Abstract
Reversible insertion of IS492 at a site within epsG on the Pseudoalteromonas atlantica chromosome controls peripheral extracellular polysaccharide production and biofilm formation by P. atlantica. High-frequency precise excision of IS492 from epsG requires 5 and 7 bp of flanking DNA, suggesting that IS492 transposition involves a site-specific recombination mechanism. The site specificity of IS492 insertion was examined in P. atlantica and shown to be specific for a 7-bp target, 5′-CTTGTTA-3′. Characterization of numerous insertion events at the target site in epsG indicated that insertion is also orientation specific. The frequency of IS492 insertion at the epsG target site (2.7 × 10−7/cell/generation), determined by quantitative PCR, is 4 to 5 orders of magnitude lower than the frequency of IS492 precise excision from the same site. Comparison of insertion sites for IS492 and the highly related ISPtu2 from Pseudoalteromonas tunicata suggests DNA sequence and/or structural features that may contribute to site recognition and recombination by the transposase of IS492.
A variety of specialized DNA recombination systems create genetic diversity and regulate gene expression in prokaryotes and eukaryotes. These specialized recombination systems fall into two categories, transposition and site-specific recombination. Transposition involves movement or copying of a mobile genetic element from a donor DNA site to a target DNA site with no requirement for DNA homology. Transposable elements drive the evolution of genomes through DNA rearrangements resulting from the process of transposition or by homologous recombination between repeated copies of the element in the genome (for a review, see reference 34). Site-specific recombination systems, which require very short, shared sequences between the recombining DNA segments, often regulate gene expression or generate antigenic diversity (for a review, see reference 16).
Specialized DNA recombinases of the DEDD motif family (also called the Piv/MooV family) mediate reactions that have features of both transposition and site-specific recombination (8, 18, 23, 28, 29, 37). The transposases of the IS110/IS492 family of insertion sequences and the Piv (pilin inversion) site-specific DNA invertases of Moraxella lacunata and Moraxella bovis are members of the DEDD family of recombinases, (8, 23, 29, 37). The recombinases of this family do not share the conserved amino acid motifs of the characterized site-specific tyrosine (Y) or serine (S) recombinase families or the classical (DDE), rolling circle, or Y or S transposase (Tnp) families (11). However, like the DDE Tnps, molecular modeling indicates that the conserved acidic residues are assembled into the catalytic pocket of an RNase H-like fold. The DEDD motif residues are located at the same linear spacing and predicted tertiary location as the DEDD catalytic amino acids of the RuvC-like Holliday junction resolvases (8, 37), which are part of the polynucleotidyl transferase superfamily (33), and all four acidic residues have been shown to be required for Piv catalysis of inversion (8).
The IS110/IS492 family, which includes almost 200 elements in Eubacteria and Archaea (IS Finder [www-is.biotoul.fr]), differs from other insertion element (IS) families in the following aspects: (i) the ISs either lack terminal inverted repeats or have subterminal inverted repeats; (ii) insertion of the IS does not necessarily generate a target site duplication; and (iii) the ISs are mobilized by the DEDD recombinases (9, 12, 15, 18, 19, 22-24, 26, 28, 29, 32, 35). These features suggest that IS110-related elements utilize a novel mechanism for DNA transposition. The products of movement for some IS110/IS492-related elements point to a site-specific mechanism for recombination, such as the restored target sites following excision of IS492 or IS4321 from donor DNA (4, 28). Genetic and molecular analyses of IS492 movement have shown that the DEDD recombinase MooV (mover of IS492 in oceanic variants) mediates high-frequency precise excision of the element from its donor site, restoring the target DNA without error (19, 29). A site-specific mechanism for excision of the 1,202-bp IS492 element is supported by the requirement for a 5-bp direct repeat that flanks the inserted element (29). Transposase-mediated precise excision is unprecedented for classical transposable elements (14, 20) and is usually observed with the Y- and S-Tnps (11).
In this work, we have focused on characterizing the insertion step in transposition of the IS492. While it is not uncommon for transposons and ISs to target particular regions of the genome or “hot spots” based on various influencing factors, such as transcription levels, DNA supercoiling, or DNA bending, most transposable elements show either no or limited sequence specificity for target sites (for reviews, see references 11 and 27), with the notable exceptions of Tn7 (30) and IS608 (from the IS200/IS605 family) (3). ISs of the IS110/IS492 family are remarkable in that many have been reported to exhibit both site and directional specificity for insertion, including IS117, IS1383, IS621, IS4321, ISPpu10, ISPst6, and ISPa21 (9, 18, 26, 28, 32, 35).
