Abstract
Spontaneous hydrolytic deamination of DNA bases represents a considerable mutagenic threat to all organisms, particularly those living in extreme habitats. Cytosine is readily deaminated to uracil, which base pairs with adenine during replication, and most organisms encode at least one uracil DNA glycosylase (UDG) that removes this aberrant base from DNA with high efficiency. Adenine deaminates to hypoxanthine approximately 10-fold less efficiently, and its removal from DNA in vivo has to date been reported to be mediated solely by alkyladenine DNA glycosylase. We previously showed that UdgB from Pyrobaculum aerophilum, a hyperthermophilic crenarchaeon, can excise hypoxanthine from oligonucleotide substrates, but as this organism is not amenable to genetic manipulation, we were unable to ascertain that the enzyme also has this role in vivo. In the present study, we show that UdgB from Mycobacterium smegmatis protects this organism against mutagenesis associated with deamination of both cytosine and adenine. Together with Ung-type uracil glycosylase, M. smegmatis UdgB also helps attenuate the cytotoxicity of the antimicrobial agent 5-fluorouracil.
Spontaneous hydrolytic deamination of cytosine (C), adenine (A), and guanine (G) gives rise to uracil (U), hypoxanthine (Hx), and xanthine, respectively (3). Because the first two reactions occur at appreciable rates (31) and because their products are mutagenic, living organisms have evolved sophisticated defense mechanisms to deal with this threat to their genomic integrity. Deamination of C to U occurs with the highest frequency. Because U base pairs with A during replication, a failure to remove U from DNA prior to the next round of DNA synthesis gives rise to C→T (or, on the opposite strand, G→A) transition mutations (1). The A-to-Hx reaction is only about 10 times slower than the reaction affecting C and has biological significance due to Hx pairing with cytosine during replication.
Similar to other modified DNA bases, U and Hx are removed from DNA by base excision repair. This process is initiated by one of several DNA glycosylases, which removes the aberrant base, giving rise to an abasic (apyrimidinic or apurinic) site (AP site). In the subsequent step, the AP site is cleaved by an AP endonuclease on its 5′ side, giving rise to a single-strand break. The baseless sugar phosphate is then removed by the 5′→3′ exonuclease activity of polymerase I in bacteria or by the N-terminal lyase activity of polymerase β in eukaryotes. The same polymerases then fill in the single nucleotide gap, and the remaining nick is sealed by a DNA ligase (20). Base excision repair can also proceed by mechanisms that differ somewhat from the canonical pathway described above, but these mechanisms do not alter the outcome of the studies described in this work and are not described in detail here.
Uracil removal is catalyzed by uracil DNA glycosylases (UDGs). These enzymes possess two highly conserved motifs; motif A activates a catalytic water molecule for nucleophilic attack on the glycosidic bond, and motif B stabilizes the protein-DNA complex (25). To date, five UDG families with somewhat different substrate specificities have been characterized (25, 30). Enzymes belonging to family 1 are the best-characterized enzymes. They can remove uracil from U·A pairs or U·G mispairs in double-stranded DNA (dsDNA) and even more efficiently from single-stranded DNA (ssDNA) substrates (32). Family 2 enzymes excise uracil from U·G mismatches in dsDNA. Because these enzymes have most likely evolved to deal with the deamination of 5-methylcytosine to thymine, they are specific for U·G and T·G mispairs and excise neither uracil nor thymine from ssDNA (21). The active site of these enzymes is altered compared to the active site of family 1 UDGs. Family 3 consists of the single-strand-selective monofunctional UDGs, which appear to be hybrids between members of families 1 and 2 (5). As their name implies, family 3 enzymes are preferentially active on uracil-containing ssDNA substrates. Family 4 enzymes have been identified in hyperthermophiles such as Thermotoga maritima (28), Archaeoglobus fulgidus (29), and Pyrobaculum aerophilum (6). These enzymes act on U opposite A or G and exhibit maximal activity at high temperatures. Family 5 uracil glycosylases (UdgB) have been identified in a limited number of organisms, such as hyperthermophilic archaea and Mycobacterium tuberculosis (7, 30, 37). Unexpectedly, motif A of UdgB enzymes contains no polar amino acid, but motif B resembles motif B of family 1 proteins (30). In vitro assays showed that UdgB removes uracil from both ssDNA and dsDNA (30). Interestingly, this enzyme could also excise Hx from oligonucleotide substrates (30, 36) and is thus the only glycosylase that removes both aberrant pyrimidines and purines from DNA in vitro. One of the goals of the present study was to learn whether this enzyme also processes U and Hx in vivo.
