Abstract
Band 7 proteins, which encompass members of the stomatin, prohibitin, flotillin, and HflK/C protein families, are integral membrane proteins that play important physiological roles in eukaryotes but are poorly characterized in bacteria. We have studied the band 7 proteins encoded by the cyanobacterium Synechocystis sp. strain PCC 6803, with emphasis on their structure and proposed role in the assembly and maintenance of the photosynthetic apparatus. Mutagenesis revealed that none of the five band 7 proteins (Slr1106, Slr1128, Slr1768, Sll0815, and Sll1021) was essential for growth under a range of conditions (including high light, salt, oxidative, and temperature stresses), although motility was compromised in an Slr1768 inactivation mutant. Accumulation of the major photosynthetic complexes in the thylakoid membrane and repair of the photosystem II complex following light damage were similar in the wild type and a quadruple mutant. Cellular fractionation experiments indicated that three of the band 7 proteins (Slr1106, Slr1768, and Slr1128) were associated with the cytoplasmic membrane, whereas Slr1106, a prohibitin homologue, was also found in the thylakoid membrane fraction. Blue native gel electrophoresis indicated that these three proteins, plus Sll0815, formed large (>669-kDa) independent complexes. Slr1128, a stomatin homologue, has a ring-like structure with an approximate diameter of 16 nm when visualized by negative stain electron microscopy. No evidence for band 7/FtsH supercomplexes was found. Overall, our results indicate that the band 7 proteins form large homo-oligomeric complexes but do not play a crucial role in the biogenesis of the photosynthetic apparatus in Synechocystis sp. strain PCC 6803.
Members of the band 7 superfamily of proteins are found throughout nature and are defined by a characteristic sequence motif, termed the SPFH domain, after the initials of the various subfamilies: the stomatins, the prohibitins, the flotillins (also known as “reggies”), and the HflK/C proteins (12, 49). The stomatins and prohibitins and to a lesser extent flotillins are highly conserved protein families and are found in a variety of organisms ranging from prokaryotes to higher eukaryotes (29, 34, 49), whereas HflK and HflC homologues are only present in bacteria.
In eukaryotes band 7 proteins are linked with a variety of disease states consistent with important cellular functions (6). In general the eukaryotic band 7 proteins tend to be oligomeric and are involved in membrane-associated processes: for example, prohibitins are involved in modulating the activity of a membrane-bound FtsH protease (17, 46) and the assembly of mitochondrial respiratory complexes (30), stomatins are involved in ion channel function (47), and flotillins are involved in signal transduction and vesicle trafficking (25).
In the case of prokaryotes, most work so far has focused on the roles of the HflK/C and YbbK (also known as QmcA, a stomatin homologue) band 7 proteins of Escherichia coli (7, 16, 17, 36) and the structure of a stomatin homologue in the archaeon Pyrococcus horikoshii (57). Much less is known about the structure, function, and physiological importance of band 7 proteins in other prokaryotes, especially the cyanobacteria (12).
The unicellular cyanobacterium Synechocystis sp. strain PCC 6803 is a widely used model organism for studying various aspects of cyanobacterial physiology and, in particular, oxygenic photosynthesis. One of the main areas of our research is to understand the mechanism by which the oxygen-evolving photosystem II (PSII) complex found in the thylakoid membrane of Synechocystis sp. strain PCC 6803 is repaired following light damage. Recent work has identified an important role for FtsH proteases in PSII repair (19, 41). Given that FtsH is known to form large supercomplexes with HflK/C in E. coli (36) and with prohibitins in Saccharomyces cerevisiae mitochondria (46), we hypothesized that one or more band 7 proteins might interact with FtsH in cyanobacteria and play a role in the selective turnover of the D1 reaction center polypeptide during PSII repair and so provide resistance to high light stress (40). This idea was given early support by the detection of both FtsH and Slr1106, a prohibitin homologue, in a His-tagged PSII preparation isolated from Synechocystis sp. strain PCC 6803 (40) and the detection of Slr1128 (a stomatin homologue), Sll1021 (a possible flotillin homologue), and FtsH in a His-tagged preparation of ScpD, a small chlorophyll a/b-like-binding protein that associates with PSII (56). Recent mutagenesis experiments have also suggested a role for Slr1128 in maintaining growth at high light intensities (53).
In this paper we have used targeted gene disruption mutagenesis and various biochemical approaches to investigate the structure and function of band 7 proteins in Synechocystis sp. strain PCC 6803, with particular emphasis on PSII function. We provide evidence that four predicted band 7 proteins in Synechocystis sp. strain PCC 6803 (Slr1106, Slr1768, Slr1128, and Sll8015) form large independent complexes, which in the case of Slr1128 forms a ring-like structure. No evidence was found for the formation of supercomplexes with FtsH. Importantly, single and multiple insertion mutants lacking up to four of the five band 7 proteins are able to grow as well as the wild type (WT) under a range of growth conditions, including high light stress. Our results suggest that band 7 proteins are not essential in Synechocystis sp. strain PCC 6803 and are not required for efficient PSII repair. Possible functions of the cyanobacterial band 7 proteins are discussed in the light of recent results from other systems.
MATERIALS AND METHODS
Databases and phylogenetic analyses.
Protein sequences were retrieved from the UniProt (release 14.6; http://www.uniprot.org/) (1) and the NCBI protein (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein) databases. Assignments of the various band 7 proteins to respective band 7 protein subfamilies were made according to the InterPro database (release 18.0; http://www.ebi.ac.uk/interpro/) (28). Phylogenetic analyses were performed in the “workbench” environment (http://workbench.sdsc.edu/) using the ClustalW module (50) for initial protein sequence alignments and the DRAWGRAM/PHYLIP module (9) to generate phylogenetic trees. The TMPRED server (http://www.ch.embnet.org/software/TMPRED_form.html) (13) was used to predict transmembrane (TM) domains of proteins of interest.
Cyanobacterial strains and growth conditions.
A glucose-tolerant (GT) (55) and WT (45) Synechocystis sp. strain PCC 6803 were used in this study. Unless stated otherwise, strains were grown in liquid BG-11 mineral medium or on solid BG-11 plates containing 1.5% (wt/vol) agar, both containing 5 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid-KOH, pH 8.2, at a light intensity of 20 μE m−2 s−1 of white fluorescent light and at 29°C. Glucose-tolerant strains were cultivated in medium supplemented with 5 mM glucose, and where applicable kanamycin (50 μg ml−1), chloramphenicol (30 μg ml−1), spectinomycin (50 μg ml−1), or erythromycin (10 μg ml−1) was added to the medium. Comparative growth analyses were performed mainly between the Synechocystis sp. strain PCC 6803 GT and a quadruple mutant (ΔQ) strain under the following suboptimal growth conditions: (i) in plate assays on BG-11 and on BG-11 supplemented with 5 mM glucose agar plates, with low (5 μE m−2 s−1) and high (100 and 400 μE m−2 s−1) light, shift from high (400 μE m−2 s−1) to low (5 μE m−2 s−1) light, and low (22°C) and high (37°C) temperatures; (ii) in liquid BG-11 medium supplemented with 5 mM glucose, low (22°C) temperature, in the presence of hydrogen peroxide (0.2 and 0.4 mM H2O2), supplemented with low (2 mM NaCl) and high (684 mM NaCl) salt (as well as a shift from low to high salt and vice versa), iron depletion (no iron source in the BG-11 medium), the presence of DCMU (10 μM), 12-h light/12-h dark cycles, and low (ambient air level) and high (3% [vol/vol]) CO2 levels, as well as a shift from high to low CO2 levels (no glucose was added to the liquid BG-11 medium when the effect of CO2 levels was assessed).
Construction of mutants.