The insertion reaction for IS492 is of particular interest because it is the counterpart to the high-frequency precise excision of this element which controls phase variation of peripheral extracellular polysaccharides (PEPS) in the gram-negative marine bacterium Pseudoalteromonas atlantica (5). Five IS492 insertion sites have been characterized in P. atlantica previously, and at each site, the element is flanked by the same 5 and 7 bp on the left and right side, respectively, suggesting a common target site (29). Precise excision of IS492 from one of these target sites in the glucosyl transferase gene (epsG) occurs at a frequency of 10−3 to 10−2 per cell per generation on solid media, depending on growth conditions, and correlates directly with the phenotypic switch from PEPS− (crenated colony morphology [C]) to PEPS+ (mucoid colony morphology [M]) (4, 19) (Fig. 1). In this paper we determine the insertion frequency at the epsG target site, using a quantitative PCR (qPCR)-based assay, and identify the sequence and directional specificity of IS492 insertion in P. atlantica. We also report the insertion sites for the closely related element ISPtu2 in Pseudoalteromonas tunicata and discuss possible common features between the target sites of IS492 and other IS110-related elements.
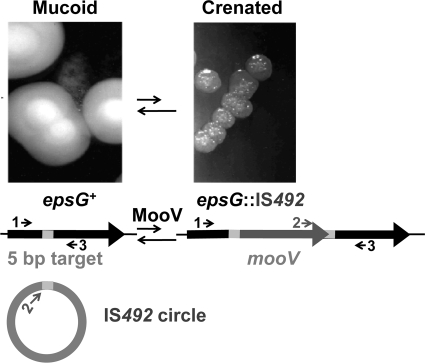
FIG. 1.
IS492 insertion at epsG. Insertion of IS492 (dark gray) into the epsG gene (black) at the 5-bp target site (light gray) that appears in direct repeat upon insertion is shown. The IS492 circle with the 5-bp target sequence linking the ends of the element is a product of precise excision of IS492, but has not been confirmed as an intermediate in insertion. The primers used to characterize insertion of IS492 at epsG are depicted as arrows 1 (L58), 2 (CJ250B), and 3 (RVEPS).
MATERIALS AND METHODS
Bacterial strains, media, growth conditions, and reagents.
P. atlantica T6c (10) and the derivative strains DB27 (hsd1 Rifr) and DB27 recA (DB50) were gifts from D. Bartlett (5). These strains were cultured at 25°C under aerobic conditions on Difco 2216 marine agar (MA) or marine broth (MB) as described in reference 29. Escherichia coli DH5α (29) was grown at 37°C on Luria-Bertani (LB) medium (FisherBiotech). Restriction and modification enzymes used in plasmid constructions and Southern blot analysis were purchased from New England Biolabs. All sequencing was performed by the University of Michigan Biomedical Research Core Facilities.
Primers for PCR and qPCR.
The sequences of all oligonucleotides utilized in this work are listed in Table 1.
TABLE 1.
Oligonucleotides used in PCR and qPCR
| Primer name | Sequence (5′ to 3′)a |
|---|---|
| AGARL | AGTATATGATGGAAGAGTTAGAC |
| AGARR | CATAACCTATGTTGGCTGGG |
| AGARPRB | HEX-TATTTTGGTCGAGATAACGGTGGC-BHQ2 |
| CJ250B | TAAGAAAGTGGCTATTATTGCGTGC |
| EPSL0 | ATGGATTATATACCTGCATGACATGC |
| EPSPRB3 | FAM-TCTATCAAGCTCTCTATTGAGCAAGG-BHQ1 |
| FWMV | ATTAGCTATTGACGCCATAGTCT |
| L58 | CGGTACTGTCTTATCATCCTAATCG |
| MOOV2 | TGCAACTAAATCACTCATAGCC |
| RVEPS | TCAATTTCTTCAACAGGAG |
FAM, 6-carboxyfluorescein; BHQ, Black Hole Quencher; HEX, hexachlorofluorescein.
Isolation of PEPS+-to-PEPS− (PEPS+→PEPS−) phase variants.
M-to-C (M→C) variants were obtained from P. atlantica strains DB27 and DB27 recA in the following manner. One M colony was used to inoculate 5 ml MB and incubated at 25°C for 18 h at 220 rpm. After 18 h of incubation, a sterile cotton swab was used to remove the biofilm at the air-broth interface, streaked onto an MA plate, and used to inoculate another 5 ml MB. This procedure was repeated until C colonies appeared on the streak from the cotton swab. C colonies were then restreaked for isolation. One M colony present in the same cotton swab streak was isolated and marked “sibling.”
In addition, the following three clonal cell lineages from mucoid DB27 recA were created: M0→C1(M0)→M1(C1)→C2(M1)→M2(C2)→C3(M2), where the parenthetical subscript indicates the parental variant from which the variant was isolated. In these cell lineages, M siblings were also isolated with each C variant.
Once isolated colonies were visible, colony PCR was performed as described previously (29), using primer pairs CJ250B/RVEPS and CJ250B/L58 to amplify the 3′ end of the element-eps junction, assaying for insertion in either orientation. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced using the CJ250B primer. Ten C variants were analyzed from DB27 and 50 C variants from DB27 recA.
