Guanine is present at low levels in mammalian blood and tissues (17, 61); however, the amount may not be sufficient for survival of the spirochete.
The limiting step in guanine nucleotide biosynthesis from adenine and hypoxanthine is the conversion of IMP to XMP, which is catalyzed by IMP dehydrogenase (IMPDH) (65). Guanine nucleotides are essential for DNA and RNA synthesis, signal transduction, and cell cycle control; thus, IMPDH activity is critical for the survival of most organisms (60). The enzymes required for the final two steps of guanine nucleotide biosynthesis, IMPDH and GMP synthase, are encoded by the guaB and guaA genes, respectively. The guaA and guaB genes and the corresponding activities of their protein products are conserved in B. burgdorferi (28, 67). These genes are typically carried on the chromosomes of bacterial species. However, in B. burgdorferi, the guaAB operon resides on a 26-kbp circular plasmid, cp26, and it shares an approximately 185-bp intergenic region with, and is transcribed divergently from, the essential virulence gene ospC (8, 12, 28, 50, 54, 62). The cp26 plasmid has been shown to harbor numerous genes important for B. burgdorferi survival in vivo and in vitro, including ospC (16, 34, 50, 53, 56) and resT (7), as well as BBB26 and BBB27 (20). Because of these critical functions, this plasmid is the only plasmid of the approximately 21 B. burgdorferi plasmids that is present in all natural isolates and has never been shown to be lost during in vitro culture (2, 7, 18, 20, 44, 52).
Here we establish that the enzymatic activities of GuaA and GuaB are critical for the survival of B. burgdorferi in the infectious cycle and highlight a potential difference in this spirochete's requirement for purine salvage in the disparate mammalian and tick environments.
MATERIALS AND METHODS
B. burgdorferi clones and growth conditions.
All low-passage infectious B. burgdorferi clones used in this study are listed in Table 1 and were derived from strain B31 clone A3, which lacks plasmid cp9 but contains all 20 additional plasmids described for the parental strain MI-B31 (10). Wild-type clone A3-M9 was derived by passage of A3 through a mouse, followed by single-colony isolation (55). A3-M9 lacks both cp9 and lp21, which have been demonstrated to be dispensable for mouse and tick infection (10, 38, 55). B. burgdorferi was grown in liquid Barbour-Stoenner-Kelly (BSK) II medium supplemented with gelatin and 6% rabbit serum (1) and was plated on solid BSK medium as previously described (40, 41). All cultures were grown at 35°C with 2.5% CO2. Kanamycin was used at a concentration of 200 μg/ml, and gentamicin was used at a concentration of 40 μg/ml.
TABLE 1.
B. burgdorferi clones used in this study
Reverse transcriptase qPCR of spirochete mRNA isolated from infected mouse tissues.
Total RNA was isolated from the heart tissue of groups of seven C3H/HeN or C3H/HeN/SCID mice infected with 1 × 105 wild-type B. burgdorferi A3-M9 cells at 2 and 4 weeks postinoculation. Mouse infection was assessed by reisolation of spirochetes from ear and bladder tissues. Heart tissues were harvested immediately following mouse sacrifice, immersed in RNApro solution (FastRNA pro Green kit; MP Biomedicals, Solon, OH), and homogenized using a FastPrep-24 instrument and lysing matrix D beads in impact-resistant 2-ml tubes. Samples were processed for 40-s intervals at setting 6.0, followed by 3 min on ice, until tissue was no longer visible. RNA was isolated using the FastRNA pro Green kit (MP Biomedicals, Solon, OH) according to the manufacturer's specifications. cDNA was synthesized using random hexamer primers and a high-capacity cDNA reverse transcriptase kit (Applied Biosystems, Branchburg, NJ). Parallel cDNA reactions were carried out in the absence of reverse transcriptase. cDNA was treated with RNase H for 1 h at 37°C to remove RNA-DNA hybrids. Samples were cleaned and concentrated using a MiniElute kit (Qiagen, Valencia, CA). cDNA samples were quantitated by spectrophotometric analysis at 260 nm and diluted to obtain a working concentration of 50 ng/μl in DNase- and RNase-free water. Real-time quantitative PCR (qPCR) mixtures were prepared using 100 ng cDNA and a TaqMan universal PCR Mastermix kit (Applied Biosystems). Using an Applied Biosystems 7900HT instrument, samples were assayed for the flaB transcript (21) and for the guaA, guaB, and ospC transcripts using primers 9 to 16 and probes 1 to 4 (Table 2). The flaB transcript was used as the endogenous reference to which all other target genes were normalized. The amounts of the flaB, guaA, guaB, and ospC transcripts were interpolated by using a standard curve for each gene target that was generated using genomic DNA isolated from 105, 104, 103, 102, and 101 spirochetes. Samples were analyzed in triplicate, and gene expression is reported below in number of gene transcripts per flaB mRNA copy. The amplification efficiencies of the target genes were as follows: flaB, 98.9%; guaA, 96.3%; guaB, 97.9%; and ospC, 98.4%. The amplification for samples lacking reverse transcriptase was similar to that for the no-template control. Data sets were compared using the nonparametric Kruskal-Wallis test with Dunn's multiple-comparison test using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego CA).
TABLE 2.
Primers and probes used in this study
| Primer or probe no. | Designation | Sequence (5′-3′)a |
|---|---|---|
| Primers | ||
| 1 | guaAF | GATTTTGGATCCCAATATAGCC |
| 2 | guaAR | CCCATTCTATGGTTGATGGAGGCTTAGA |
| 3 | guaA16159-SalIF | GTCGACGGATGGAATTGTAGGCCG |
| 4 | guaA15926-SalIRC | GTCGACGTAAACACTGGATTGTTGCGC |
| 5 | guaAcompF-SalI | GCGTCGACGGTTTATAGCTAGATCTTTTGATTTGGC |
| 6 | guaAcompRC-SalI | GCGTCGACTCAGCAGAATTTGCAGATGTATTCCC |
| 7 | flgBPo-XhoI | TAATACTCGAGCTTCAAGGAAGATTT |
| 8 | 3′ aacC1-NheI | GCTAGCCGATCTCGGCTTGAACG |
| 9 | flaB TaqMan FWD | TCTTTTCTCTGGTGAGGGAGCT |
| 10 | flaB TaqMan REV | TCCTTCCTGTTGAACACCCTCT |
| 11 | guaA TaqMan FWD | GAAGAATTAGAGAAATTGGCGTTTATAC |
| 12 | guaA TaqMan REV | GGTAGGAGCTTCTTTTGAATAAACAG |
| 13 | guaB TaqMan FWD | AGGAATAGGTCCGGGTAGTATATGC |
| 14 | guaB TaqMan REV | TCATAGACATCGCAGATTGCTGTT |
| 15 | ospC1-F | ACGGATTCTAATGCGGTTTTACCT |
| 16 | ospC1-R | CAATAGCTTTAGCAGCAATTTCATCT |
| 17 | guaA 3′ KpnI | GGGGTAACCTTATTATTCCCATTCTATGGTTGATGGAGGC |
| Probes | ||
| 1 | flaB TaqMan probe | 6-FAM-AAACTGCTCAGGCTGCACCGGTTC-TAMRA |
| 2 | guaA TaqMan probe | 6-FAM-AGCAGGACTTCCACTTAGTATTATTCCTGAGATATTCAT-TAMRA |
| 3 | guaB TaqMan probe | 6-FAM-AACTCCAACTCCCGCAACGATTCTTGTT-TAMRA |
| 4 | ospC1 probe | 6-FAM-CTGTGAAAGAGGTTGAAGCGTTGCTGTCAT-TAMRA |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Mutation of guaA.