As discussed above, uracil is generated in DNA by spontaneous hydrolytic deamination of cytosine. However, it can also be directly incorporated into DNA during replication in the form of dUMP. This process is not mutagenic, because dUMP is incorporated opposite adenine. However, it can be cytotoxic, particularly in cells with an imbalanced dUTP-dTTP pool, such as mutants lacking dUTPase, or in cells in which thymidylate synthase is inhibited by, e.g., 5-fluorodeoxyuridine (5FdUrd) treatment. Deoxyuridine incorporated during DNA synthesis is processed primarily by family 1 UDGs, which have been shown to associate with replicating polymerase complexes. Under normal circumstances, this repair process affects only the newly synthesized strand. However, at high dUTP/dTTP ratios, uracil repair might become saturated, so that some uracil remains in the DNA until the following round of DNA replication. In this scenario, processing of uracil in both template and nascent strands would give rise to double-strand breaks and thus to genomic instability (14, 15). This hypothesis is supported by the finding that organisms lacking an efficient repair system can tolerate considerable replacement of thymine with uracil in their DNA (40). The second goal of the present study was to test whether UdgB, like family 1 UDGs, contributed to the cytotoxicity associated with a dUTP-dTTP pool imbalance.
We wanted to study whether the substrates processed by UdgB in vitro are also addressed in vivo by this enzyme. As we were unable to do this with P. aerophilum, we set out to find an organism encoding UdgB that would be more amenable to genetic manipulation. M. tuberculosis encodes two potential UDGs, one belonging to family 1 (ung, M. tuberculosis Rv2976c) and one belonging to family 5 (udgB, M. tuberculosis Rv1259) (30). However, to avoid complications linked to the pathogenicity of this organism, we decided to investigate the role of Ung and UdgB in the nonpathogenic organism Mycobacterium smegmatis, which is, unlike P. aerophilum, amenable to genetic manipulation. Our results provide novel insights into the coordinated action of family 1 and family 5 glycosylases.
MATERIALS AND METHODS
Pattern search, sequences, alignment, and phylogeny.
A pattern search for conserved sequence motif A (GLAPA/G) and motif B (HPS) of the UdgB family of glycosylases was performed using the myHits web server (http://myhits.isb-sib.ch) (24) and standard protein sequence databases, including the Swiss-Prot and TrEMBL databases. Multiple-sequence alignments were obtained by using clustalW with default parameters and were visualized with Boxshade 3.2.1. Protein sequences of the following organisms were obtained from http://tigr.org: M. tuberculosis H37Rv (Rv1259), Mycobacterium bovis subsp. bovis AF2122/97 (Mb1289), M. tuberculosis CDC1551 (MT_1297.1), Mycobacterium leprae TN (ML1105), Mycobacterium avium 104 (MAV_1407), Mycobacterium sp. strain MCS (Mmcs_3957), M. smegmatis mc2 (MSMEG_5031), Frankia alni ACN14a (FRAAL6229), Frankia sp. strain CcI3 (Francci3_3919), Actinomyces naeslundii MG1 (ANA_1938), Propionibacterium acnes KPA171202 (PPA1235), Rubrobacter xylanophilus DSM 9941 (Rxyl_2949), Myxococcus xanthus DK 1622 (MXAN_3589), Streptomyces coelicolor A3 (SCO1990), Streptomyces avermitilis MA-4680 (SAV6242), Rhodococcus sp. strain RHA1 (RHA1_ro05941), Synechococcus sp. strain OS Type B prime (GYMBORF2457), Sulfolobus solfataricus P2 (SSO2733), Sulfolobus acidocaldarius DSM 639 (Saci_1756), Thermoplasma acidophilum DSM 1728 (Ta0477), “Candidatus Pelagibacter ubique” HTCC1062 (SAR11_0873), and P. aerophilum (PAE1327).