Southern blotting.
The restriction endonuclease used for the Southern blot analyses (BanII) was chosen based on the sizes of restriction fragments from the P. atlantica chromosomal DNA containing IS492, which were determined using Pattern Locator software (25) on the P. atlantica genome sequence (GenBank NC 008228). BanII has no recognition site in IS492. Digested P. atlantica chromosomal DNA was transferred to Magnagraph nylon membranes (GE Osmonics, Minnetonka, MN), using standard techniques described by Ausubel et al. (1) or the Bio-Rad vacuum blotter. DNA was cross-linked to the membrane by using a Bio-Rad GS gene linker. Hybridization and detection steps were performed using DIG high prime DNA labeling and detection starter kit I (Roche Applied Science, Indianapolis, IN). Hybridization was performed at 68°C, followed by one low-stringency wash in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 10 min, one moderate-stringency wash in 0.5× SSC-0.1% sodium dodecyl sulfate for 10 min, and two high-stringency washes in 0.1× SSC for 10 min each (all washes were performed at 68°C). The probe was prepared using the PCR DIG probe synthesis kit (Roche); DIG-dUTP was incorporated into a 744-bp PCR product in reaction mixtures containing primers EPSL0 and MooV2 and the plasmid template pAG949 encoding IS492 (29). The left-most sequence of IS492 was used to generate the probe to minimize hybridization with partial copies of IS492 on the P. atlantica chromosome, all of which are missing a significant portion of the left end.
qPCR to determine the insertion frequency at epsG and colony morphology assay to obtain PEPS+→PEPS− phase variation frequency.
The following protocol was carried out with three independent P. atlantica DB27 recA mucoid colonies. Each colony was resuspended in 40 μl MB, serially diluted and plated onto MA to give approximately 500 colonies per plate, and incubated for 7 days at 25°C. One hundred colonies from a plate for each of the three original colonies were pooled, washed once in 1 ml marine broth, and resuspended in 1 ml marine broth. One aliquot of 100 μl was removed for serial dilutions and platings for enumeration of mucoid to crenated switching within the cell population. Genomic DNA was isolated using standard protocols (1) from an aliquot of each sample correlating to ∼7 × 109 cells. Each genomic sample was digested with NgoMIV, which does not have a recognition site within the agr or IS492::epsG sequences, before determining the DNA concentration (based on absorbance at 260/280 nm; Eppendorf BioPhotometer).
qPCRs were performed using three replicates from each of the three genomic DNA samples. The forward and reverse primers for the IS492::eps target site and for agr are designated FWMV/RVEPS and AGARL/AGARR, respectively; the probes for the IS492::epsG site and for agr are EPSPRB3 (FAM-490 fluorophore on the 5′ end and Black Hole Quencher 1 on the 3′ end) and AGARPRB (HEX-530 fluorophore on the 5′ end and Black Hole Quencher 2 on the 3′ end), respectively. qPCR mixtures (50 μl) contained the following: 100 ng digested chromosomal DNA, 1× Taq polymerase PCR buffer B (Fisher), 2 U Taq polymerase, 400 nM each deoxynucleoside triphosphate (Amersham), 6 mM MgCl2, 300 nM of AGARL and AGARR, 500 nM of FWMV and RVEPS, 5 nM of AGARPRB, and 200 nM of EPSPRB3. To generate a standard curve for quantification of the template in the reactions, a standard plot was generated for each target using pAG957 (epsG::IS492) and pBHG116 (agr). Plasmid pAG957 contains IS492 with 357 bp of mooV replaced with the chloramphenicol acetyltransferase (cat) gene (IS492Δmoov::cat) inserted at the target site within 134 bp of the epsG sequence (29). Plasmid DNA was isolated, quantitated, and used in a dilution series corresponding to 100 pg to 1 fg target DNA (4 replicates of each dilution were used for the standard curve in each experiment). Cycling conditions for standards and experimental reactions were as follows: 50°C for 2 min, 95°C for 3 min, followed by 30 cycles of 95°C for 30 s and 57°C for 2 min. Standard plots of log starting quantity versus threshold cycles (CT) were generated with the Bio-Rad iCycler iQ software v3.0, using the data from the dilution series corresponding to the amplification of agr and IS492::epsG; the starting quantities (SQ) of chromosomes with an IS492 insertion at eps and total chromosomal copies (agr) in each genomic sample were calculated.
The frequency of IS492 insertion at eps (PINS) was calculated using the formula: PINS = 1 − (1 − XINS)1/n, where n is the number of generations in the sample population (13). XINS was determined by dividing the total amount of IS492::eps DNA (in fg) present in each sample by the total amount of agr locus (in fg) present in the same sample. When calculating the number of generations (n), the inoculum was 100 cells (each colony in the pooled samples started from a single cell). M→C phase variation frequency (PPV′) could not be determined due to lack of C colonies on the plates after serial dilutions of pooled samples.