Synechocystis sp. strain PCC 6803 single or multiple band 7 gene disruption mutants were generated by individual or successive transformations of WT or GT cells with plasmids containing the disrupted genes (see Fig. S1 in the supplemental material): slr1106 was disrupted by a kanamycin resistance cassette inserted at the NaeI site 236 bp into the coding sequence; slr1768 was disrupted by a chloramphenicol resistance cassette inserted at the EcoNI site 256 bp into the coding sequence; slr1128 was disrupted by a spectinomycin resistance cassette inserted at the MscI site 238 bp into the coding sequence; sll0815 was disrupted by an erythromycin resistance cassette inserted at the HindIII site 317 bp into the coding sequence; sll1021 was disrupted by an erythromycin resistance cassette inserted at the HpaI site 836 bp into the coding sequence. Two distinct quadruple band 7 gene disruption mutants were generated, wherein one (ΔQ) the slr1106, slr1768, slr1128, and sll0815 genes were disrupted and in the other (ΔQ*) the slr1106, slr1768, slr1128, and sll1021 genes had been disrupted. Unless stated otherwise, data on a quadruple mutant presented in this paper refer to the first mutant (ΔQ). The genotypes of the mutants were verified by PCR analysis using the following gene-specific primers: slr1106-Fw, 5′-GGGCTCGAGATGAGCAAACAATCCTTT-3′; slr1106-Rev, 5′-GGGGGATCCCTAGTTAGCCAGGTCAGTTAG-3′; slr1768-Fw, 5′-GCCCATATGGGTGCTGTTATCTCGGCGATC-3′; slr1768-Rev, 5′-GGGGGATCCTTAGGGGTCCGACATGATAATGGG-3′; slr1128-Fw, 5′-GCCCATATGGAAGCCTTTTTTCTCTTCTTTCTCGTC-3′; slr1128-Rev, 5′-GGGGGATCCCTAAACTGCCCGATGGCGGTCTAC-3′; sll0815-Fw, 5′-GCCCATATGGCACGACAAGCTCGCTATCAA-3′; sll0815-Rev, 5′-GGGGGATCCTCAACTATCCGATGTTATTTTTTC-3′; sll1021-Fw, 5′- GCCCATATGCAAAGTAAATTTTGGTTTGAATTTCTCC-3′; sll1021-Rev, 5′-GGGAGATCTCTAAATTTCCTCCGGGGAAAAATT-3′.
Preparation of crude thylakoid membranes.
Crude Synechocystis sp. strain PCC 6803 thylakoid membranes for one-dimensional (1-D) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were prepared by glass bead (212 to 300 μm in diameter) breakage at 4°C followed by differential centrifugation as described by Komenda et al. (20) and for 1-D Blue native polyacrylamide gel electrophoresis (BN-PAGE) membranes were prepared according to the methods of Zhang et al. (59). Chlorophyll a (Chl a) content was determined by extraction into methanol and absorption measurement at 666 and 750 nm (18). Protein concentrations were determined using a DC protein kit according to the manufacturer's instructions (Bio-Rad Laboratories, United Kingdom).
Antigen production and generation of polyclonal and affinity-purified antibodies.
Band 7 genes were cloned as either full-length or truncated versions into the pET16b expression vector (i.e., slr1106, Δ1-225slr1768, Δ1-150slr1128, and sll0815). After the expression of recombinant His6-tagged proteins in BL21-Gold(DE3)pLysS E. coli cells (Stratagene, United Kingdom), antigens were affinity purified via a batch method using chelating Sepharose (GE Healthcare, United Kingdom) coupled with Ni2+ and sent to Seqlab (Germany) for a 3-month immunization procedure in rabbits. Samples from final bleeds were affinity purified in a batch method using CNBr-activated Sepharose (GE Healthcare, United Kingdom) coupled with the respective antigens. Attempts to recombinantly express the Sll1021 protein either as full-length or truncated (Δ1-300sll1021) versions failed.
Protein analysis and immunoblotting.
Membrane protein samples were either separated on 12.5% (vol/vol) denaturing 1-D SDS-PAGE gels containing 6 M urea (23) or on 5 to 12.5% (vol/vol) linear gradient native 1-D BN-PAGE gels (37, 38) as described by Kügler et al. (22) with modifications described by Cline and Mori (8) and Herranen et al. (11). For 2-D BN/SDS-PAGE, a lane of the 1-D BN-PAGE gel was incubated in Laemmli SDS sample buffer containing 5% (vol/vol) β-mercaptoethanol and 6 M urea for 1 h at room temperature (RT). Subsequently, this gel strip was placed onto a 1-mm-thick 14% (vol/vol) SDS-PAGE gel with 6 M urea (23). 2-D BN/SDS-PAGE analyses of pulse-labeled samples were performed as described by Sobotka et al. (43). Gels were either stained with Coomassie blue R-250 or by silver staining (3) or electroblotted onto nitrocellulose membrane (0.2-μm pore size; Bio-Rad Laboratories, United Kingdom). Immunoblotting analyses were performed using specific primary antibodies and a horseradish peroxidase-conjugated secondary antibody (GE Healthcare, United Kingdom). Signals were visualized using a chemiluminescence kit (SuperSignal West Pico; Pierce). Primary antibodies used in this study were the following: (i) a purified, polyclonal Slr1106-specific antiserum from rabbit, (ii) a purified, polyclonal Slr1768-specific antiserum from rabbit, (iii) a purified, polyclonal Slr1128-specific antiserum from rabbit, (iv) a purified, polyclonal Sll0815-specific antiserum from rabbit, (v) a polyclonal PsbO-specific antiserum from rabbit, kindly provided by James Barber (Imperial College London, United Kingdom), (vi) a polyclonal C-terminal D1-specific antipeptide antiserum from rabbit (34), (vii) a polyclonal CP43-specific antiserum from rabbit (P6), (viii) a polyclonal SbtA-specific antipeptide (residues 184 to 203) antiserum from rabbit, kindly provided by Teruo Ogawa (Institute of Physical and Chemical Research [Riken], Japan), (ix) a polyclonal antipeptide antibody specific for E. coli FtsH, which is potentially cross-reactive with all Synechocystis sp. strain PCC 6803 FtsH homologues, kindly provided by Teru Ogura (University of Kumamoto, Japan).
PSII activity measurements and photoinhibition analysis.
The activity of PSII of Synechocystis sp. strain PCC 6803 was assessed by measuring the light-saturated (800 μE m−2 s−1 of actinic light illumination) rate of oxygen evolution from whole cells using 1 mM 2,6-dichloro-p-benzoquinone (DCBQ) and 2 mM potassium ferricyanide as artificial electron acceptors. The amount of oxygen dissolved in a liquid culture was determined using a Hansatech DW2 oxygen electrode (Hansatech Instruments Ltd., United Kingdom) linked to a computer managed by an Oxy-Lab 1 control box (Hansatech Instruments Ltd., United Kingdom). For photoinhibition analyses, 1-liter cultures of Synechocystis sp. strain PCC 6803 were grown under routine growth conditions to an optical density at 730 nm of approximately 0.8. Cells were then harvested and resuspended in fresh BG-11 medium to a concentration of 20 μg of Chl a ml−1 and subjected to a white light intensity of 100 μE m−2 s−1 for 1 h and then 1,200 μE m−2 s−1 for 6 h at the normal growth temperature (29°C), with or without lincomycin (100 μg ml−1), a protein synthesis inhibitor. The oxygen evolution rates were calculated in terms of μmol of oxygen per mg of Chl a per h, and the normalized rates (t = 0 corresponds to 100%) were plotted as a function of time.
l-[35S]methionine-cysteine pulse-labeling of Synechocystis sp. strain PCC 6803 cells.
l-[35S]methionine-cysteine pulse-labeling analyses of Synechocystis sp. strain PCC 6803 cells were performed as described by Sobotka et al. (43).
Differential membrane protein extraction.
Crude membrane isolations containing 20 μg of Chl a were resuspended in 100 μl of buffer: extraction buffer (EB; 20 mM Tricine, pH 8.0), EB with 2 M NaCl, or 20 mM CAPS, pH 12.0. After two freeze-thaw cycles (30 min at −80°C, 20 min at RT), samples were pelleted in a benchtop ultracentrifuge (Beckman Coulter T-100; TLA 120.1; 100,000 × g, 20 min, 4°C). The supernatant was kept as the soluble fraction, whereas the pellet was resuspended in 100 μl of EB. Five microliters of each fraction was analyzed by 1-D SDS-PAGE and immunoblotting.
Two-phase partitioning of plasma and thylakoid membranes.
Purified plasma and thylakoid membrane fractions of Synechocystis sp. strain PCC 6803 were prepared by aqueous polymer two-phase partitioning (32).
Preparation of covalently cross-linked antibodies to protein A-Sepharose and immunoprecipitation procedure.
Antibodies of a polyclonal antiserum of interest were covalently cross-linked to protein A-Sepharose (PC-A10; Generon, United Kingdom) using the cross-linker dimethylpimelimidate as described by Schneider et al. (39). Proteins or protein complexes of Synechocystis sp. strain PCC 6803 from freshly prepared crude membrane extracts corresponding to an amount of 40 μg Chl a, resuspended in 450 μl ACA buffer (750 mM ɛ-amino caproic acid, 50 mM bis-Tris HCl, pH 7.0, 0.5 mM EDTA) and solubilized by the addition of 50 μl of a 10% (wt/vol) solution of the nonionic detergent β-dodecyl maltoside (β-DM; final concentration, 1% [wt/vol]) were immunoprecipitated using 30 μl of protein A-Sepharose with covalently coupled antibodies as described by Komenda et al. (21) with the following modifications. The incubation period was reduced to 1 h at 4°C, and the captured protein complexes were eluted under native conditions (incubation of the protein A beads with 40 μl of 0.2 M glycine, pH 2.4, 0.1% [wt/vol] β-DM for 5 min at RT, and subsequent centrifugation of the liquid phase into 40 μl 1 M bis-Tris, pH 7.0, 20% [vol/vol] glycerol for neutralization). The samples were then used for 1-D SDS-PAGE and immunoblotting analyses as well as for visualization under the electron microscope and subsequent single-particle analysis.