Media, chemicals, oligonucleotides, plasmids, and bacterial strains.
Middlebrook 7H9 broth and Middlebrook 7H10 agar were purchased from Difco. When appropriate, antibiotics were added at the following concentrations: hygromycin, 100 μg/ml; streptomycin, 100 μg/ml; and rifampin (rifampicin), 150 μg/ml. 5FdUrd was purchased from Sigma-Aldrich. Oligonucleotides were synthesized by Microsynth AG (Balgach, Switzerland) and are listed in Table 1. Plasmids and bacterial strains that were used in this study are listed in Table 2.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| ung1 | 5′GGAATTCCATATGCGATATGGCCGAGGACACAG3′ |
| ung2 | 5′GGAAGATCTGGCGAGTTCGGTGCGCAG3′ |
| ung3 | 5′GGAAGATCTGCGACGCTGAAGCCGATG3′ |
| ung4 | 5′TGCATGCATCGCCGAGCGCATCGTGAAC3′ |
| 5031_1 | 5′GGAATTCCATATGGCGGGCATCCCGACCGAG3′ |
| 5031_2 | 5′GGAAGATCTGCCGAGGCCGCCAGCC3′ |
| 5031_3 | 5′GGAAGATCTCTCGAACTCATCGCCGACC3′ |
| 5031_4 | 5′TGCATGCATGTTGATCATGACCTCGTCGAC3′ |
| rpoB-fwd | 5′CATCGACCACTTCGGCAAC3′ |
| rpoB-rev | 5′CTCCTCGTCGGCGGTCAG3′ |
| Sequencing primer | 5′GACCACCCAGGACGT3′ |
TABLE 2.
Plasmids and M. smegmatis strains used in this study
| Plasmid or strain | Source or reference |
|---|---|
| Plasmids | |
| pMCS5 | MoBiTec |
| pBluescript-hyg | This study |
| pMCS5-rpsL | This study |
| pMCS5-rpsL-hyg | This study |
| pMCS5-rpsL-hyg-Δung | This study |
| pMCS5-rpsL-hyg-Δ5031 | This study |
| Bacterial strains | |
| mc2155 | 34 |
| Msm SMR5 | 27 |
| Msm SMR5 ΔrrnB | 26 |
| Msm SMR5 ΔrrnB-Δung | This study |
| Msm SMR5 ΔrrnB-Δ5031 | This study |
| Msm SMR5 ΔrrnB-Δung/Δ5031 | This study |
Generation of mutants.
Wild-type rpsL from M. tuberculosis with ∼500 bp of 5′ flanking sequence was ligated into the XbaI site of pMCS5. The hygromycin resistance cassette from pBluescript-hyg was cloned into the SnaBI site of pMCS5-rpsL, resulting in pMCS5-rpsL-hyg.
Allelic replacement techniques were used to generate M. smegmatis knockout mutants (4). Using genomic DNA, 1- to 1.5-kb fragments upstream (5′) and downstream (3′) of the target genes ung (MSMEG_2399) and udgB (MSMEG_5031) were amplified by PCR and cloned into pMCS5-rpsL-hyg. To amplify the ung upstream region, primers ung1 and ung2 were used. For amplification of the ung downstream region, primers ung3 and ung4 were used. The resulting deletion allele lacked bp 156 to 561 of the 729-bp ung open reading frame.