RESULTS
The IS492::epsG junction is identical in PEPS+→PEPS− phase variants.
In order to determine the specificity of target site selection by IS492 within the eps locus, 60 independent PEPS− (C) variants were isolated from PEPS+ (M) cells of DB27 and DB27 recA strains by using a biofilm enrichment technique that is described in Materials and Methods. The epsG locus of each variant was examined for the presence of IS492 by PCR analyses (Fig. 1). Using an outwardly directed IS492-specific primer (CJ250B) and epsG-specific reverse primers (L58 and RVEPS), the epsG junction with the 3′ end of IS492 was amplified and sequenced. Fifty-eight of the 60 PEPS+→PEPS− phase variants that were examined exhibited insertion of IS492 at the same site and in the same orientation, giving identical epsG::IS492 junction sequences (Fig. 2). The insertion orientation bias places mooV and epsG transcription in the same direction. The two PEPS− isolates that did not give a PCR product in the screen for insertion of IS492 at the epsG site were shown by PCR to have part of the epsG gene deleted (data not shown). Southern blot analysis showed that these two PEPS− (C) variants have the same pattern of IS492 insertions as does the mucoid parent, suggesting that IS492 is not inserted in epsG or at a new location on the chromosome and, thus, the deletion in epsG is responsible for the PEPS− phenotype (data not shown).
FIG. 2.
Chromosomal sequence flanking IS492 and ISPtu2 insertions in P. atlantica and P. tunicata. Approximately 70 bp and 36 bp of chromosomal sequence flanking the left and right ends, respectively, of the seven complete copies of IS492 found in P. atlantica and the three copies of ISPtu2 found in P. tunicata are shown. The right flank sequence for the three partial elements in P. atlantica is also depicted; the partial copies have various amounts of the right end of IS492 and are missing the left 5-bp flank sequence, as well as the left end of IS492. Gray boxes highlight the bases that are conserved in at least 8 of the 10 elements. The bold italic sequences are the completely conserved 5-bp and 7-bp flank sequence. The position of each IS within the chromosomal sequence is shown by the boldface IS. The arrows below the sequence indicate perfect inverted repeats within the conserved sequence.
IS492 selection of target sites on the P. atlantica chromosome is site specific.
Sequencing of the complete P. atlantica T6c genome in collaboration with the Joint Genome Institute and the Department of Energy (GenBank NC 008228) revealed seven complete copies of IS492 at different chromosomal sites in this strain, all of which have the same 5- and 7-bp flanking sequences, and three partial sequences, which have the right end of IS492 with the 7-bp flank sequence (Fig. 2). Excision of IS492 from the chromosome removes one copy of the 5-bp sequence, leaving 5′-CTTGTTA-3′ (19). Earlier work showed that these conserved 5-bp and 7-bp flanking sequences are sufficient for precise excision of IS492 from a plasmid in E. coli (2, 29); thus, the number of potential IS492 target sites may be quite high if the same sequence is also a minimum target site. The number of nonoverlapping sites on the P. atlantica chromosome containing the conserved 5-bp or 7-bp flanking sequences was determined to be 11,102 and 783, respectively, using Pattern Locator software from the University of Georgia (UGA) Computational Microbiology Laboratory (25). The potential insertion of IS492 at new sites on the chromosome was assayed by Southern blotting.
The chromosomal DNA used in the Southern blot analyses was from two DB27 M-cell lineages proceeding M0→C1(M0)→M1(C1)→C2(M1)→M2(C2)→C3(M2), which included mucoid cells [designated mucoid siblings MS(M0), MS(M1), etc.] that were isolated with the C variants from M0, M1, and M2. As seen in the representative blot shown in Fig. 3A, IS492 is observed only to move into the epsG locus in C cells and out of the epsG locus in M cells; no new insertion sites were apparent. All of these variants were characterized by PCR for site-specific insertion and precise excision of IS492 at the single epsG site (Fig. 3B). Three additional M-cell lineages, which contained M0→C1(M0) plus MS(M0), and 10 independent M0→C1(M0) variants were examined by Southern blot analysis and also showed movement of IS492 only into or out of the single epsG site (data not shown).
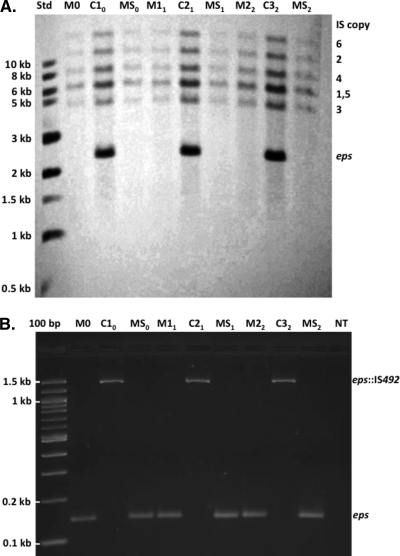
FIG. 3.