Electron microscopy and single-particle analysis.
Samples were negatively stained using 2% (wt/vol) uranyl acetate, pH 4.5, on carbon-coated copper electron microscope grids via the droplet method devised by Harris (10). In order to obtain an even distribution of protein complexes across the carbon support surface, a dilution series was made for each sample over several grids. Imaging was performed on Kodak SO-163 film (Eastman Kodak Company) at RT using a Philips-FEI Tecnai 12 electron microscope (EM; http://www.fei.com/contact-us.aspx) operating at 120 kV and 27,500× magnification, as maintained by the Centre of Biomolecular Electron Microscopy at Imperial College London. Only the micrographs with minimal astigmatism and drift, based on postscanning image analysis after scanning into the computing environment using a Nikon LS9000 CoolScan densitometer (Nikon Ltd., United Kingdom) operating at a step size of 6.35 μm, were chosen. Finally, after calculating each micrograph's power spectrum, seven micrographs, for which the first minimum was calculated to be in the 20- to 22.5-Å spatial resolution range, remained and were chosen for further image analysis. A data set consisting of all possible single particles (3,708 image locations, each a pixel box of 160 by 160 pixels) was compiled using automatic selection in Boxer, a module of the EMAN software package (version 1.7) (27). All subsequent image processing was performed using the Imagic-5 environment (Image Science GmbH, Berlin, Germany) with protein visualized as white and stain as black. No correction was made for the contrast transfer function (CTF). The single-particle image locations were analyzed at a sampling frequency of 2.31 Å on the specimen scale. Reference-free alignment using a soft-edged circular mask applied to each image, followed by multivariate statistical analyses, gave initial 2-D class averages that were then iteratively refined in order to obtain the final averages shown (35, 52). A clear subpopulation, consisting of 499 particles, was identified and assigned as the top views, i.e., the largest 2-D projection observed. These data were then processed de novo to give further refined class averages.
RESULTS
Identification and analysis of potential cyanobacterial band 7 proteins.
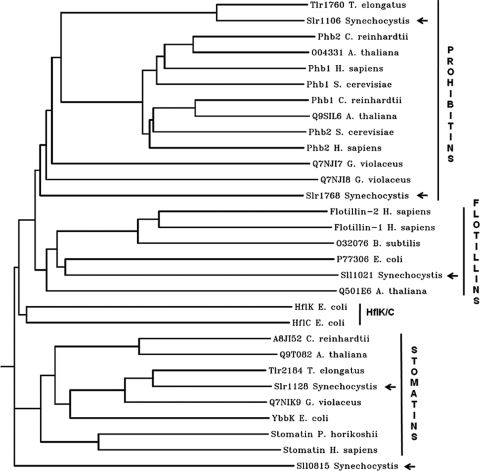
To assess the prevalence of band 7 proteins in cyanobacteria, an InterPro database search was performed (run in January 2009), and 149 potential cyanobacterial band 7 proteins from 45 different strains were listed under the band 7 protein family database entry IPR001107. According to the InterPro database, most of these cyanobacterial band 7 proteins were assigned to the prohibitin (58 entries; IPR000163) and stomatin (40 entries; IPR0001972) subfamilies, whereas a significant number of proteins (51 entries) could not be reliably assigned to any protein subfamily. The phylogenetic analysis of selected cyanobacterial and other band 7 proteins showed that the different band 7 protein subfamilies branch early on (Fig. 1). This suggests low sequence similarity among the various band 7 protein subfamilies and might be explained by either a distant common ancestor (49) or as a result of convergent evolution of the SPFH domain (34). There is also variability in the number of potential band 7 proteins encoded in cyanobacteria. For example, five were identified in Synechocystis sp. strain PCC 6803 and only two were found in the thermophilic cyanobacterium Thermosynechococcus elongatus (Table 1).
FIG. 1.
Phylogenetic analysis of selected band 7 proteins. Selected protein sequences were aligned using CLUSTALW, and a rooted phylogenetic tree was generated using PHYLIP/DRAMGRAM. Band 7 proteins of the following organisms were included in this analysis (in alphabetical order): Arabidopsis thaliana, Bacillus subtilis (subsp. subtilis strain 168), Chlamydomonas reinhardtii, Escherichia coli (strain K-12), Gloeobacter violaceus, Homo sapiens, Pyrococcus horikoshii, Saccharomyces cerevisiae (baker's yeast), Synechocystis sp. strain PCC 6803, Thermosynechococcus elongatus BP-1. Arrows indicate the band 7 proteins of Synechocystis sp. strain PCC 6803.
TABLE 1.
Band 7 proteins of selected organisms used for the phylogenetic analysis shown in Fig. 1
| Organism | Band 7 proteins (S/P/F/H/O)a | UniProt no.b | Proteins included in phylogenetic analysis
|
||
|---|---|---|---|---|---|
| NCBI no. | Protein name(s) | Annotationc | |||
| A. thaliana | 7/11/0/0/4 | O49460 | NP_194580 | Prohibitin 1/At4g28510 | P |
| O04331 | NP_198893 | Prohibitin 3/At5g40770 | P | ||
| Q9T082 | NAd | At4g27580 | S | ||
| Q501E6e | NP_197907e | At5g25250 | None | ||
| B. subtilis | 0/0/1/0/0 | O32076 | NP_390979 | YuaG | F |
| C. reinhardtii | 1/2/0/0/1 | A8I8V6 | XP_001701753 | Prohibitin 1 | P |
| A8J633 | XP_001696940 | Prohibitin 2 | P | ||
| A8JI52 | XP_001703625 | None | S | ||
| E. coli | 1/0/0/2/1 | P0AA53 | NP_415022 | YbbK/QmcA | S |
| P0ABC3 | NP_418596 | HflC | H | ||
| P0ABC7 | NP_418595 | HflK | H | ||
| P77306 | NP_417523 | YqiK | None | ||
| G. violaceus | 1/1/0/0/1 | Q7NJI7 | NP_924791 | Gll1845 | P |
| Q7NIK9 | NP_925120 | Gll2174 | S | ||
| Q7NJI8 | NP_924790 | Gll1844 | None | ||
| H. sapiensf | 10/9/24/0/6 | P35232 | NP_002625 | Prohibitin 1 | P |
| Q99623 | NP_009204 | Prohibitin 2 | P | ||
| P27105 | NP_004090 | Tomatin | S | ||
| NP_937837 | |||||
| O75955 | NP_005794 | Flotillin 1 | F | ||
| Q6FG43 | NA | Flotillin 2 | F | ||
| P. horikoshii | 2/0/0/0/0 | O59180 | NP_143371 | Stomatin/PH1511 | S |
| S. cerevisiae | 0/2/0/0/0 | P40961 | NP_011648 | Prohibitin 1 | P |
| P50085 | NP_011747 | Prohibitin 2 | P | ||
| Synechocystis sp. strain | 1/1/0/0/3 | P72754 | NP_440089 | Slr1106 | P |
| PCC 6803 | P73049 | NP_440390 | Slr1768 | None | |
| P72655 | NP_439977 | Slr1128 | S | ||
| P74042 | NP_441437 | Sll0815 | None | ||
| P72929 | NP_440266 | Sll1021 | None | ||
| T. elongatus | 1/1/0/0/0 | Q8DI32 | NP_682550 | Tlr1760 | P |
| Q8DGX8 | NP_682974 | Tlr2184 | S | ||
S, stomatins; P, prohibitins; F, flotillins; H, HflK/C; O, other proteins.
According to the UniProt database (version 14.6).
Annotations as stomatin (S), prohibitin (P), flotillin (F), or HflK/C (H) are according to the InterPro database (release 18.0).
NA, not available.
Q501E6 of A. thaliana was included in this analysis because it has been identified in a detergent-resistant membrane fraction (4) and as a plant flotillin homologue (34).
Sixteen of the 24 flotillins of H. sapiens listed in the InterPro database are fragments of flotillin-1.
The band 7 proteins are not essential in Synechocystis sp. strain PCC 6803.