To generate the M. smegmatis udgB knockout mutant, the 5031_1 and 5031_2 primers were used to amplify the udgB downstream region, and primers 5031_3 and 5031_4 were used to amplify the udgB upstream region. The resulting deletion allele lacked bp 213 to 696 of the 906-bp M. smegmatis udgB open reading frame. The integrity of the mutant vectors was verified by sequence analysis.
The vectors were transformed into M. smegmatis mc2 155 derivative SMR5 rrnB as previously described (4).
For generation of the M. smegmatis ung udgB double mutant, the udgB knockout vector was transformed into the M. smegmatis ung mutant, and selection procedures described above were used. Disruption of the targeted genes and absence of merodiploidy were ascertained by Southern blot analysis.
Determination of spontaneous mutation frequencies.
Three independent experiments with three sets of cultures for each strain were performed. Cultures were grown until late log phase in Middlebrook 7H9 medium, and the number of viable cells was around 109 cells/ml. Subsequently, the cultures were diluted to obtain 2 × 103 cells/ml and incubated for 2 days at 37°C. One hundred microliters of each culture was plated on freshly prepared Middlebrook 7H10 agar plates containing 150 μg/ml rifampin, and serial dilutions were plated on nonselective medium. After 3 to 6 days of incubation at 37°C, the numbers of CFU were determined. The number of CFU obtained on agar plates containing rifampin was divided by the number of CFU obtained on nonselective medium. The mean mutation frequency for a set of cultures was used to calculate the mutation frequency.
Mutation spectra.
For mutational analyses of rifampin-resistant clones, single colonies were picked, and a 500-bp rpoB fragment was amplified by PCR performed with primers rpoB-fwd and rpoB-rev. Subsequently, the coding strand was sequenced using an ABI sequencer. Mutations leading to rifampin resistance were analyzed by inspection of a 300-bp region of the rpoB gene located between bp 1200 and 1500.
MIC and growth kinetics.
A 1:1,000 dilution of a saturated culture was grown in Middlebrook 7H9 medium. The optical density at 600 nm was determined every 10 min for 48 h using a microtiter plate spectrophotometer. Generation times were calculated during the exponential growth phase.
The MIC was determined by visual inspection of microtiter plates. Cultures were grown in synthetic Middlebrook 7H9 medium, since this medium does not contain any pyrimidines or pyrimidine derivatives. 5FdUrd was added to a 200-μl culture having an optical density at 600 nm of 0.0025. The MIC of a drug was defined as the concentration at which growth was completely inhibited after 3 days of incubation at 37°C.
RESULTS
P. aerophilum UdgB is conserved in Actinomycetales and hyperthermophilic Archaea.
The uracil- and Hx-excising enzyme P. aerophilum UdgB was first identified and biochemically characterized using two hyperthermophilic archaea: P. aerophilum (30) and Thermus thermophilus (37). Unfortunately, the in vivo biological role of this enzyme could not be studied, as these organisms are not amenable to genetic manipulation. We therefore searched for other organisms that encode proteins belonging to the same glycosylase family with which we could carry out genetic experiments.
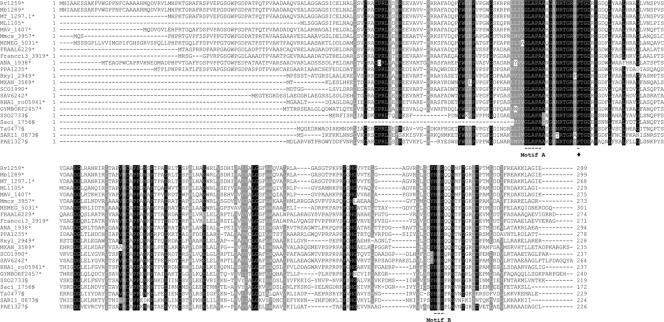
We performed a pattern search with conserved amino acid motifs A (GLAPA/G) and B (HPS) using a public database. Twenty-two organisms carrying family 5 UDG orthologues were detected (Fig. 1). Phylogenetic analyses revealed that UdgB is restricted to Actinomycetales and Thermoprotei, organisms that are either hyperthermophilic archaea or contain G+C-rich genomes. We chose to work with M. smegmatis, a fast-growing and nonpathogenic mycobacterial species.