The guaA gene was amplified from B31 genomic DNA using Taq polymerase and primers 1 and 2 (Table 2) and was cloned into the vector pCR-XL-TOPO (Invitrogen), yielding pguaA. A 233-bp region of guaA (encoding amino acids 185 to 264) was removed from pguaA by inverse PCR using the Expand Long PCR system (Roche) and primers 3 and 4 (Table 2), yielding linear plasmid pguaAmut with SalI sites at its ends. The kanamycin resistance cassette, flgBp-kan (5), was amplified from pBSV2 (49) with XhoI ends by using primers 7 and 8 and Taq polymerase (Table 2) and was cloned into the pCR2.1-TOPO vector (Invitrogen). The flgBP-kan gene cassette was removed from the pCR2.1-TOPO vector by XhoI digestion and ligated into inactivation constructs digested with SalI to create pguaA::flgBP-kan. Twenty micrograms of pguaA::flgBP-kan plasmid DNA purified from Escherichia coli was transformed into A3-M9 as previously described (10, 13, 41), and recombinants were selected on solid BSK medium containing kanamycin. Colonies were screened by PCR for the presence of the kanamycin resistance cassette in the guaA locus, using primers 1 and 2 (Table 2). Total genomic DNA was prepared from PCR-positive A3-M9 guaA::flgBP-kan clones and screened with a panel of primers for the presence of all B. burgdorferi plasmids (10). A clone that had the plasmid content of the parent clone was used in further experiments (Table 1).
Construction of pBSV2G guaAB.
The guaA::flgBP-kan mutant was complemented with pBSV2G guaAB, a shuttle vector carrying a wild-type copy of the guaAB operon and native promoter. Plasmid pBSV2G guaAB was constructed by PCR amplifying a 3.4-kbp DNA fragment containing the guaAB operon and its putative promoter region with SalI ends from wild-type B. burgdorferi genomic DNA, using Vent polymerase (Invitrogen) and primers 5 and 6 (Table 2). TA ends were added to this DNA fragment using Taq polymerase, and the product was cloned into the vector TOPO-XL (Invitrogen). The 3.4-kbp guaAB DNA fragment was removed from TOPO-XL by SalI digestion and cloned into the B. burgdorferi shuttle vector pBSV2G (9) digested with SalI. The plasmid structure and sequence were analyzed and verified by restriction digestion and sequence analysis. Transformation of plasmid pBSV2G guaAB DNA isolated from E. coli into the low-passage infectious clone A3-M9 guaA::flgBp-kan was attempted five times without success. To facilitate transformation of pBSV2G guaAB into A3-M9 guaA::flgBp-kan, the E. coli purified plasmid was first transformed into the high-passage, noninfectious clone B31-A (5). B31-A lacks lp25 but contains lp56 (24, 58), both of which carry putative restriction-modification systems (22, 24). Purification of pBSV2G guaAB from lp56+ B31-A should result in partially modified B. burgdorferi DNA that may be less likely to be targeted for degradation upon transformation into the lp25+ lp56+ low-passage, infectious clone. B. burgdorferi clone A3-M9 guaA::flgBp-kan was transformed with pBSV2G guaAB purified from B31-A or with pBSV2G purified from E. coli (10, 13, 41). Transformants were recovered using this method. Transformants were screened by PCR for the presence of the guaAB and aacC1 genes or the aacC1 gene alone by using primer pairs 1/2 and 7/8 (Table 2). PCR-positive transformants were analyzed to determine their plasmid contents, and clones that retained the plasmids found in the parent clone were selected and used for further experiments (Table 1).
Functional analysis of the GuaA protein.
The wild-type and mutant alleles of guaA, along with their native promoters, were PCR amplified from B. burgdorferi A3-M9 and A3-M9 guaA::flgBp-kan genomic DNA, respectively, using the Expand Long PCR system (Roche) and primers 6 and 17 (Table 2). The resulting PCR products were cloned into the pCR2.1 TOPO vector (Invitrogen), yielding pCR2.1 Bb guaA and pCR2.1 Bb guaA::flgBp-kan. The structures and sequences of the plasmids were analyzed and verified by restriction digestion and sequence analysis. Plasmids pCR2.1 Bb guaA and pCR2.1 Bb guaA::flgBp-kan and the pCR2.1 vector were transformed into E. coli guaA::Tn10 strain ght1 (59). Transformants were selected on LB plates supplemented with 100 μg/ml ampicillin and individually tested to determine their abilities to grow on M63 agar (27) supplemented with 0.2% glucose, 0.1% Casamino Acids, 1 mM MgSO4·7H2O, 0.00005% thiamine (vitamin B1), and 100 μg/ml ampicillin and either lacking or containing 20 mg/liter guanine (28).
Immunoblot analysis of the GuaB protein.
B. burgdorferi clones were grown in 100 ml of BSK II medium containing the appropriate antibiotic to a density of 1 × 107 spirochetes/ml. Cells were harvested, washed twice in HN buffer (50 mM HEPES, 50 mM NaCl; pH 7.6), and resuspended in 1 ml of the buffer at a final concentration of 1 × 106 spirochetes/μl. Cells were disrupted by sonication, and 10 μl (1 × 107 spirochetes) of total cell lysates was resolved by electrophoresis on 12.5% sodium dodecyl sulfate gels and blotted onto nitrocellulose. Immunoblots were hybridized with a rabbit polyclonal antibody that was raised against purified recombinant B. burgdorferi GuaB protein (1:1,000 dilution of anti-IMPDH) and with a monoclonal antibody that recognizes B. burgdorferi flagellin protein (1:200 dilution of H9724, provided by T. Schwan) (3). The conditions, secondary antibodies, and detection methods used have been described previously (16).
In vitro growth analysis.
Cultures of B. burgdorferi were inoculated using frozen stocks into 5 ml of BSK II medium containing the appropriate antibiotic and grown to a density of approximately 1 × 107 spirochetes/ml. Clones were subsequently diluted in triplicate to obtain 1 × 105 spirochetes/ml in 5 ml of BSK II medium containing the appropriate antibiotic. Spirochete density was determined every 24 h for 120 to 140 h using a Petroff-Hausser counting chamber.
Experimental tick-mouse infection cycle.