In order to probe the physiological importance of the band 7 proteins, five single mutants were constructed in the GT strain of Synechocystis sp. strain PCC 6803, which is commonly used to study photosynthesis. In each of these single mutants, the conserved SPFH domain found in each band 7 protein was disrupted by the insertion of an antibiotic resistance cassette, and PCR analysis was used to confirm that each of the mutants had segregated (data not shown). To test whether there might be overlap of function between these genes two quadruple mutants (ΔQ and ΔQ*) were also generated in which four genes (slr1106, slr1768, slr1128, and sll0815 in the ΔQ strain and slr1106, slr1768, slr1128, and sll1021 in the ΔQ* strain) were disrupted. Again, PCR confirmed that the mutants had segregated (see Fig. S2A and S3A in the supplemental material). From these data we can conclude that intact band 7 proteins are not essential for cell viability under the growth conditions used. Electron microscopy further confirmed that there was no obvious defect in the quadruple mutant (ΔQ) with regard to size of cell or structure of the cytoplasmic and thylakoid membranes (see Fig. S4 in the supplemental material). Segregated mutants could also be created in the WT strain of Synechocystis sp. strain PCC 6803 under photoautotrophic conditions. It was noticeable that the Slr1768 inactivation mutant in the genetic background of the WT strain showed reduced motility on agar plates (see Fig. S5 in the supplemental material), despite the retention of pili on the cell surface as visualized by negative stain electron microscopy (data not shown).
To test whether growth might be affected under suboptimal growth conditions, growth of the single mutants and the quadruple mutant (ΔQ) created in the glucose-tolerant strain was compared to that of the GT strain under a range of different conditions (described in Materials and Methods), including low (5 μE m−2 s−1) and high (100 and 400 μE m−2 s−1) light, a shift from high (400 μE m−2 s−1) to low (5 μE m−2 s−1) light, low (22°C) and high (37°C) temperatures (these experiments were performed as plate assays under both photoautotrophic and photoheterotrophic growth conditions), the presence of hydrogen peroxide (0.2 and 0.4 mM H2O2) to test for enhanced susceptibility to oxidative stress, low (2 mM NaCl) and high (684 mM NaCl) salt to test for a possible role in osmotic stress, iron depletion, the presence of DCMU (10 μM), 12-h light/12-h dark cycles (these experiments were performed in liquid culture under photoheterotrophic growth conditions), and low (ambient air level) and high (3% [vol/vol]) CO2, as well as a shift from high to low CO2 levels (the experiments performed to assess the effects of different CO2 levels were performed in liquid culture under photoautotrophic growth conditions). Under all conditions tested there was no clear difference between the growth of the quadruple mutant (ΔQ) and the GT strain (data not shown).
PSII activity and repair in the band 7 quadruple mutant.
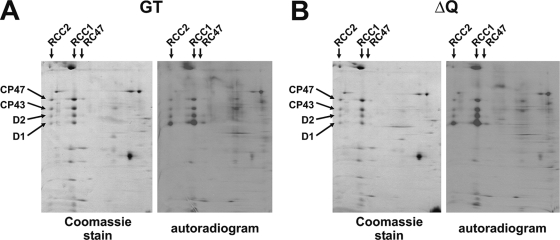
The main aim of this work was to assess the possible role of band 7 proteins in PSII assembly and repair, particularly of those in the prohibitin subfamily for which there is evidence for a role in membrane quality control (24). Oxygen evolution (Fig. 2) and immunoblotting experiments (data not shown) revealed that the quadruple mutant (ΔQ) was able to accumulate WT levels of PSII complexes. Importantly, PSII repair was functional in the quadruple mutant (Fig. 2), and rates of D1 turnover were similar in the quadruple mutant and WT as assessed in pulse-chase experiments using l-[35S]methionine (see Fig. S6 in the supplemental material). In addition the overall assembly state of the major protein complexes in the thylakoid membrane and dynamics of protein synthesis and turnover in the light were similar in the mutant and WT as judged from the analysis by 2-D BN/SDS-PAGE of 35S-pulse-labeled proteins (Fig. 3). Overall, these data did not support a crucial role for the Slr1106, Slr1768, Slr1128, and Sll0815 proteins in PSII assembly or repair.
FIG. 2.
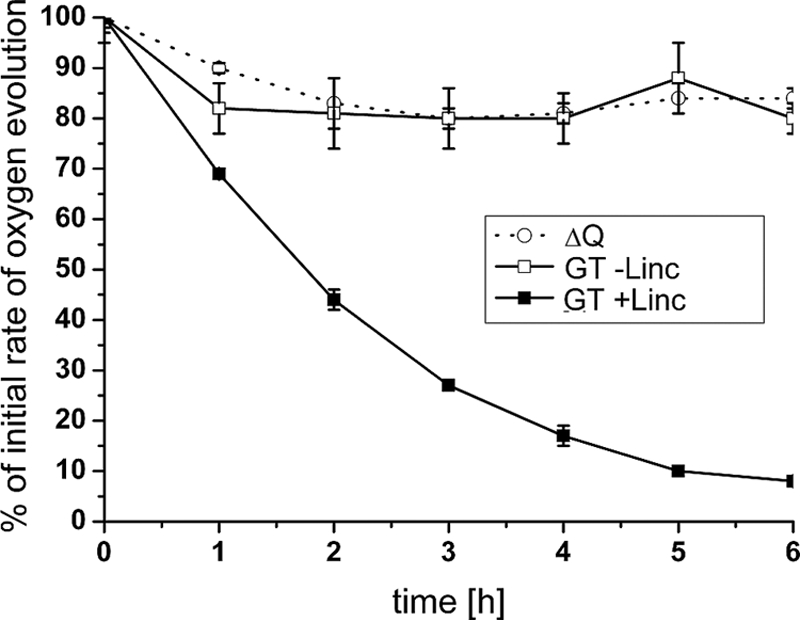
Photoinhibition analyses of the Synechocystis sp. strain PCC 6803 GT and the ΔQ quadruple mutant. Cells at a Chl a concentration of 20 μg ml−1 were treated with high light (1,200 μE m−2 s−1) at 29°C over a period of 6 hours. Oxygen evolution of whole cells was assessed for the Synechocystis sp. strain PCC 6803 GT and the ΔQ quadruple mutant strains in the presence or absence of 100 μg ml−1 lincomycin at the indicated time points in the presence of 2 mM DCBQ and 1 mM K3Fe(CN) using a Hansatech DW2 oxygen electrode (Hansatech Instruments Ltd., United Kingdom). Oxygen evolution rates (in μmol oxygen per mg of Chl a per h) were normalized (value at t = 0 was 100%) and plotted as a function of time. Error bars represent standard deviation from the means of three measurements. The initial, absolute rates of oxygen evolution for the respective strains were as follows: 324 (ΔQ without Linc), 316 (GT without Linc), and 350 (GT +Linc) (all in μmol oxygen per mg of Chl a per h).
FIG. 3.
Comparative 2-D BN/SDS-PAGE analysis of pulse-labeled crude membrane extracts isolated from the Synechocystis sp. strain PCC 6803 GT and the quadruple mutant strain, ΔQ. Coomassie blue-stained gels and autoradiograms of the 2-D BN/SDS-PAGE analysis of pulse-labeled samples (an amount of 6 μg of Chl a was loaded per sample) were prepared as described by Sobotka et al. (43). The positions of PSII dimers (RCC2), PSII monomers (RCC1), and RC47 protein complexes (RC47, PSII core complexes lacking CP43) as well as those of the D1, D2, CP43, and CP47 PSII subunits are indicated.
Membrane association of band 7 proteins in Synechocystis sp. strain PCC 6803.
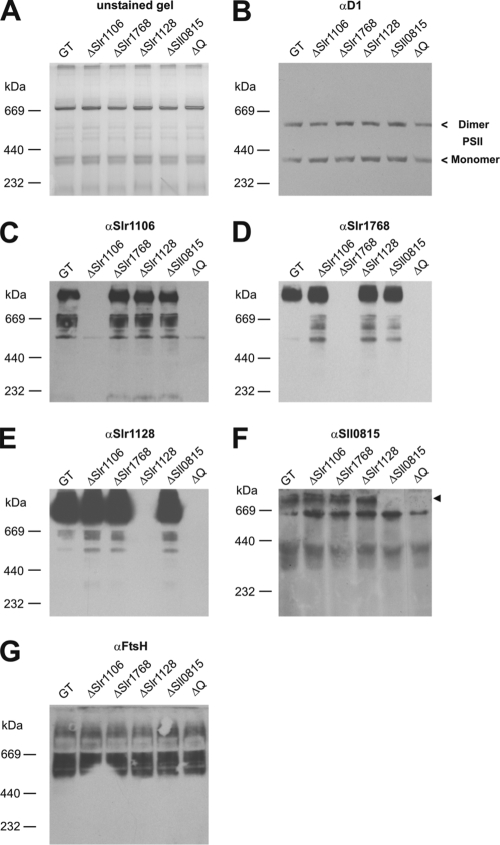
Experiments were also performed to clarify the cellular location, size, and structure of the band 7 protein complexes. The TMPRED server predicted one N-terminal TM domain for each of the Slr1106, Slr1128, and Sll1021 band 7 proteins, two N-terminal TM domains for Slr1768, and no TM domains for Sll0815. Immunoblotting experiments using antibodies specific for Slr1106, Slr1768, Slr1128, and Sll0815 confirmed that all four proteins were membrane associated (Fig. 4A). In addition these four band 7 proteins were not washed from the membrane by 2 M NaCl and were only partially released by treatment with 20 mM CAPS, pH 12.0. In contrast the peripheral PsbO protein of PSII was released under all conditions and the integral D1 PSII subunit could not be extracted. In conclusion, the Slr1106, Slr1768, Slr1128, and Sll0815 band 7 proteins of Synechocystis sp. strain PCC 6803 are tightly associated membrane proteins.