Southern blot and PCR analyses of PEPS+→PEPS− phase variants. (A) Genomic DNA from a representative DB27 M-cell lineage (M0→C10→M11→C21→M22→C32) and the sibling mucoid cells isolated with the C variants (MS0, MS1, and MS2) was probed with a 744-bp digoxigenin-labeled PCR product that is specific for the complete copies of IS492 on the P. atlantica chromosome. The chromosome fragments that hybridized with the probe are labeled to the right of the Southern blot with the number of the IS492 copy that each fragment contains; note that fragments 1 and 5 are of similar sizes and comigrated in gel electrophoresis before transfer to the nylon membrane. The sizes of the labeled molecular-size standards (Std) are indicated to the left of the blot. (B) The DNA samples used for the Southern blot in panel A were also analyzed in PCR assays with the primers 1 and 3 that are shown in Fig. 1. The PCR products showing insertion of IS492 at the eps site are indicated as eps::IS492, and the products corresponding to no insertion are labeled eps.
A variant of C cells called translucent (T) cells can also arise during growth on MA. This variant, like C cells, is defective for expression of PEPS (5) and, thus, potentially contained IS492 inserted at a new site affecting PEPS production. T variants and M siblings were isolated from three independent C colonies, and the location of IS492 copies was determined by Southern blot analyses. The chromosomal DNA fragments containing insertions were found to be the same for C and the T variants (data not shown); there is no apparent movement of the element to a new location in the T cells. PCR analysis of the seven IS492 copies in the T cells also showed that all copies were in their previously identified locations and, thus, had not moved to a new site within a chromosomal fragment (data not shown).
Insertion of IS492 at the epsG locus is a low-frequency event.
Bartlett et al. (5) estimated that the frequency of PEPS+→PEPS− phase variation (PPV′) is less than 1 × 10−5, based on the inability to detect PEPS− variants arising in M colonies by using colony phase variation assays. As described above, PEPS− variants can be isolated from biofilms that form at the air-broth interface during growth in MB.
To measure PPV′ and the insertion frequency at the epsG site (PINS), DB27 M cells were grown on MA, as described in Materials and Methods, and assayed for PINS and PPV′. PINS was determined using genomic DNA from each of three independent samples in qPCRs designed to measure the total number of chromosomes and the number of chromosomes in which IS492 is inserted into the epsG target site. Two different dual-labeled fluorogenic probes (TaqMan technology) (17) were used to measure amplification of the β-agarase gene, agrA (6), and the epsG::IS492 junction (Fig. 4A). Quantification of agrA copies reflects the number of chromosomes in each sample. The 5′ end of the TaqMan probe, EPSPRB3, is 4 bp downstream of the 5′-CTTGT-3′ sequence that is targeted by IS492 in epsG. The forward primer used, FWMV, anneals to the last 23 bases of IS492. When insertion occurs, the forward primer and probe are brought together in close proximity, resulting in Taq-mediated cleavage of the 5′ fluorophore (FAM-490) from EPSPRB3. The fluorophore fluorescence is detected once it is liberated from the 3′ Black Hole Quencher. No signal is detected when epsG has no IS492 insert, as the forward primer FWMV is IS492 specific. The agrA probe (AGARPRB) anneals 2 bp downstream of the forward primer (AGARL) so that the HEX-530 fluorophore is released with every extension reaction. The amount of these two loci in each sample is quantitated (Fig. 4B), and a frequency of insertion is calculated as described in Materials and Methods. The PINS was determined to be 2.7 × 10−7/cell/generation. Platings of aliquots from the same M-cell samples that were used in the determination of PINS gave no PEPS+→PEPS− phase variants after incubation. In light of the low value for PINS, our inability to determine a value for PPV′ is not surprising and is consistent with the results of Bartlett et al. (5).
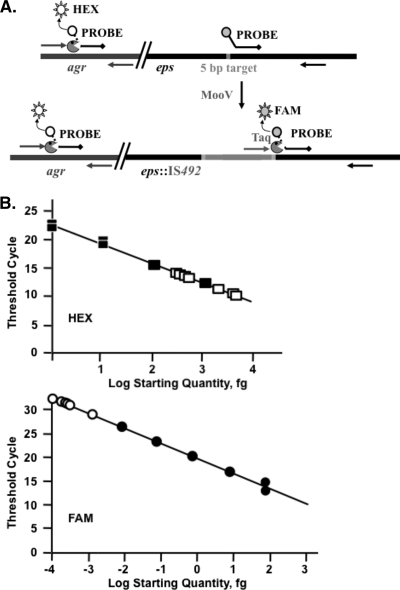
FIG. 4.
qPCR assay for IS492 insertion frequency into epsG. (A) Two TaqMan probes with the different fluorophores (HEX, white circle; FAM, gray circle) and Black Hole Quenchers (black diamonds) were designed to separately measure amplification by the forward and reverse primers for agr (dark-gray arrows) and for the junction of IS492 (light-gray arrow) with epsG (black arrow). Release of the fluorophore (sun) from the probe by the 5′-to-3′ exonuclease activity of Taq polymerase ( ) is measured (Bio-Rad iCycler qPCR system). (B) The plot of the log starting quantity in femtograms for standard DNA (closed squares, agr; closed circles, epsG::IS492) versus the qPCR threshold cycle is used to determine the amount of DNA that has the agr and eps::IS492 loci in the starting reaction for each unknown (open squares, agr; open circles, eps::IS492). From these data, the insertion frequency can be calculated. The correlation coefficients for these plots are 0.995 (HEX) and 0.997 (FAM).