The Rocky Mountain Laboratories (RML) are accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for all animal experiments were prepared according to the guidelines of the National Institutes of Health and were approved by RML's Animal Care and Use Committee. Two different strains of mice were used in experiments. Mice from an outbred colony of Swiss-Webster mice maintained at RML (called RML mice) represented a genetically heterogeneous rodent population, and C3H/HeN mice (Harlan Sprague-Dawley, Indianapolis, IN) represented a uniform, inbred rodent population. For needle inoculation of mice, 250 μl of an inoculum containing either 5 × 103 or 1 × 107 spirochetes was used; 200 μl of the inoculum was delivered intraperitoneally, and 50 μl was delivered subcutaneously. Groups of three to five mice were inoculated for each B. burgdorferi clone. The number of spirochetes inoculated into mice was determined using a Petroff-Hausser counting chamber and was verified by subsequently counting CFU on solid BSK medium; for each inoculum 10 to 20 colonies were screened by PCR to determine the presence of the virulence plasmids lp25 and lp28-1, which confirmed that >90% of the individuals in the population contained the plasmids essential for mouse infectivity. The total plasmid contents of all in vivo inoculum cultures were confirmed using a panel of primers that amplify specific DNA targets on all B. burgdorferi plasmids (10). Mouse infection was assessed 4 to 6 weeks postinoculation by immunoblot analysis of mouse sera and reisolation of spirochetes from ear, bladder, and joint tissues, as previously described (10, 13, 14, 42).
Cohorts of 100 to 200 4- to 5-month-old Ixodes scapularis tick larvae (from a colony maintained at RML, Hamilton, MT) were experimentally infected with comparable, exponential-phase cultures of various B. burgdorferi clones as previously described (15, 36). Each immersion was performed in duplicate, and the entire experiment was repeated twice. One hundred to 200 larvae or 15 to 25 nymphs were applied to each mouse and were allowed to feed to repletion. Infection of RML mice on which infected ticks fed was assessed 6 to 11 weeks after tick feeding, as described above.
Determination of spirochete densities in ticks.
The spirochete densities in ticks were assessed 7 to 25 days after feeding to repletion by plating dilutions of triturated whole ticks on solid BSK medium, as previously described (15, 21), and performing a qPCR analysis of tick DNA, as described below. Genomic DNA was isolated from batches of 40 unfed larvae, 20 fed larvae, 4 to 7 unfed nymphs, or 1 to 3 fed nymphs infected with B. burgdorferi. Ticks were homogenized with a plastic pestle in an Eppendorf tube, and genomic DNA was isolated using a NucleoSpin tissue kit (Clontech Laboratories, Inc., Mountain View, CA) according to the manufacturer's specifications. qPCR was performed using 100 ng of total genomic DNA, as previously described (21, 48), primers 9 and 10, and probe 1 (Table 2). The number of spirochete genomes per tick was calculated as follows: (number of flaB copies amplified/ng total genomic DNA) × (ng total genomic DNA isolated/tick).
RESULTS
The guaAB operon is expressed during mammalian infection.
B. burgdorferi requires host-derived nucleotide precursors for production of nucleic acids. Surprisingly, the interconversion of purine bases to DNA occurs in the absence of a ribonucleotide reductase (4, 23, 35). It has recently been demonstrated that, in the absence of a ribonucleotide reductase, purine salvage and DNA synthesis by B. burgdorferi involve a combination of salvage of deoxynucleosides directly from the host and interconversion of purine bases to deoxynucleosides by a deoxyribosyl transferase (23) (Fig. 1). Moreover, it has been demonstrated that the GuaB and GuaA enzymes, the final two enzymes in the B. burgdorferi purine salvage pathway required for guanine nucleotide synthesis (28, 67), are able to use dIMP and dXMP, respectively, as substrates to produce dGMP (23), in addition to the well-characterized substrates IMP and XMP, respectively, to generate GMP (28, 67) (Fig. 1). Because of the pivotal roles that these enzymes play in the purine salvage pathway of B. burgdorferi for generation of both guanine nucleotides and deoxynucleotides (23), we hypothesized that the enzymatic activities of GuaA and GuaB contribute to the ability of spirochetes to survive in host tissues whose guanine, guanosine, and/or deoxyguanosine concentrations may be insufficient to support spirochete growth.
To begin to address the role(s) of the GuaAB enzymes in a mammalian host, we sought to determine the pattern of gene expression of the guaAB operon during an active infection. The guaAB operon is divergently transcribed from the ospC gene on cp26, and this operon and gene share an approximately 185-bp intergenic region (8, 12, 28). Expression of the ospC gene is upregulated during tick feeding (19) and remains high throughout establishment of an early infection in a mammal (25, 56). However, the OspC protein ultimately elicits a robust humoral immune response, and repression of ospC expression is vital for B. burgdorferi persistence in the mouse (56, 62, 64). A duplicated palindromic sequence consisting of 20-bp inverted repeats is located in the intergenic region between the ospC and guaA open reading frames (28, 62, 63). This DNA sequence, termed the ospC operator, has been shown to be important for repression of ospC expression (62, 63); however, the effect of this region, if any, on guaAB expression is unknown.
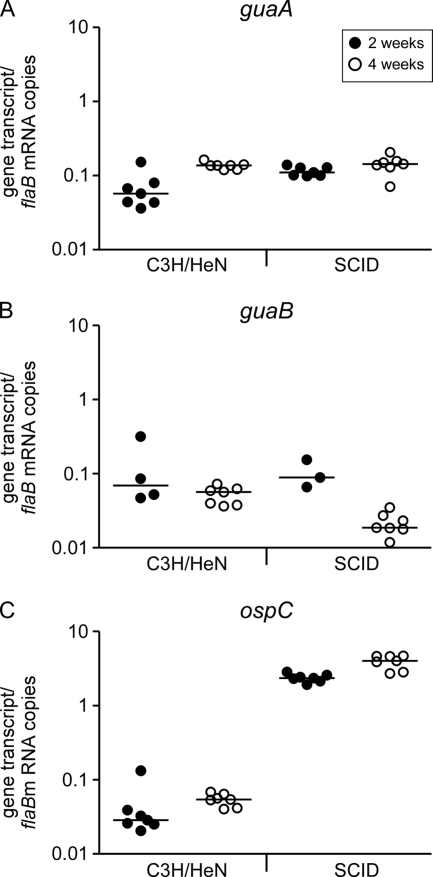
Groups of C3H/HeN and C3H/HeN/SCID mice were inoculated with 1 × 105 wild-type B. burgdorferi cells. Mice were sacrificed 2 and 4 weeks postinoculation, and total RNA was extracted from heart tissue and analyzed for guaA, guaB, ospC, and flaB mRNA transcripts by reverse transcriptase qPCR. All mice were confirmed to be positive for infection, as assessed by reisolation of spirochetes from bladder and joint tissues. The guaA and guaB transcripts were present at detectable levels in spirochetes isolated from infected mouse tissue at 2 and 4 weeks postinoculation (Fig. 2A and 2B). Comparison of the median levels of the guaA transcript in spirochetes isolated from infected C3H/HeN and C3H/HeN/SCID mouse tissues at 2 and 4 weeks using the Kruskal-Wallis test suggested that there was a significant difference in the mean rank sums of the transcript levels for all in vivo growth conditions (H = 11.77; df = 3; P = 0.0082). Similarly, analysis of the guaB expression data revealed a significant difference in the mean rank sums of the transcript levels for all in vivo growth conditions (H = 14.47; df = 3; P = 0.0023), suggesting that expression of these two genes may be regulated in vivo. These data indicate that the guaA and guaB genes are expressed by spirochetes during mouse infection, suggesting that the biochemical functions that they encode are important for mammalian infection. In addition, unlike expression of ospC, expression of the guaA and guaB genes did not exhibit a dramatic 100-fold increase in the absence of an acquired humoral immune response. These data suggest that the guaAB operon is not coregulated with the divergently transcribed ospC gene.