FIG. 4.
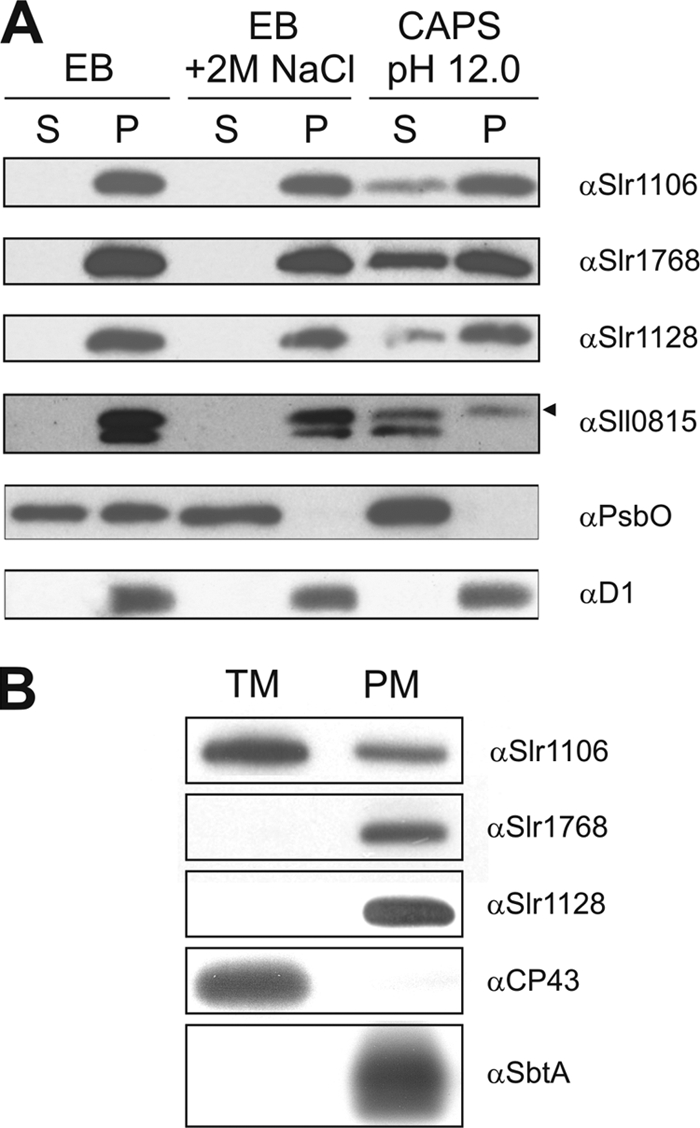
Membrane association and localization of band 7 proteins of Synechocystis sp. strain PCC 6803. (A) Differential membrane protein extraction from Synechocystis sp. strain PCC 6803 GT crude membrane isolations. After two consecutive freeze-thaw cycles (30 min at −80°C and 20 min at RT) in the indicated buffers (EB, 20 mM Tricine, pH 8.0; EB with 2 M NaCl; or 20 mM CAPS, pH 12.0), soluble (S) and pellet (P) fractions were generated by ultracentrifugation. An amount corresponding to 1 μg of Chl a was loaded per lane. The arrowhead indicates the antibody signal that corresponds to the Sll0815 protein. (B) Purified thylakoid membrane (TM) and plasma membrane (PM) fractions were generated by aqueous polymer two-phase partitioning, and 5 μg of protein was loaded in each lane. All samples in panels A and B were analyzed by 1-D SDS PAGE followed by immunoblotting with the indicated antibodies. (A) PsbO and D1 were used as markers for peripheral and integral membrane proteins, respectively. (B) CP43 and SbtA were used as markers for the purity of the thylakoid and plasma membrane fractions, respectively.
Distribution of band 7 proteins in the thylakoid and cytoplasmic membranes.
Synechocystis sp. strain PCC 6803, like most cyanobacteria, contains an internal thylakoid membrane system in addition to a surrounding cytoplasmic membrane. The aqueous two-phase partitioning technique developed by Norling and colleagues (32) was used to separate these two types of membranes. Subsequent immunoblotting experiments revealed that the Slr1106 protein could be found in both the thylakoid and cytoplasmic membrane fractions, whereas the Slr1768 and Slr1128 proteins were found in the cytoplasmic membrane fraction only (Fig. 4B). As expected the PSII CP43 protein was detected only in the thylakoid membrane, whereas SbtA (a sodium-dependent bicarbonate transporter) was located in the cytoplasmic membrane (32, 59).
The band 7 proteins form large complexes.
1-D BN-PAGE in conjunction with immunoblotting was used to assess the sizes of four band 7 proteins (Slr1106, Slr1768, Slr1128, and Sll0815) in Synechocystis sp. strain PCC 6803 (Fig. 5). In all cases the band 7 proteins were found predominantly in large complexes of sizes significantly greater than 669 kDa, although several smaller less-abundant complexes were also detected. In yeast mitochondria the prohibitin homologues Phb1p and Phb2p form large hetero-oligomeric protein complexes, in which both subunits display an interdependency on each other and where the deletion of one of the subunits leads to the destabilization of the other (2, 30). However, analysis of the single band 7 mutants of Synechocystis sp. strain PCC 6803 suggested that each of the band 7 protein complexes could accumulate to the same level in the absence of the other band 7 subunits, consistent with the formation of independent complexes. This conclusion was supported by immunoprecipitation experiments performed on GT extracts (Fig. 6). Antibodies specific for the Slr1106, Slr1768, and Slr1128 band 7 proteins of Synechocystis sp. strain PCC 6803 immunoprecipitated only their cognate subunit and not the other band 7 proteins under investigation.
FIG. 5.
Band 7 protein complexes of Synechocystis sp. strain PCC 6803. Crude membrane isolations (samples contained 1 μg of Chl a) of the Synechocystis sp. strain PCC 6803 GT and various band 7 single and multiple gene disruption mutant strains were analyzed by 1-D BN-PAGE on a 5 to 12.5% (wt/vol) linear gradient polyacrylamide gel and immunoblotted with the indicated antibodies. (B) The positions of monomeric and dimeric PSII protein complexes are indicated. (F) The arrowhead indicates the antibody signal that corresponds to the Sll0815 protein.
FIG. 6.
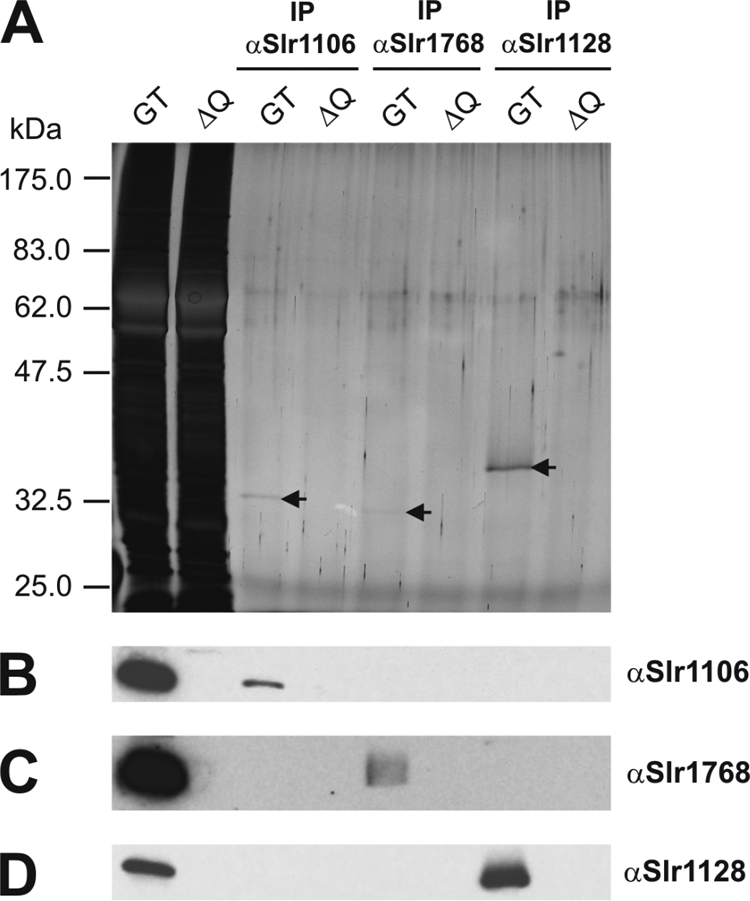
Immunoprecipitation of band 7 proteins of Synechocystis sp. strain PCC 6803. Crude membrane isolations of the Synechocystis sp. strain PCC 6803 GT and the quadruple mutant strain ΔQ (initial samples contained 40 μg of Chl a) were used for immunoprecipitation with anti-Slr1106, anti-Slr1768, or anti-Slr1128 antibodies coupled to protein A-Sepharose beads. Crude membranes of both strains (0.25 μg Chl a) and eluted fractions (7.5 μl of a total volume of 80 μl) were analyzed by 1-D SDS-PAGE (A) and immunoblotted with indicated antibodies (B to D).