FIG. 1.
Multiple-sequence alignment of P. aerophilum UdgB with the following homologues: Rv1259 of M. tuberculosis H37Rv (genomic G+C content, 65.60%), Mb1289 of M. bovis subsp. bovis AF2122/97 (65.63%), MT_1297.1 of M. tuberculosis CDC1551 (65.60%), ML1105 of M. leprae TN (57.79%), MAV_1407 of M. avium 104 (68.98%), Mmcs_3957 of Mycobacterium sp. strain MCS (68.38%), MSMEG_5031 of M. smegmatis mc2 (67.40%), FRAAL6229 of F. alni ACN14a (72.82%), Francci3_3919 of Frankia sp. strain CcI3 (70.07%), ANA_1938 of A. naeslundii MG1 (68.46%), PPA1235 of P. acnes KPA171202 (60.01%), Rxyl_2949 of R. xylanophilus DSM 9941 (70.47%), MXAN_3589 of M. xanthus DK 1622 (61.86%), SCO1990 of S. coelicolor A3 (71.99%), SAV6242 of S. avermitilis MA-4680 (70.70%), RHA1_ro05941 of Rhodococcus sp. strain RHA1 (66.98%), GYMBORF2457 of Synechococcus sp. strain OS Type B prime (58.45%), SSO2733 of S. solfataricus P2 (35.78%), Saci_1756 of S. acidocaldarius DSM 639 (36.70%), Ta0477 of T. acidophilum DSM 1728 (45.99%), SAR11_0873 of “Candidatus Pelagibacter ubique” HTCC1062 (29.68%), and PAE1327 of P. aerophilum IM2 (51.36%). Multiple-sequence alignment was performed with ClustalW, and the alignment was visualized with Boxshade 3.2.1. Organisms with high G+C contents are indicated by asterisks, and archaea are indicated by section signs. Motif A is responsible for activation of the catalytic water molecule. Motif B presumably stabilizes the DNA-protein complex once the base is flipped out of the double helix. The conserved phenylalanine, indicated by a diamond, interacts with the flipped-out base in the binding pocket.
Generation of unmarked M. smegmatis udgB and ung knockout mutants.
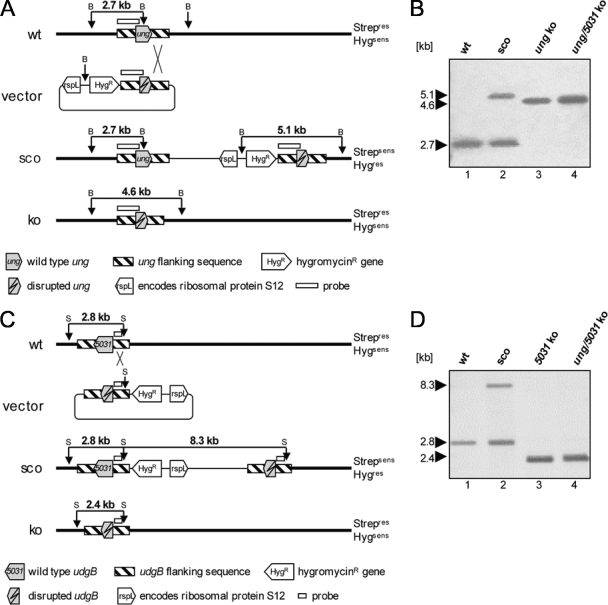
We targeted genes implicated in the processing of uracil. To survey the biological effects of the two different UDGs in mycobacteria, we generated knockout mutants with mutations in the ung gene encoding Ung belonging to family 1 and in M. smegmatis udgB encoding the family 5 protein UdgB. Further, a double-knockout mutant lacking both genes was constructed (Fig. 2).
FIG. 2.