FIG. 2.
guaA and guaB genes are expressed during mammalian infection. Groups of C3H/HeN and C3H/HeN/SCID (SCID) mice were inoculated with 1 × 105 A3-M9 cells. Mice were sacrificed 2 and 4 weeks postinoculation. RNA samples were prepared from heart tissues, and flaB, guaA (A), guaB (B), and ospC (C) expression was quantified by reverse transcriptase qPCR. The data are expressed as the number of gene transcripts per flaB mRNA copy. Each data point indicates the average of triplicate measurements for the tissue RNA of an individual infected mouse. •, samples obtained 2 weeks postinoculation; ○, samples obtained 4 weeks postinoculation. The bars indicate the median values of the data sets. Data sets for each gene transcript were compared using the Kruskal-Wallis nonparametric rank test followed by Dunn's multiple-comparison test.
The GuaAB proteins are required for mouse infectivity by needle inoculation.
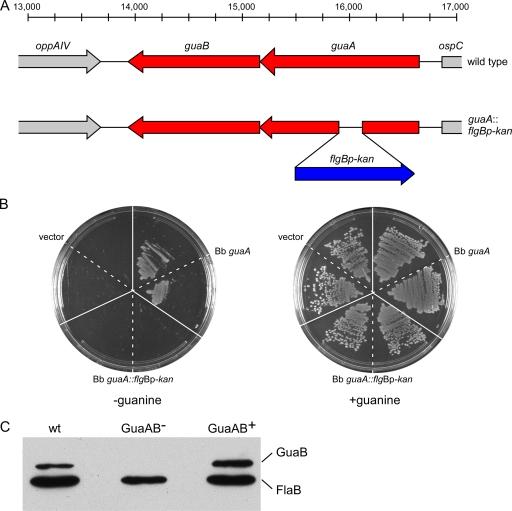
The GuaA and GuaB proteins have been successfully disrupted in high-passage, noninfectious B. burgdorferi clones (20, 57), indicating that these enzymes are not essential for growth of B. burgdorferi in vitro. The expression of guaAB in vivo (Fig. 2) suggests that the enzymes that these genes encode may be important for spirochete growth and survival in the mouse. In order to examine the roles of these enzymes in the B. burgdorferi infection cycle, a deletion-insertion mutation in guaA was constructed in the low-passage, infectious clone A3-M9 (23) (Table 1 and Fig. 3A). Attempts to generate an antibody to the GuaA protein were unsuccessful. Therefore, in order to test the loss of function of the mutant GuaA protein (GMP synthase activity) encoded by the guaA mutant allele, the wild-type and mutant guaA alleles from B. burgdorferi A3-M9 and A3-M9 guaA::flgBp-kan were used to functionally complement an E. coli guaA mutant. E. coli guaA::Tn10 strain ght1 is unable to grow on minimal media lacking guanine due to its lack of GMP synthase activity and therefore its inability to synthesize guanine nucleotides de novo (28, 59). Growth of this mutant is restored by supplementing the medium with guanine or providing a functional guaA gene in trans (28, 59). Functional complementation of this E. coli mutant with a wild-type copy of the B. burgdorferi guaA gene has been shown previously to allow growth of the E. coli mutant in the absence of guanine (28). Similar to the results for the control with the vector alone, a plasmid containing the mutant B. burgdorferi guaA allele was unable to functionally complement the E. coli guaA mutant for growth in the absence of exogenous guanine, whereas the E. coli guaA mutant harboring the wild-type B. burgdorferi guaA allele did not require supplemental guanine for growth (Fig. 3B). These data demonstrate that the insertion-deletion mutation in B. burgdorferi guaA resulted in a mutant allele that does not encode a functional GuaA protein.
FIG. 3.
guaA mutant spirochetes do not produce functional GuaA or GuaB protein. (A) Diagram of the wild-type and mutant guaAB loci on cp26. A total of 233 bp was deleted from the guaA gene and replaced with an flgBp-kan antibiotic resistance cassette. (B) Functional complementation of an E. coli guaA mutant with the B. burgdorferi guaA wild-type and mutant alleles. E. coli guaA::Tn10 strain ght1 harboring the pCR2.1 vector alone (vector), pCR2.1 Bb guaA (Bb guaA), or pCR2.1 Bb guaA::flgBp-kan (Bb guaA::flgBp-kan) was streaked on M63 agar without (−guanine) and with (+guanine) 20 mg/liter guanine. (C) Protein lysates from A3-M9 (wt), A3-M9 guaA::flgBp-kan/pBSV2G (GuaAB−), and A3-M9 guaA::flgBp-kan/pBSV2G guaAB (GuaAB+) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting using antisera against GuaB and FlaB.
Because guaA and guaB appear to be cotranscribed, we anticipated that disruption of guaA would result in the loss of guaB expression. As expected, no GuaB protein was detected by immunoblot analysis of spirochetes in which guaA was inactivated (Fig. 3C). Complementation of the guaA mutant clone with the B. burgdorferi shuttle vector pBSV2G (9) containing both the guaA and guaB genes under the control of their own promoter (23, 28) (Table 1) restored GuaB expression (Fig. 3C), demonstrating that the mutation in guaA was polar on guaB. Moreover, we recently showed that the B. burgdorferi guaA mutant was unable to convert IMP or dIMP to GMP or dGMP and incorporate the products into RNA or DNA. In these experiments, the wild-type and complemented mutant strains incorporated GMP and dGMP into both RNA and DNA (23). Thus, in this mutant, the pathway for the conversion of IMP or dIMP to GMP or dGMP was successfully disrupted, and the mutant is therefore null for GuaA and GuaB.
Spirochetes functionally lacking GuaAB and the isogenic complemented clone exhibited no growth defect in vitro (data not shown), confirming that any in vivo phenotypic differences were not due to an in vitro growth difference. Finally, because the flgBp-kan resistance cassette disrupting guaA is transcribed in the same direction as ospC (Fig. 3A), we examined expression of the OspC protein in in vitro-grown B. burgdorferi clones A3-M9, A3-M9 guaA::flgBp-kan/pBSV2G, and A3-M9 guaA::flgBp-kan/pBSV2G guaAB by immunoblotting. The presence of the flgBp-kan cassette in guaA had no effect on OspC protein production (data not shown). These data demonstrated that the flgBp-kan cassette did not alter expression of OspC in any way.