Does FtsH form supercomplexes with band 7 proteins in Synechocystis sp. strain PCC 6803?
FtsH has been shown to form large supercomplexes with a prohibitin heterocomplex in yeast mitochondria (46) and with an HflK/C complex in E. coli (17, 36). To test whether FtsH might also form a stable complex with a band 7 protein complex in Synechocystis sp. strain PCC 6803, in particular to the prohibitin homologue Slr1106, 1-D BN-PAGE gels of 1% (wt/vol) β-DM-solubilized crude membrane extracts from WT and the various band 7 mutants were immunoblotted using an antibody that recognizes all four FtsH subunits of Synechocystis sp. strain PCC 6803 (Fig. 5G). In all cases, immunodetectable FtsH migrated as a diffuse band with an apparent molecular mass of approximately 550 to 670 kDa. No signal could be detected when the same antibody was used to probe immunoprecipitated band 7 proteins (data not shown). Overall there was no evidence from this 1-D BN-PAGE and immunoblotting analysis of band 7 mutant and GT strains for the significant accumulation of stable FtsH/band 7 complexes in Synechocystis sp. strain PCC 6803. The thermophilic cyanobacterium Thermosynechococcus elongatus was also examined, as it is known that supercomplexes are more stable in this organism and are more easily detected by 1-D BN-PAGE (60). Again, no evidence was obtained for the formation of significant levels of a FtsH/band 7 supercomplex (data not shown).
Interaction of Slr1106 with NdhI (Sll0520).
Previous work has shown that human mitochondrial prohibitins are able to interact with subunits of complex I (30) and that prohibitin might have a chaperone function in the assembly of complex I (5). In cyanobacteria the analogous complex to the mitochondrial complex I is the NDH-1 complex, which is found in the thylakoid membrane as several different forms (59). We found that polyclonal antiserum specific for the NdhI protein was able to coimmunoprecipitate Slr1106 and NdhI from WT extracts, whereas in the M55 mutant lacking NDH-1 complexes (33) neither subunit was immunoprecipitated (see Fig. S7 in the supplemental material). These data suggested a possible interaction between the NdhI subunit of the NDH-1 complex and the Slr1106 band 7 protein of Synechocystis sp. strain PCC 6803 in vivo. 2-D BN/SDS-PAGE analysis revealed that the NDH-1L and NDH-1 M complexes, which both contain NdhI (59), were still able to accumulate in the quadruple band 7 mutant to wild-type levels at both low (air level) and high (3% [vol/vol]) CO2 levels, as well as when the culture was shifted from high to low CO2 levels (see Fig. S8 in the supplemental material). Therefore, Slr1106 is not required for the accumulation of these or the other major complexes of the thylakoid membrane under our standard laboratory growth conditions.
The Slr1128 stomatin homologue of Synechocystis sp. strain PCC 6803 forms a ring-like complex.
To gain insights into the 3-D structure of band 7 proteins, immunoaffinity-purified Slr1128 protein complexes (Fig. 6) were visualized by negative stain and transmission electron microscopy (Fig. 7). The images of 3,708 individual Slr1128 protein complexes were collected and analyzed by single-particle analysis (see Materials and Methods). After the initial calculation of the 2-D class averages from all 3,708 single particles by using reference-free alignment techniques, a subpopulation of 499 single-particle top views, i.e., the largest Slr1128 particle observed in terms of projection surface area, was identified. Subsequent to a de novo iterative refinement of these 499 single particles, seven different class averages of the Slr1128 protein complexes were distinguished by this in silico method. The Slr1128 complex appeared to have a ring-like structure with an approximate diameter of about 16 nm. However, no definitive conclusions can be drawn concerning the number of individual Slr1128 subunits in each ring, given the small subpopulation of the extracted data set.
FIG. 7.
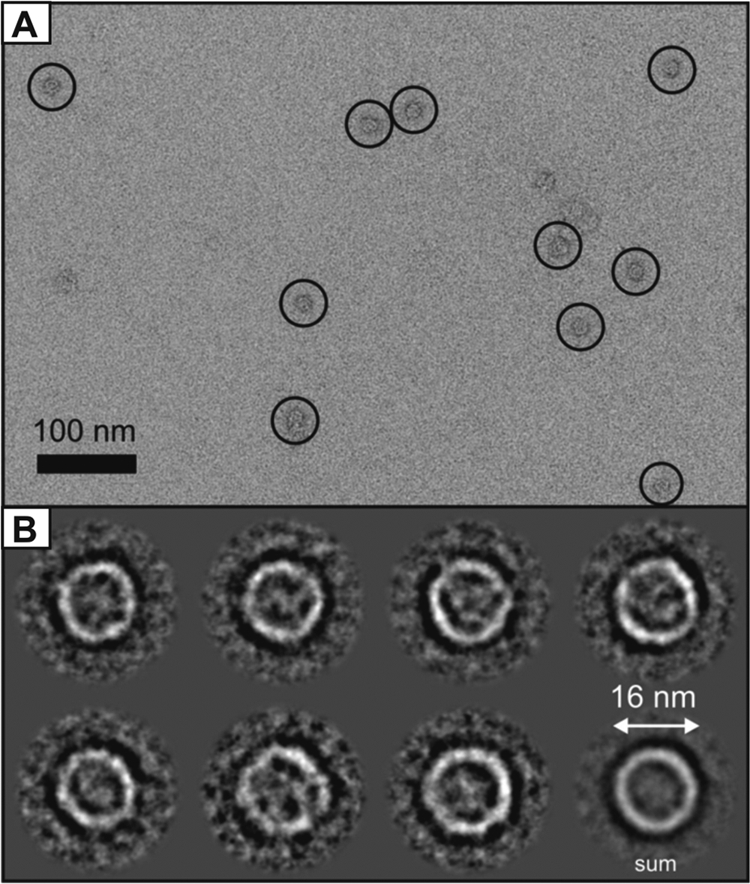
Single-particle analysis of Slr1128 protein complexes of Synechocystis sp. strain PCC 6803. (A) Selected region of an electron micrograph that shows Slr1128 protein complexes (circled). Bar, 100 nm. (B) A subpopulation of 499 particles was classified as being the largest projections observed within a total data set of 3,708, and these were assigned as a top views. These were further classified into seven classes, as shown, with a total average (last image) of ∼16 nm in diameter. Protein is appears as white, and stain is black.
DISCUSSION
Despite the widespread occurrence of band 7 proteins in prokaryotes (12), relatively little work has been done to assess their structure and physiological importance. We have shown that four (Slr1106, Slr1768, Slr1128, and Sll0815) out of the five band 7 proteins encoded by the cyanobacterium Synechocystis sp. strain PCC 6803 are expressed as large membrane-bound complexes. In the case of Sll0815, sequence analyses had suggested that it was potentially soluble (12). For the fifth band 7 protein (Sll1021), we were unable to generate antibodies to allow its study. However, Sll1021 protein was recently detected in the cytoplasmic membrane fraction (14). We have also been able to show that immunopurified Slr1128 forms a large ring-like structure with an approximate diameter of 16 nm (Fig. 7). This result is highly significant, as it is the first determined structure of a native protein complex, albeit at low resolution, for a member of the stomatin subfamily of band 7 proteins. Recent work has shown that the E. coli-expressed soluble domain of the p-stomatin (PH1511) from the hyperthermophilic archaeon Pyrococcus horikoshii is able to form a trimer (57). Whether a trimer represents the building block for the formation of a higher-order structure in vivo remains uncertain. A ring-like structure with an approximate diameter of 20 nm was shown recently for the mitochondrial prohibitin Phb1/Phb2 hetero-oligomeric complex (48). Thus, the ability of band 7 proteins to form ring-like structures might be a common feature. 1-D BN-PAGE analysis revealed some heterogeneity in the size of the Slr1106, Slr1768, and Slr1128 protein complexes. The smaller complexes might reflect the presence of assembly complexes, as suggested for Phb1/Phb2 in yeast (48), bona fide functional complexes, or artifactual complexes generated during the solubilization or electrophoresis procedures.