) is measured (Bio-Rad iCycler qPCR system). (B) The plot of the log starting quantity in femtograms for standard DNA (closed squares, agr; closed circles, epsG::IS492) versus the qPCR threshold cycle is used to determine the amount of DNA that has the agr and eps::IS492 loci in the starting reaction for each unknown (open squares, agr; open circles, eps::IS492). From these data, the insertion frequency can be calculated. The correlation coefficients for these plots are 0.995 (HEX) and 0.997 (FAM).
An IS492-related element, ISPtu2, utilizes the same 7-bp target sequence as does IS492.
The genome sequence for P. tunicata recently became available (36), and we used TBLASTN to search for DEDD motif recombinases. A 316-amino-acid recombinase having 76% amino acid identity with MooV was identified, and three genes encoding this recombinase were found within nearly identical 1,201-bp DNA segments at different positions on the chromosome. We submitted the 1,201-bp sequence to IS Finder (www-is.biotoul.fr), and it has been assigned the name ISPtu2. The sequence at each of the ISPtu2 insertion sites is the same 5-bp left flank sequence and 7-bp right flank sequence observed for IS492 insertion in P. atlantica (Fig. 2). In addition, comparison of 100 bp of sequence flanking each side of IS492 and ISPtu2, using Clustal W, indicated significant sequence homology among most of the left flank sequences but no significant homology among the right flank sequences (Fig. 2). Using 200-bp sequences that flank the 7-bp target for each element, the Mfold program (38) predicts an energetically stable bulged stem-loop structure in the left flank sequences. The 7-bp target sequence immediately follows the stem of this extensive bulged stem-loop or is within a short stem-loop that follows the bulged stem-loop (data not shown). Interestingly, the only two IS492 copies in P. atlantica that are or appear to be inserted within coding sequence, the eps copy of IS492 and copy 3 (inserted in opposing orientation within a predicted gene for an immunoglobulin-like group 2 protein), do not have the extensive sequence homology at the left flank, but they maintain a predicted bulged stem-loop structure as seen for the five other IS492 copies and three ISPtu2 copies. This observation and the A-rich sequences and strings of T residues in the conserved left flank sequence prompted us to look for a rho-independent terminator encoded in this region, using the prediction parameters of Kingsford et al. (21), but none of the potential structures fit a consensus rho-independent terminator.
DISCUSSION
Our initial assay for IS492 insertion specificity and directionality focused on the movement of the element in P. atlantica M→C variants, since there are multiple gene targets whose inactivation would result in loss of PEPS production, and the change in colony morphology provides an easy screen for movement of IS492. Examination of 60 M→C variants by PCR and sequencing showed that all the IS492 insertion events targeted the same site in epsG and the orientation of the element relative to the gene was consistently the same. Using Southern blot analysis, movement of the element was followed in C variants and M colonies that were isolated at the same time from parent M colonies and in T and M variants from parent C colonies. Under the growth conditions utilized (low colony density on MA), IS492 excises at high frequency (10−2/cell/generation) from the epsG site (19), and there are hundreds of potential target sites in the genome based on the conserved target sequence at the 10 IS492 (partial and full element) insertion sites in the P. atlantica genome, which is also the minimal sequence required for IS492 precise excision (29). In multiple independent cell lineages, IS492 moved into and out of the epsG site, but no new insertion sites were detected, suggesting that IS492 is highly specific in selection of insertion sites. Measurement of the frequency of insertion at the epsG site by qPCR revealed a very low level of insertion, 2.7 × 10−7/cell/generation. Obviously, if IS492 moves into new sites at this low frequency, we cannot assay enough colonies by Southern blotting to conclude that IS492 inserts only at the previously identified sites. However, based on the sites identified for IS492 insertion in P. atlantica and for the highly related ISPtu2 in P. tunicata, it safe to say that IS492 inserts with unusual site specificity, targeting the 7-bp sequence 5′-CTTGTTA-3′, and it is likely that other aspects of the surrounding sequence contribute to the target specificity.