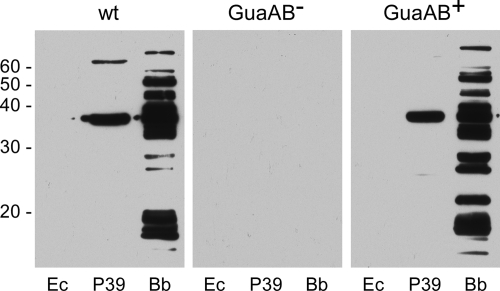
The requirement for the GuaAB enzymes in the mammalian environment was assessed by needle inoculation of mice with 5 × 103 or 1 × 107 spirochetes functionally lacking or containing GuaAB. No mice became infected when spirochetes lacking GuaAB were used at either dose, as assessed by the lack of serological conversion to B. burgdorferi proteins and reisolation of spirochetes from mouse tissues (Fig. 4 and Table 3). In contrast, seven of eight mice inoculated with 5 × 103 GuaAB+spirochetes and four of four mice inoculated with 1 × 107 GuaAB+ spirochetes were seroreactive and positive for reisolation of spirochetes from tissues (Fig. 4 and Table 3). The complementation data demonstrated that the deletion-insertion in guaA had no deleterious effect on expression of the adjacent essential virulence gene, ospC. Together, these data demonstrate that the GuaAB proteins are essential for survival of B. burgdorferi in the mouse.
FIG. 4.
Serological responses of mice inoculated with wild-type and GuaAB-deficient B. burgdorferi: immunoblot analysis of sera collected 6 weeks postinoculation from either C3H/HeN mice needle inoculated with 5 × 103 or 1 × 107 spirochetes or RML mice on which infected ticks fed. The protein lysates tested for seroreactivity included a lysate of E. coli carrying a cloning vector (Ec), a lysate of E. coli carrying a cloning vector expressing the B. burgdorferi P39 protein (45) (also known as BmpA [43]) (P39), and a lysate of infectious B. burgdorferi strain B31 (Bb). Representative results for the serum of one mouse infected with B. burgdorferi clone A3-M9 (wt), A3-M9 guaA::flgBp-kan/pBSV2G (GuaAB−), or A3-M9 guaA::flgBp-kan/pBSV2G guaAB (GuaAB+) are shown. Neither the inoculum nor the route of infection had an effect on the serological responses of the mice to the different B. burgdorferi clones. Molecular masses (in kilodaltons) are indicated on the left. Comparable serum dilutions and exposure times were used for all immunoblots.
TABLE 3.
Infection of mice with B. burgdorferi lacking or containing GuaAB
| Clone | No. of mice infected with 5 × 103 spirochetes/ total no. analyzeda | No. of mice infected with 1 × 107 spirochetes/ total no. analyzeda | Total no. of mice infected/total no. analyzedb |
|---|---|---|---|
| A3-M9 guaA::flgBp-kan/pBSV2G | 0/8 | 0/5 | 0/13 |
| A3-M9 guaA::flgBp-kan/pBSV2G guaAB | 7/8 | 5/5 | 12/13 |
The number of mice infected was assessed by immunoblot analysis with cell lysates of B. burgdorferi and E. coli producing P39 recombinant protein at 3 and 6 weeks postinoculation and by reisolation of spirochetes from ear, bladder, and joint tissues harvested 6 weeks postinoculation.
Based on total number of mice infected with 5 × 103 and 1 × 107 spirochetes, the P value calculated by Fisher's exact test was 0.002.
Consistent with these data, analysis of B. burgdorferi reisolated from mice infected with guaAB mutant spirochetes complemented with the guaAB genes on a shuttle vector indicated that all reisolated bacteria retained the complementing plasmid (data not shown). Although the shuttle vector alone is stable during in vitro propagation of spirochetes, the frequency of maintenance of the shuttle vector by spirochetes during growth in the mouse is significantly less than 100% (56). These data suggest that synthesis of the GuaA and GuaB enzymes is required throughout infection and indicate that the cytoplasmic GuaA and GuaB proteins are not neutralizing targets of the mammalian immune response. In contrast, it has been demonstrated that the shuttle vector encoding the surface-localized OspC protein is actively selected against during infection of immunocompetent mice and is not present in B. burgdorferi recovered from mouse tissue bacteria reisolated after 10 weeks of persistent infection (56).
Spirochetes lacking GuaAB are attenuated for replication in the tick.
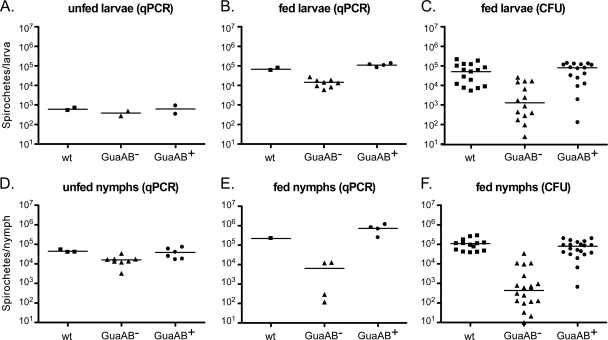
Spirochetes lacking GuaAB were unable to survive in mice following needle inoculation (Table 3). To determine whether the guaAB gene products are also required for B. burgdorferi colonization, replication, and/or persistence in ticks, we artificially infected I. scapularis larvae with spirochetes functionally lacking or containing GuaAB. Larval ticks were immersed in B. burgdorferi cultures that were the same density. qPCR using total DNA isolated from unfed infected larvae demonstrated that the spirochete loads were equivalent (∼500 spirochetes/tick) prior to feeding on mice regardless of the presence of the guaAB gene products (Fig. 5A) (H = 2.0; df = 2; P = 0.38), indicating that all ticks received comparable inocula and suggesting that GuaAB are not involved in spirochete colonization of the tick midgut. Comparably infected groups of larvae were allowed to feed to repletion on naive mice in three separate experiments. The spirochete densities in ticks were determined by qPCR and by plating dilutions of triturated whole ticks on solid BSK medium 7 to 21 days following the blood meal. Both methods of analysis demonstrated that there was an approximately 100-fold decrease in the median number of GuaAB− spirochetes per larval tick, resulting in a GuaAB-dependent statistical difference in the mean rank sums of the spirochete load per larval tick (Fig. 5B and 5C) (for qPCR, H = 10.29, df = 2, and P = 0.0058; for CFU determination, H = 17.49, df = 2, and P = 0.002).
FIG. 5.