We could find no evidence for the accumulation of large supercomplexes between FtsH protease complexes (assumed to be hexameric) and the various band 7 complexes. This is in contrast to the situation in yeast mitochondria and E. coli, where such supercomplexes can be isolated by detergent solubilization and sucrose density gradient centrifugation (36, 46). However, recent work has shown that a second band 7 protein in E. coli, termed YbbK or QmcA, is able to interact with FtsH but that the resulting complex is much less stable than the FtsH/HflKC supercomplex (7). It therefore remains possible that similar weak interactions between band 7 proteins and FtsH might still be present in Synechocystis sp. strain PCC 6803.
Our data suggest that band 7 proteins are not required for photoprotection in Synechocystis sp. strain PCC 6803 and in particular the assembly and repair of the photosystem II complex, which is considered the primary target for photodamage in vivo (31). This raises the question as to the significance of the presence of Slr1106 in His-tagged PSII preparations (40). We still cannot exclude the possibility that Slr1106 plays a noncrucial and redundant role in PSII assembly and degradation. However, the low levels of Slr1106 detected in the His-tagged PSII preparation might reflect adventitious binding of Slr1106 to the lipoprotein complex, especially if Slr1106 has lipid-binding properties. Likewise, we have been able to detect an interaction between Slr1106 and the NdhI (Sll0520) subunit of the NDH-1 complex (see Fig. S7 in the supplemental material). More work is required to confirm the physiological relevance of this interaction, especially as assembly of NDH-1 is not drastically affected in the absence of Slr1106 (see Fig. S8 in the supplemental material).
Mutants with defects in the assembly and maintenance of quality control in the membrane often show impaired growth under stress conditions (15, 41). Somewhat surprisingly, the growth of single and multiple band 7 insertion mutants was not significantly compromised under a range of adverse conditions as judged in growth experiments (see Materials and Methods). Microarray data (kindly provided by Iwane Suzuki, University of Tsukuba, Japan) generated for WT Synechocystis sp. strain PCC 6803 indicated changes in transcript level for the Sll0815 protein (4.48-fold increase upon a shift from 34 to 22°C) or the Slr1106 and Slr1128 proteins (2.89-fold and 3.13-fold increases, respectively, upon exposure to 0.25 mM H2O2). However, when the quadruple mutant (ΔQ) was grown photoheterotrophically under these stress conditions in liquid culture (growth at 22°C or exposure to 0.2 and 0.4 mM H2O2), no difference in growth could be observed in comparison to the Synechocystis sp. strain PCC 6803 GT strain (data not shown). This suggests that the observed changes in transcript levels might be compensated for in vivo or that the lack of the Slr1106, Slr1128, or Sll0815 protein does not have a noticeable effect on the ability of the quadruple mutant to grow under these conditions.
Very recently, Wang et al. (53) reported that Slr1128 formed complexes with His-tagged HliA (also called ScpC) and His-tagged HliB (or ScpD) and that these complexes interacted with trimeric PSI complexes. Our data are not readily reconciled with these results. First, in agreement with earlier proteomics data (11), we have shown here that Slr1128 is predominantly located in the cytoplasmic membrane fraction of Synechocystis sp. strain PCC 6803 (Fig. 4B), which is considered to lack Scps (53, 56), and also trimeric PSI as assessed by native gel electrophoresis and the lack of the PsaE subunit (58). In addition in coimmunoprecipitation experiments we were unable to pull down ScpC (HliA) with Slr1128 (see Fig. S9 in the supplemental material). In our experience significant amounts of the Slr1128 protein are retained by immobilized Ni-nitrilotriacetic acid columns even when non-His-tagged crude membrane extracts are chromatographed (see Fig. S10 in the supplemental material). This raises the possibility that the presence of Slr1128 in His-tagged Scp fractions might be artifactual. Wang and colleagues (53) also indicated that growth of an Slr1128 mutant is sensitive to high light illumination (400 μE m−2 s−1), in contrast to the phenotype of the slr1128 gene disruption mutants described here. To test the role of Slr1128 further, we also examined the phenotype of a slr1128 disruption mutant created in the Synechocystis sp. strain PCC 6803 WT background. We found that the mutant strain was able to grow as well as the WT in BG-11 liquid culture at similar light irradiances (40, 200, and 400 μE m−2 s−1) to those used by Wang and colleagues (53) (data not shown). The disparity in the growth data on the Slr1128 mutants might reflect subtle differences in the growth conditions used or differences in the mutants examined. In both types of mutant the conserved SPFH domain was disrupted. However, the Slr1128 mutants constructed here could potentially express the first 79 out of 321 amino acid residues (the N-terminal TM domain is predicted to lie between residues 68 and 86), whereas Wang and colleagues constructed a mutant only able to express the first 36 residues (53). We therefore cannot totally exclude the possibility that a functional truncated Slr1128 subunit might still accumulate in the mutant described here, although work on other band 7 proteins has emphasized the importance of the C-terminal region for formation of higher-order oligomers (42, 48). This same possibility also extends to the other band 7 proteins investigated here.
The question arises as to the possible function of band 7 proteins both in cyanobacteria and other organisms. The emerging view is that prohibitins, flotillins, and stomatins might have a scaffold function in the organization of the membrane (6) and consequently have wide-ranging effects on membrane structure and function, including vesicle trafficking, assembly of complexes, the operation of ion channels, and transmembrane signaling. Such diverse roles might explain why cyanobacteria contain different numbers of band 7 proteins (Fig. 1). In the case of Synechocystis sp. strain PCC 6803 evidence is now emerging that the thylakoid membrane is heterogeneous (51) with regions preferentially involved in membrane protein synthesis and presumably the assembly of large membrane complexes. In addition recent proteomics data suggest heterogeneity also in the cytoplasmic membrane (44). In addition there might be routes, so far unclear, by which proteins and lipids in the cytoplasmic membrane are trafficked to the thylakoid membrane (26, 54). Thus, proteins with membrane scaffolding functions would be predicted to be present in cyanobacteria. Our work suggests that if band 7 proteins do play such a role, it is not a crucial one.
Supplementary Material
Acknowledgments
We are grateful to the Biotechnology and Biological Sciences Research Council for funding this research. M.B. is grateful to the Imperial College London for the award of a Ph.D. bursary, and J.K. is grateful to the Academy of Sciences of the Czech Republic (projects no. AV0Z50200510 and IAA400200801) and to the Ministry of Education, Youth and Sports of the Czech Republic (project no. MSM6007665808) for support. J.N. currently holds a Royal Society University Fellowship.
We are also grateful to Iwane Suzuki for providing the microarray data and to Uwe Kahmann for the whole-cell electron microscopy of the Synechocystis sp. strain PCC 6803 GT and the quadruple mutant (ΔQ) strains. We also thank Mark Wass, who gave input on the phylogenetic analysis of the band 7 proteins, and to Marianne Kristiansen and Ingeborg van Knippenberg, who contributed to early work on Slr1106.