Potential reasons for the low frequency of insertion into the epsG target site include the very low level of excision of IS492 elements at non-eps chromosomal sites and the high level of transcription through the eps target site (19). PCR analysis shows that IS492 precise excision does occur at a low frequency from all of the non-epsG sites (20; our unpublished data). The element at one of these non-eps sites must be the source for IS492 inserted at eps. However, in the Southern blot analyses of M cells and their C variants, where IS492 moves into the eps target site, excision of IS492 from one of the non-eps chromosomal sites was not detected. These results suggest that homologous recombination with a sister chromosome frequently restores the element at the donor sites or that transposition from non-eps sites can be replicative, unlike in excision from the epsG site. The nature of the IS492 transposition intermediate and the source of the element that inserts at epsG in P. atlantica are currently being investigated.
As described in the introduction, it is very rare for transposable elements, particularly ISs, to exhibit site-specific insertion. However, elements of the IS110/IS492 family exhibit both site specificity and directionality in insertion. For example, the Streptomyces coelicolor elements IS110 and IS117 insert into a single specific site on the phage φC31 and on the Streptomyces lividans chromosome, respectively (7, 18). IS621 and ISPpu10 target repetitive extragenic palindromic sequences in Escherichia coli and Pseudomonas putida, respectively (9, 32), and IS1111-attC group elements target the pseudopalindromes of integron attC sites (35). IS1383 of P. putida strain H inserts into the inverted repeats of IS1384, which is a member of the IS5 family (26), and IS4321 and IS5075 (members of the IS1111 subgroup of the IS110/IS492 family) target the terminal inverted repeats of Tn21 family transposons (28). All of these elements were also shown to insert into their preferred site in one direction. In comparison to these members of the IS110/IS492 family, IS492 also exhibits insertion directionality and shows remarkable sequence-based site specificity, consistently targeting the same 7-bp sequence, which is often adjacent to sequence that encodes potential stem-loop structures. Although two IS492 insertion sites (copies 4 and 5) in P. atlantica are adjacent to an IS4-related element, designated ISPat1 (IS Finder [www-is.biotoul.fr]), IS492 does not target the inverted repeats of the element as seen with other members of the family; copies 4 and 5 are inserted 73 and 74 bp, respectively, downstream of a copy of ISPat1.
What determines the site specificity and directionality for the IS110/IS492 elements? This question is difficult to answer for the family as a whole. There is no sequence or structural feature that is consistently found at all the insertion sites for members of the IS110/IS492 family, although most have a stem-loop structure predicted at various distances from the actual insertion site. Some of these elements have been shown to form circles upon precise excision in which the junction of the element ends has tantalizing sequence homology with the target site, including IS492 (5-bp junction sequence; Fig. 1), IS1383, and IS117 (3-bp junction sequences), suggesting a site-specific recombination event that also confers directionality. The requirement for the 5-bp flanking sequence by IS492 supports this mechanism. However, most of the IS110/IS492 elements do not share homology with the target site. Post and Hall (31) have suggested, based on comparison of IS1111-attC group elements and their target sites, that a sequence within the element directs insertion specificity and directionality, perhaps through an encoded RNA that interacts with the transposase. This mechanism certainly is appealing in light of the recent work of Barabas et al. (3) that demonstrates the role of IS608 DNA as a component of the transposase complex, directing both site-specific insertion and precise excision of IS608. IS608, a member of the IS200/IS605 family, encodes a Y-Tnp that is structurally and, most likely, mechanistically different from the DEDD transposases of the IS110/IS492 family. However, this does not preclude the possibility that the IS110/IS492 elements also utilize IS DNA in directing the specificity of the transposase and, like IS608, have a single-stranded DNA transposition intermediate. We are currently employing both genetic and in vitro assays for identifying the DNA and protein determinants of IS492 insertion specificity.
Acknowledgments
This work was supported by National Science Foundation grant MCB-0004123 (to A.C.K.) and the University of Georgia Office of the Vice President for Research.
We thank Steven Trau and Ashley Currier for assistance with Southern blot analyses of phase variants and Tim Hoover and Dave Samuels for productive discussions and comments on the manuscript.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl (ed.). 1993. Current protocols in molecular biology. Wiley, New York, NY.
- 2.Balding, D. P. 2000. Transposition of IS492: in vivo and in vitro characterization of a member of an atypical group of insertion sequences. Dissertation. Emory University, Atlanta, GA.
- 3.Barabas, O., D. R. Ronning, C. Guynet, A. B. Hickman, B. Ton-Hoang, M. Chandler, and F. Dyda. 2008. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell 132:208-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett, D. H., and M. Silverman. 1989. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J. Bacteriol. 171:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, D. H., M. E. Wright, and M. Silverman. 1988. Variable expression of extracellular polysaccharide in the marine bacterium Pseudomonas atlantica is controlled by genome rearrangement. Proc. Natl. Acad. Sci. USA 85:3923-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belas, R. 1989. Sequence analysis of the agrA gene encoding β-agarase from Pseudomonas atlantica. J. Bacteriol. 171:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruton, C. J., and K. F. Chater. 1987. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolor A3(2). Nucleic Acids Res. 15:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchner, J. M., A. E. Robertson, D. J. Poynter, S. S. Denniston, and A. C. Karls. 2005. Piv site-specific invertase requires a DEDD motif analogous to the catalytic center of the RuvC Holliday junction resolvases. J. Bacteriol. 187:3431-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S., S. Ohta, and E. Ohtsubo. 2003. A novel IS element, IS621, of the IS110/IS492 family transposes to a specific site in repetitive extragenic palindromic sequences in Escherichia coli. J. Bacteriol. 185:4891-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corpe, W. A. 1980. In G. Bitton and K. C. Marshall (ed.), Adsorption of microorganisms to surfaces, p. 105-144. John Wiley and Sons, Inc., New York, NY.