Spirochetes lacking GuaAB are attenuated for growth in fed ticks. I. scapularis larvae were artificially infected by immersion in liquid cultures of B. burgdorferi clone A3-M9 (wt), A3-M9 guaA::flgBp-kan/pBSV2G (GuaAB−), or A3-M9 guaA::flgBp-kan/pBSV2G guaAB (GuaAB+). (A) Total DNA was isolated from unfed larvae, and the spirochete density was assessed by qPCR. The symbols indicate the average numbers of spirochetes per larval tick calculated from the number of genome equivalents in the pooled DNA isolated from separate groups of 40 unfed larval ticks. (B and C) Seven to 21 days after the ticks were allowed to feed to repletion, the number of spirochetes per larval tick was determined by qPCR (B) and by plating dilutions of triturated whole fed larvae on solid BSK medium and determining the number of CFU (C). The symbols indicate the number of spirochetes per fed larval tick calculated from the number of genome equivalents in the pooled DNA isolated from separate groups of 20 larval ticks (B) or the number of colonies per individual fed larval tick (C). (D) Following molting, the spirochete densities in unfed nymphs were determined by qPCR. The symbols indicate the number of spirochetes per unfed nymph, calculated from the number of genome equivalents in the pooled DNA isolated from separate groups of four to seven unfed nymphs. (E and F) Seven to 21 days after the ticks were allowed to feed to repletion, the number of spirochetes per nymphal tick was determined by qPCR (E) and by plating dilutions of individual triturated whole fed nymphs on solid BSK medium and determining the number of CFU (F). The symbols indicate the number of spirochetes per fed nymphal tick calculated from the number of genome equivalents in the pooled DNA isolated from separate groups of one to three nymphal ticks (E) or the number of colonies per individual fed nymphal tick (F). The bars indicate the median values of the data sets. The data were analyzed using the Kruskal-Wallis nonparametric rank test followed by Dunn's multiple-comparison test.
Following molting, spirochete loads in unfed nymphal ticks were determined by qPCR. In the absence of active replication, the median number of spirochetes functionally lacking guaAB per unfed nymph was approximately twofold less than the number per unfed nymph harboring GuaAB+ spirochetes (Fig. 5D) (H = 8.647; df = 2; P = 0.0013). Further analysis of the spirochete load per nymph by qPCR and plating of triturated whole nymphs was carried out 7 to 21 days following feeding to repletion on naive mice. As observed for infected larval ticks, the median spirochete load in fed nymphs infected with spirochetes lacking GuaAB was 100-fold less than the median spirochete load in nymphs infected with the complemented GuaAB+ clone or with the wild-type control, resulting in a GuaAB-dependent statistical difference in the mean rank sums of the spirochete load per nymph (Fig. 5E and 5F) (for qPCR, H = 6.667, df = 2, and P = 0.0357; CFU determination, H = 34.86, df = 2, and P < 0.0001). Together, these data indicate that in contrast to the findings for the mammalian environment, the guaAB gene products are important, but not essential, for spirochete replication in the tick. The biochemical activities of the GuaA and GuaB enzymes contribute to the growth of B. burgdorferi following a tick blood meal.
GuaAB are essential for mouse infection by tick bite.
The naive mice on which infected ticks fed were assessed to determine whether they were infected with B. burgdorferi. None of the mice on which either larvae or nymphs infected with spirochetes lacking GuaAB fed became infected (Table 4). In contrast, three of four mice on which larval ticks infected with GuaAB+ spirochetes fed and four of four mice on which nymphal ticks infected with GuaAB+ spirochetes fed were seropositive and reisolation positive (Table 4). The difference in infectivity was not likely due to a difference in the starting inoculum, as there was little GuaAB-dependent difference in spirochete densities in unfed larval or nymphal ticks just preceding attachment and feeding (Fig. 5A and 5D). However, the observed difference in infectivity may have been affected by the inability of the GuaAB-deficient spirochetes to replicate to densities equal to the densities of GuaAB+ spirochetes in the tick midgut during the blood meal. Together, our data indicate that the guaAB gene products are essential for survival of B. burgdorferi in the mammal when the bacteria are introduced by needle inoculation, as well as by tick bite, the natural route of infection (Tables 3 and 4), and that these gene products contribute to the ability of spirochetes to replicate to high densities in the tick midgut following a blood meal (Fig. 5).
TABLE 4.
Tick bite infection of mice with B. burgdorferi lacking or containing GuaAB
| Clone | No. of mice infected by larval tick bite/total no. analyzeda | No. of mice infected by nymphal tick bite/total no. analyzeda | Total no. of mice infected/total no. analyzedb |
|---|---|---|---|
| A3-M9 guaA::flgBp-kan/pBSV2G | 0/4 | 0/4 | 0/8 |
| A3-M9 guaA::flgBp-kan/pBSV2G guaAB | 3/4 | 4/4 | 7/8 |
The data are pooled data from two separate tick feeding experiments. Infection was assessed by immunoblot analysis with cell lysates of B. burgdorferi and E. coli producing P39 recombinant protein at 3, 6, and 9 weeks postinoculation and by reisolation of spirochetes from ear, bladder, and joint tissues at 9 weeks postinoculation.
Based on total number of mice infected by larval and nymphal tick bites, the P value calculated by Fisher's exact test was 0.02.
DISCUSSION
The B. burgdorferi strain B31 genome is divided into as many as 24 genetic elements, including a single 910-kbp linear chromosome, 11 circular plasmids, and 12 linear plasmids (8, 12, 31, 46, 47). It is currently not known why the genome of this spirochete is divided into a diverse set of replicons. The cp26 replicon has been referred to as a mini-chromosome because it carries genes essential for bacterial growth, as well as mammalian infectivity, and is present in all natural isolates (2, 7, 18, 20, 44, 52). It has recently been suggested that linkage of ospC with the constitutively required functions on cp26 encoded by resT, BBB26, and BBB27 provides a mechanism for the maintenance of ospC in environments where this gene is not required or is disadvantageous (20, 56). Here we establish that two additional genes present on the cp26 plasmid, guaA and guaB, and the enzymes that they encode, GMP synthase and IMPDH, respectively, are essential for mammalian infectivity (Fig. 4 and Tables 3 and 4) and provide a growth advantage to spirochetes in the tick (Fig. 5). Moreover, these data provide additional examples of genes vital to the survival of B. burgdorferi in vivo that are located on plasmids, reinforcing the conclusion that genetic linkage of critical physiological and virulence functions on the cp26 plasmid is significant for stable maintenance of this plasmid throughout the infection cycle of B. burgdorferi.
A deletion or insertion in guaA effectively resulted in a GuaAB-deficient clone due to the inability of the mutant guaA allele to functionally complement an E. coli guaA mutant, the polar effect of the mutation on guaB, and the inability of the mutant spirochetes to convert IMP or dIMP to GMP or dGMP for RNA and DNA synthesis (Fig. 3) (23). Spirochetes lacking GuaAB were unable to infect mice after needle inoculation of up to 1 × 107 spirochetes (Fig. 4 and Table 3) and after a tick bite, the natural route of infection (Fig. 4 and Table 4); however, complementation with guaAB in trans restored infectivity, confirming that the in vivo defect resulted from loss of GuaAB. The median densities in fed ticks of spirochetes functionally lacking guaAB were up to 100-fold less than those of spirochetes containing these genes (Fig. 5). Together, these data demonstrate that, similar to the pncA gene on lp25, these genes encode physiological functions that are critical for the survival of B. burgdorferi throughout its infection cycle (15, 37, 39; M. W. Jewett, unpublished data). Consistent with our findings, a previous study utilizing random transposon mutagenesis in an infectious B. burgdorferi genetic background identified a transposon mutant with a mutation in the guaB gene that was defective in infection of a mouse (6). However, since the mutation was not complemented, the data suggested but did not prove that the infectivity defect resulted from the guaB mutation.