Footnotes
Published ahead of print on 14 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bairoch, A., R. Apweiler, C. H. Wu, W. C. Barker, B. Boeckmann, S. Ferro, E. Gasteiger, H. Huang, R. Lopez, M. Magrane, M. J. Martin, D. A. Natale, C. O'Donovan, N. Redaschi, and L. S. Yeh. 2005. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33:D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, K. H., and M. P. Yaffe. 1998. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 4.Borner, G. H., D. J. Sherrier, T. Weimar, L. V. Michaelson, N. D. Hawkins, A. Macaskill, J. A. Napier, M. H. Beale, K. S. Lilley, and P. Dupree. 2005. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137:104-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourges, I., C. Ramus, B. Mousson de Camaret, R. Beugnot, C. Remacle, P. Cardol, G. Hofhaus, and J. P. Issartel. 2004. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 383:491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browman, D. T., M. B. Hoegg, and S. M. Robbins. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell. Biol. 17:394-402. [DOI] [PubMed] [Google Scholar]
- 7.Chiba, S., K. Ito, and Y. Akiyama. 2006. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol. Microbiol. 60:448-457. [DOI] [PubMed] [Google Scholar]
- 8.Cline, K., and H. Mori. 2001. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154:719-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 10.Harris, J. R. 2007. Negative staining of thinly spread biological samples. Methods Mol. Biol. 369:107-142. [DOI] [PubMed] [Google Scholar]
- 11.Herranen, M., N. Battchikova, P. Zhang, A. Graf, S. Sirpio, V. Paakkarinen, and E. M. Aro. 2004. Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134:470-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinderhofer, M., C. A. Walker, A. Friemel, C. A. Sturmer, H. M. Moller, and A. Reuter. 2009. Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann, K., and W. Stoffel. 1993. TMbase: a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 14.Huang, F., I. Parmryd, F. Nilsson, A. L. Persson, H. B. Pakrasi, B. Andersson, and B. Norling. 2002. Proteomics of Synechocystis sp. strain PCC 6803: identification of plasma membrane proteins. Mol. Cell. Proteomics 1:956-966. [DOI] [PubMed] [Google Scholar]
- 15.Kamata, T., H. Hiramoto, N. Morita, J. R. Shen, N. H. Mann, and Y. Yamamoto. 2005. Quality control of photosystem II: an FtsH protease plays an essential role in the turnover of the reaction center D1 protein in Synechocystis PCC 6803 under heat stress as well as light stress conditions. Photochem. Photobiol. Sci. 4:983-990. [DOI] [PubMed] [Google Scholar]
- 16.Kihara, A., Y. Akiyama, and K. Ito. 1997. Host regulation of lysogenic decision in bacteriophage l: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc. Natl. Acad. Sci. USA 94:5544-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara, A., Y. Akiyama, and K. Ito. 1996. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 15:6122-6131. [PMC free article] [PubMed] [Google Scholar]
- 18.Komenda, J., and J. Barber. 1995. Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34:9625-9631. [DOI] [PubMed] [Google Scholar]
- 19.Komenda, J., M. Barker, S. Kuvikova, R. de Vries, C. W. Mullineaux, M. Tichy, and P. J. Nixon. 2006. The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281:1145-1151. [DOI] [PubMed] [Google Scholar]
- 20.Komenda, J., L. Lupinkova, and J. Kopecky. 2002. Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur. J. Biochem. 269:610-619. [DOI] [PubMed] [Google Scholar]
- 21.Komenda, J., M. Tichy, and L. A. Eichacker. 2005. The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46:1477-1483. [DOI] [PubMed] [Google Scholar]
- 22.Kügler, M., L. J.änsch, V. Kruft, U. K. Schmitz, and H.-P. Braun. 1997. Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth. Res. 53:35-44. [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Langer, T., M. Kaser, C. Klanner, and K. Leonhard. 2001. AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem. Soc. Trans. 29:431-436. [DOI] [PubMed] [Google Scholar]
- 25.Langhorst, M. F., A. Reuter, and C. A. Stuermer. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62:2228-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberton, M., R. Howard Berg, J. Heuser, R. Roth, and H. B. Pakrasi. 2006. Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227:129-138. [DOI] [PubMed] [Google Scholar]
- 27.Ludtke, S. J., D.-H. Chen, J.-L. Song, D. T. Chuang, and W. Chiu. 2004. Seeing GroEL at 6Å resolution by single particle electron cryomicroscopy. Structure 12:1129-1136. [DOI] [PubMed] [Google Scholar]
- 28.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nijtmans, L. G., S. M. Artal, L. A. Grivell, and P. J. Coates. 2002. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life Sci. 59:143-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nijtmans, L. G., L. de Jong, M. Artal Sanz, P. J. Coates, J. A. Berden, J. W. Back, A. O. Muijsers, H. van der Spek, and L. A. Grivell. 2000. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 19:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nixon, P. J., M. Barker, M. Boehm, R. de Vries, and J. Komenda. 2005. FtsH-mediated repair of the photosystem II complex in response to light stress. J. Exp. Bot. 56:357-363. [DOI] [PubMed] [Google Scholar]
- 32.Norling, B., E. Zak, B. Andersson, and H. Pakrasi. 1998. 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 436:189-192. [DOI] [PubMed] [Google Scholar]
- 33.Ohkawa, H., G. D. Price, M. R. Badger, and T. Ogawa. 2000. Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:2591-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivera-Milla, E., C. A. Stuermer, and E. Malaga-Trillo. 2006. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell. Mol. Life Sci. 63:343-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruprecht, J., and J. Nield. 2001. Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction. Prog. Biophys. Mol. Biol. 75:121-164. [DOI] [PubMed] [Google Scholar]
- 36.Saikawa, N., Y. Akiyama, and K. Ito. 2004. FtsH exists as an exceptionally large complex containing HflKC in the plasma membrane of Escherichia coli. J. Struct. Biol. 146:123-129. [DOI] [PubMed] [Google Scholar]
- 37.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 38.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, C., R. A. Newman, D. R. Sutherland, U. Asser, and M. F. Greaves. 1982. A one-step purification of membrane proteins using a high efficiency immunomatrix. J. Biol. Chem. 257:10766-10769. [PubMed] [Google Scholar]
- 40.Silva, P., and P. J. Nixon. 2001. Identification of possible assembly and repair factors in photosystem two preparations of Synechocystis sp. PCC 6803: a new model for D1 turnover. Proc. 12th Int. Congr. Photosynthesis. CSIRO Publishing, Brisbane, Australia. www.publish.csiro.au/paper/SA0403246.htm.
- 41.Silva, P., E. Thompson, S. Bailey, O. Kruse, C. W. Mullineaux, C. Robinson, N. H. Mann, and P. J. Nixon. 2003. FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp. PCC 6803. Plant Cell 15:2152-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyers, L., E. Umlauf, and R. Prohaska. 1998. Oligomeric nature of the integral membrane protein stomatin. J. Biol. Chem. 273:17221-17226. [DOI] [PubMed] [Google Scholar]
- 43.Sobotka, R., J. Komenda, L. Bumba, and M. Tichy. 2005. Photosystem II assembly in CP47 mutant of Synechocystis sp. PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J. Biol. Chem. 280:31595-31602. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava, R., N. Battchikova, B. Norling, and E. M. Aro. 2006. Plasma membrane of Synechocystis PCC 6803: a heterogeneous distribution of membrane proteins. Arch. Microbiol. 185:238-243. [DOI] [PubMed] [Google Scholar]
- 45.Stanier, R. Y. 1973. The biology of blue-green algae. Autotrophy and heterotrophy in unicellular blue-green algae. Bot. Monogr. 9:501-518. [Google Scholar]
- 46.Steglich, G., W. Neupert, and T. Langer. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart, G. W., A. C. Argent, and B. C. Dash. 1993. Stomatin: a putative cation transport regulator in the red cell membrane. Biochim. Biophys. Acta 1225:15-25. [DOI] [PubMed] [Google Scholar]
- 48.Tatsuta, T., K. Model, and T. Langer. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16:248-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tavernarakis, N., M. Driscoll, and N. C. Kyrpides. 1999. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24:425-427. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van de Meene, A. M., M. F. Hohmann-Marriott, W. F. Vermaas, and R. W. Roberson. 2006. The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184:259-270. [DOI] [PubMed] [Google Scholar]
- 52.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Q., S. Jantaro, B. Lu, W. Majeed, M. Bailey, and Q. He. 2008. The high light-inducible polypeptides stabilize trimeric photosystem I complex under high light conditions in Synechocystis PCC 6803. Plant Physiol. 147:1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westphal, S., J. Soll, and U. C. Vothknecht. 2003. Evolution of chloroplast vesicle transport. Plant Cell Physiol. 44:217-222. [DOI] [PubMed] [Google Scholar]
- 55.Williams, J. G. K. 1988. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167:766-778. [Google Scholar]
- 56.Yao, D., T. Kieselbach, J. Komenda, K. Promnares, M. A. Prieto, M. Tichy, W. Vermaas, and C. Funk. 2007. Localization of the small CAB-like proteins in photosystem II. J. Biol. Chem. 282:267-276. [DOI] [PubMed] [Google Scholar]
- 57.Yokoyama, H., S. Fujii, and I. Matsui. 2008. Crystal structure of a core domain of stomatin from Pyrococcus horikoshii Illustrates a novel trimeric and coiled-coil fold. J. Mol. Biol. 376:868-878. [DOI] [PubMed] [Google Scholar]
- 58.Zak, E., B. Norling, R. Maitra, F. Huang, B. Andersson, and H. B. Pakrasi. 2001. The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 98:13443-13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, P., N. Battchikova, T. Jansen, J. Appel, T. Ogawa, and E. M. Aro. 2004. Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16:3326-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, P., N. Battchikova, V. Paakkarinen, H. Katoh, M. Iwai, M. Ikeuchi, H. B. Pakrasi, T. Ogawa, and E. M. Aro. 2005. Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem. J. 390:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.