- 11.Curcio, M. J., and K. M. Derbyshire. 2003. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4:865-877. [DOI] [PubMed] [Google Scholar]
- 12.Fang, Z., C. Doig, N. Morrison, B. Watt, and K. J. Forbes. 1999. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex, and its association with IS6110. J. Bacteriol. 181:1021-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy, V. G., and M. S. Fox. 2000. Transposon stability and a role for conjugational transfer in adaptive mutability. Proc. Natl. Acad. Sci. USA 97:7393-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grindley, N. D., K. L. Whiteson, and P. A. Rice. 2006. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 75:567-605. [DOI] [PubMed] [Google Scholar]
- 17.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, D. J., D. J. Lydiate, and D. A. Hopwood. 1989. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolor A3(2). Mol. Microbiol. 3:1307-1318. [DOI] [PubMed] [Google Scholar]
- 19.Higgins, B. P., C. D. Carpenter, and A. C. Karls. 2007. Chromosomal context directs high-frequency precise excision of IS492 in Pseudoalteromonas atlantica. Proc. Natl. Acad. Sci. USA 104:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidwell, M. G., and D. R. Lisch. 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15:95-99. [DOI] [PubMed] [Google Scholar]
- 21.Kingsford, C. L., K. Ayanbule, and S. L. Salzberg. 2007. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulakov, L. A., G. J. Poelarends, D. B. Janssen, and M. J. Larkin. 1999. Characterization of IS2112, a new insertion sequence from Rhodococcus, and its relationship with mobile elements belonging to the IS110 family. Microbiology 145:561-568. [DOI] [PubMed] [Google Scholar]
- 23.Lenich, A. G., and A. C. Glasgow. 1994. Amino acid sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J. Bacteriol. 176:4160-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mrazek, J., and S. Xie. 2006. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 22:3099-3100. [DOI] [PubMed] [Google Scholar]
- 26.Müller, C., U. Lauf, and H. Hermann. 2001. The inverted repeats of IS1384, a newly described insertion sequence from Pseudomonas putida strain H, represent the specific target for integration of IS1383. Mol. Genet. Genomics 265:1004-1010. [DOI] [PubMed] [Google Scholar]
- 27.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 28.Partridge, S. R., and R. M. Hall. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185:6371-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins-Balding, D., G. Duval-Valentin, and A. C. Glasgow. 1999. Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol. 181:4937-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters, J. E., and N. L. Craig. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2:806-814. [DOI] [PubMed] [Google Scholar]
- 31.Post, V., and R. M. Hall. 2009. Insertion sequences in the IS1111 family that target the attC recombination sites of integron-associated gene cassettes. FEMS Microbiol. Lett. 290:182-187. [DOI] [PubMed] [Google Scholar]
- 32.Ramos-González, M. I., M. J. Campos, J. L. Ramos, and M. Espinosa-Urgel. 2006. Characterization of the Pseudomonas putida mobile genetic element ISPpu10: an occupant of repetitive extragenic palindromic sequences. J. Bacteriol. 188:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, P., R. Craigie, and D. R. Davies. 1996. Retroviral integrases and their cousins. Curr. Opin. Struct. Biol. 6:76-83. [DOI] [PubMed] [Google Scholar]
- 34.Siguier, P., J. Filee, and M. Chandler. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9:526-531. [DOI] [PubMed] [Google Scholar]
- 35.Tetu, S. G., and A. J. Holmes. 2008. A family of insertion sequences that impacts integrons by specific targeting of gene cassette recombination sites, the IS1111-attC group. J. Bacteriol. 190:4959-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, T., F. F. Evans, D. Schleheck, A. Mai-Prochnow, C. Burke, A. Penesyan, D. S. Dalisay, S. Stelzer-Braid, N. Saunders, J. Johnson, S. Ferriera, S. Kjelleberg, and S. Egan. 2008. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS ONE 3:e3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tobiason, D. M., J. M. Buchner, W. H. Thiel, K. M. Gernert, and A. C. Karls. 2001. Conserved amino acid motifs from the novel Piv/MooV family of transposases and site-specific recombinases are required for catalysis of DNA inversion by Piv. Mol. Microbiol. 39:641-651. [DOI] [PubMed] [Google Scholar]
- 38.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]