Our data demonstrate that the guaAB genes are expressed during infection of both immunocompetent and immunocompromised mice (Fig. 2), suggesting that the encoded biochemical activities are important during all stages of infection. This expression pattern contrasts with that of ospC, which is adjacent to the guaAB operon on cp26 and shares a 185-bp intergenic region with guaA (8, 12, 28, 62) and whose expression is dramatically increased in immunocompromised mice (Fig. 2). These data indicate that although guaA and ospC share the same intergenic region that harbors inverted repeats demonstrated to be important for downregulation of ospC expression following early infection (62, 63), expression of the guaAB operon is not coregulated by this region of DNA.
In E. coli, the gua operon is controlled by guanine and adenine nucleotide pools, resulting in induction by excess AMP and repression by excess GMP (30). It is unlikely that expression of the guaAB operon is regulated by AMP in B. burgdorferi, given the absence of the purAB genes from the genome, resulting in the inability of this spirochete to convert IMP to AMP. Instead, IMP in B. burgdorferi is fated for conversion to GMP by GuaA and GuaB (Fig. 1). Because the concentration of guanine in mammalian tissues does not appear to be sufficient to support growth of the spirochete, B. burgdorferi seems to rely primarily on adenine and hypoxanthine salvage during mammalian infection, which therefore requires expression of guaA and guaB for the final steps of guanine nucleotide biosynthesis (Fig. 1). Indeed, hypoxanthine incorporation assays demonstrated that spirochetes lacking GuaAB failed to synthesize guanine nucleotides (23). Moreover, unlike what has been shown for the shuttle vector alone (56), the shuttle vector carrying guaAB was stably retained by guaAB mutant spirochetes reisolated from infected mice, suggesting that the presence of these genes is essential for survival of the spirochetes in the guanine-starved environment of the mammalian host. Our data suggest that in the absence of the guaAB gene products, B. burgdorferi may not be able to synthesize guanine nucleotides, resulting in a survival defect in vivo.
The guaAB gene products are important, but not essential, for spirochete replication in the tick (Fig. 5). The fact that spirochetes lacking GuaAB were able to survive in the tick, albeit at an attenuated level relative to GuaAB-containing bacteria, suggests that a source of guanine nucleosides may be available in a tick blood meal, which allows bypass of the guaAB-encoded functions and therefore spirochete survival (Fig. 1). The degraded cellular material in the digested blood meal may provide a source of guanine nucleosides and guanine deoxynucleosides for salvage by the spirochetes. A direct source of guanine nucleosides and guanine deoxynucleosides from the blood meal may circumvent the requirement for GMP/dGMP synthesis by GuaA and GuaB (Fig. 1), although it may not be sufficient to allow a wild-type level of spirochete replication in the absence of the guaAB gene products. The ability of B. burgdorferi to transport guanine ribonucleosides and deoxyribonucleosides, presumably following dephosphorylation, and its ability upon rephosphorylation to incorporate these precursors into RNA and DNA have recently been demonstrated (23), suggesting a possible mechanism for the attenuated survival in the tick of spirochetes functionally lacking guaAB (Fig. 1). An analogous mechanism likely accounts for the lack of a growth defect of GuaAB-deficient spirochetes in vitro compared to GuaAB+ spirochetes. Similar to a digested tick blood meal, components in the rich, complex BSK II medium may be a source of excess nucleosides or nucleotides for salvage for the spirochetes. There is currently no defined medium available for in vitro culture of B. burgdorferi to test this hypothesis.
Together, the data demonstrate that the GuaAB enzymes are critical for the survival of B. burgdorferi in environments that appear to lack sufficient amounts of guanine, guanosine, and/or deoxyguanosine to support spirochete growth, such as mammalian host tissues, and thereby contribute to the enzootic infectious lifestyle and biological success of the Lyme disease spirochete.
Acknowledgments
We thank Lizbeth Hedstrom, Brandeis University, for the generous gift of recombinant IMPDH. We also thank Tom Schwan, RML, for the generous gift of anti-FlaB antibody H9724; K. Tilly for technical support; and T. Jewett, L. Knodler, and K. Tilly for critical reading of the manuscript. We gratefully acknowledge the graphic talents of Gary Hettrick and Anita Mora, who prepared the figures.
This research was supported by the Intramural Research Program of the NIH NIAID.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 1988. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J. Clin. Microbiol. 26:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, A. G., S. F. Hayes, R. A. Heiland, M. E. Schrumpf, and S. L. Tessier. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G., A. D. Putteet-Driver, and J. Bunikis. 2005. Horizontally acquired genes for purine salvage in Borrelia spp. causing relapsing fever. Infect. Immun. 73:6165-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botkin, D. J., A. Abbott, P. E. Stewart, P. A. Rosa, H. Kawabata, H. Watanabe, and S. J. Norris. 2006. Identification of potential virulence determinants by HimarI transposition of infectious Borrelia burgdorferi B31. Infect. Immun. 74:6690-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byram, R., P. E. Stewart, and P. A. Rosa. 2004. The essential nature of the ubiquitous 26-kb circular replicon of Borrelia burgdorferi. J. Bacteriol. 186:3561-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 9.Elias, A. F., J. L. Bono, J. J. Kupko, P. E. Stewart, J. G. Krum, and P. A. Rosa. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 10.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 13.Grimm, D., C. H. Eggers, M. J. Caimano, K. Tilly, P. E. Stewart, A. F. Elias, J. D. Radolf, and P. A. Rosa. 2004. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 72:5938-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm, D., A. F. Elias, K. Tilly, and P. A. Rosa. 2003. Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect. Immun. 71:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, D., K. Tilly, D. M. Bueschel, M. A. Fisher, P. F. Policastro, F. C. Gherardini, T. G. Schwan, and P. A. Rosa. 2005. Defining plasmids required by Borrelia burgdorferi for colonization of tick vector Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 42:676-684. [DOI] [PubMed] [Google Scholar]
- 16.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 101:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartwick, R. A., A. M. Krstulovic, and P. R. Brown. 1979. Identification and quantitation of nucleosides, bases and other UV-absorbing compounds in serum, using reversed-phase high-performance liquid chromatography. II. Evaluation of human sera. J. Chromatogr. 186:659-676. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch, J., and A. G. Barbour. 1992. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J. Bacteriol. 174:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodzic, E., S. Feng, K. J. Freet, D. L. Borjesson, and S. W. Barthold. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 70:3382-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jewett, M. W., R. Byram, A. Bestor, K. Tilly, K. Lawrence, M. N. Burtnick, F. Gherardini, and P. A. Rosa. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66:975-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jewett, M. W., K. Lawrence, A. C. Bestor, K. Tilly, D. Grimm, P. Shaw, M. VanRaden, F. Gherardini, and P. A. Rosa. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence, K., M. W. Jewett, P. A. Rosa, and F. Gherardini. 2009. Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol. Microbiol. 72:1517-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect. Immun. 70:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang, F. T., J. Yan, M. L. Mbow, S. L. Sviat, R. D. Gilmore, M. Mamula, and E. Fikrig. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 72:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Margolis, N., D. Hogan, K. Tilly, and P. A. Rosa. 1994. Plasmid location of Borrelia purine biosynthesis gene homologs. J. Bacteriol. 176:6427-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFarland, W. C., and B. A. D. Stocker. 1987. Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3:129-141. [DOI] [PubMed] [Google Scholar]
- 30.Mehra, R. K., and W. T. Drabble. 1981. Dual control of the gua operon of Escherichia coli K12 by adenine and guanine nucleotides. J. Gen. Microbiol. 123:27-37. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. C., J. L. Bono, K. Babb, N. El-Hage, S. Casjens, and B. Stevenson. 2000. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J. Bacteriol. 182:6254-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681-706. [DOI] [PubMed] [Google Scholar]
- 33.Noriega, F. R., G. Losonsky, C. Lauderbaugh, F. M. Liao, J. Y. Wang, and M. M. Levine. 1996. Engineered ΔguaB-A ΔvirG Shigella flexneri 2a strain CVD 1205: construction, safety, immunogenicity, and potential efficacy as a mucosal vaccine. Infect. Immun. 64:3055-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersson, J., M. E. Schrumpf, S. J. Raffel, S. F. Porcella, C. Guyard, K. Lawrence, F. C. Gherardini, and T. G. Schwan. 2007. Purine salvage pathways among Borrelia species. Infect. Immun. 75:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Policastro, P. F., and T. G. Schwan. 2003. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J. Med. Entomol. 40:364-370. [DOI] [PubMed] [Google Scholar]
- 37.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 38.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, J. de la Fuente, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 102:6972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosa, P. A., and D. Hogan. 1992. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants, p. 95-103. In U. G. Munderloh and T. J. Kurtti (ed.), Proceeding of the First International Conference on Tick Borne Pathogens at the Host-Vector Interface. University of Minnesota, St. Paul, MN.
- 41.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson, W. J., W. Burgdorfer, M. E. Schrumpf, R. H. Karstens, and T. G. Schwan. 1991. Antibody to a 39-kilodalton Borrelia burgdorferi antigen (P39) as a marker for infection in experimentally and naturally inoculated animals. J. Clin. Microbiol. 29:236-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson, W. J., W. Cieplak, M. E. Schrumpf, A. G. Barbour, and T. G. Schwan. 1994. Nucleotide sequence and analysis of the gene in Borrelia burgdorferi encoding the immunogenic P39 antigen. FEMS Microbiol. Lett. 119:381-388. [DOI] [PubMed] [Google Scholar]
- 44.Simpson, W. J., C. F. Garon, and T. G. Schwan. 1990. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb. Pathog. 8:109-118. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, W. J., M. E. Schrumpf, S. F. Hayes, and T. G. Schwan. 1991. Molecular and immunological analysis of a polymorphic periplasmic protein of Borrelia burgdorferi. J. Clin. Microbiol. 29:1940-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiol. 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, P. E., A. Bestor, J. N. Cullen, and P. A. Rosa. 2008. Tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infect. Immun. 76:1970-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 50.Stewart, P. E., X. Wang, D. M. Bueschel, D. R. Clifton, D. Grimm, K. Tilly, J. A. Carroll, J. J. Weis, and P. A. Rosa. 2006. Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect. Immun. 74:3547-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Straley, S. C., and P. A. Harmon. 1984. Growth in mouse peritoneal macrophages of Yersinia pestis lacking established virulence determinants. Infect. Immun. 45:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terekhova, D., R. Iyer, G. P. Wormser, and I. Schwartz. 2006. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J. Bacteriol. 188:6124-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tilly, K., A. Bestor, M. W. Jewett, and P. Rosa. 2007. Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect. Immun. 75:1517-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tilly, K., S. Casjens, B. Stevenson, J. L. Bono, D. S. Samuels, D. Hogan, and P. Rosa. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol. 25:361-373. [DOI] [PubMed] [Google Scholar]
- 55.Tilly, K., D. Grimm, D. M. Bueschel, J. G. Krum, and P. Rosa. 2004. Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis. 4:159-168. [DOI] [PubMed] [Google Scholar]
- 56.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tilly, K., L. Lubke, and P. Rosa. 1998. Characterization of circular plasmid dimers in Borrelia burgdorferi. J. Bacteriol. 180:5676-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tourand, Y., T. Bankhead, S. L. Wilson, A. D. Putteet-Driver, A. G. Barbour, R. Byram, P. A. Rosa, and G. Chaconas. 2006. Differential telomere processing by Borrelia telomere resolvases in vitro but not in vivo. J. Bacteriol. 188:7378-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Lookeren Campagne, M. M., J. Franke, and R. H. Kessin. 1991. Functional cloning of a Dictyostelium discoideum cDNA encoding GMP synthetase. J. Biol. Chem. 266:16448-16452. [PubMed] [Google Scholar]
- 60.Weber, G. 1983. Biochemical strategy of cancer cells and the design of chemotherapy: G. H. A. Clowes Memorial Lecture. Cancer Res. 43:3466-3492. [PubMed] [Google Scholar]
- 61.Wishart, D. S., D. Tzur, C. Knox, R. Eisner, A. C. Guo, N. Young, D. Cheng, K. Jewell, D. Arndt, S. Sawhney, C. Fung, L. Nikolai, M. Lewis, M. A. Coutouly, I. Forsythe, P. Tang, S. Shrivastava, K. Jeroncic, P. Stothard, G. Amegbey, D. Block, D. D. Hau, J. Wagner, J. Miniaci, M. Clements, M. Gebremedhin, N. Guo, Y. Zhang, G. E. Duggan, G. D. Macinnis, A. M. Weljie, R. Dowlatabadi, F. Bamforth, D. Clive, R. Greiner, L. Li, T. Marrie, B. D. Sykes, H. J. Vogel, and L. Querengesser. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35:D521-D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, Q., K. McShan, and F. T. Liang. 2007. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol. Microbiol. 64:220-231. [DOI] [PubMed] [Google Scholar]
- 63.Xu, Q., K. McShan, and F. T. Liang. 2008. Verification and dissection of the ospC operator by using flaB promoter as a reporter in Borrelia burgdorferi. Microb. Pathog. 45:70-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, Q., S. V. Seemanapalli, K. McShan, and F. T. Liang. 2006. Constitutive expression of outer surface protein C diminishes the ability of Borrelia burgdorferi to evade specific humoral immunity. Infect. Immun. 74:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zalkin, H., and P. Nygaard. 1996. Biosynthesis of purine nucleotides, p. 561-579. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 66.Zhong, J., S. Skouloubris, Q. Dai, H. Myllykallio, and A. G. Barbour. 2006. Function and evolution of plasmid-borne genes for pyrimidine biosynthesis in Borrelia spp. J. Bacteriol. 188:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou, X., M. Cahoon, P. Rosa, and L. Hedstrom. 1997. Expression, purification, and characterization of inosine 5′-monophosphate dehydrogenase from Borrelia burgdorferi. J. Biol. Chem. 272:21977-21981. [DOI] [PubMed] [Google Scholar]