Deletion strategy and Southern blot analysis. (A) Schematic diagram of the ung locus. The organizations of the wild-type region, the knockout vector that contains the ung deletion allele, the 3′ single-crossover region, and the mutated genomic ung region in the knockout mutant are shown. Probe locations and BamHI fragments detected by the probe are indicated. (B) Southern blot analysis of the Δung strain. Genomic DNA from parental strain M. smegmatis mc2155 (lane 1), the 3′ single-crossover mutant (lane 2), the Δung mutant (lane 3), and the Δung Δ5031 double-knockout mutant (lane 4) were digested with BamHI and probed with a 1.2-kb ung gene fragment. The presence of a single 4.6-kb fragment in Δung and Δung Δ5031 mutants instead of the single 2.7-kb fragment in the parental strain demonstrates that there is inactivation of ung and that the wild-type allele is not present. (C) Schematic diagram of the M. smegmatis udgB locus. The organizations of the wild-type region, the knockout vector that contains the udgB deletion allele, the 5′ single-crossover region, and the mutated genomic udgB region in the knockout mutant are shown. Probe locations and SexAI fragments detected by the probe are indicated. (D) Southern blot analysis of the Δ5031 strain. Inactivation of the M. smegmatis udgB gene and the absence of the wild-type allele are indicated by the presence of a single 2.4-kb fragment in the Δ5031 (lane 3) and Δung Δ5031 (lane 4) mutants compared to the single 2.8-kb fragment in the parental strain (lane 1) when preparations were probed with a 340-bp M. smegmatis udgB gene fragment. The results for a 5′ single crossover are shown in lane 2. Abbreviations: B, BamHI restriction site; S, SexAI restriction site; wt, wild type; ko, knockout; sco, single crossover.
The growth kinetics of the mutant M. smegmatis strains were comparable to those of the wild type; the generation times were 155 min for the wild type, 130 min for the ung knockout mutant (designated the Δung mutant), 152 min for the M. smegmatis udgB knockout mutant (designated the Δ5031 mutant), and 143 min for the double-knockout mutant (designated the Δung Δ5031 mutant).
General mutation frequency is increased in UDG knockout strains.
The general mutation frequency reflects an organism's ability to process spontaneous DNA damage. In this study, we determined the mutation frequencies of the M. smegmatis strains using a forward mutation screen with the rpoB gene that leads to rifampin resistance.
The general mutation frequencies of the single-knockout mutants were seven- to eightfold higher than those of the wild type. The mutation frequency of the double-knockout mutant was 56-fold higher, which indicates that there was a dramatic decrease in the ability of this mutant to repair spontaneous DNA damage (Table 3).
TABLE 3.
General mutation frequencies of UDG mutantsa
| Strain | General frequency of Rifr mutation | Increase compared with wild type (fold) |
|---|---|---|
| Wild type | 2.7 × 10−8 | 1.0 |
| Δung mutant | 2.0 × 10−7b | 7.4 |
| Δ5031 mutant | 2.2 × 10−7b | 8.1 |
| Δung Δ5031 mutant | 1.5 × 10−6b | 55.6 |
The values are the means of three independent experiments.
Significant increase in the mutation frequency compared to the wild type (P < 0.005, Student's t test; n = 3).
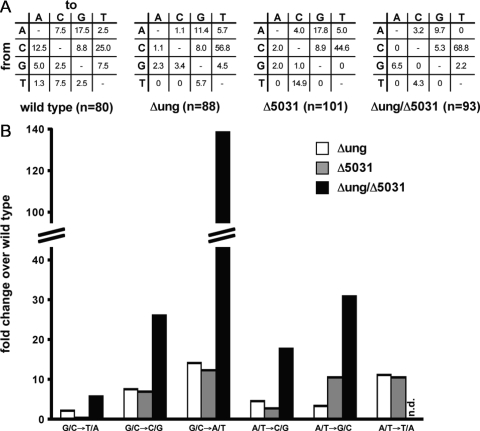
Sequence analysis of the rpoB gene in the Rifr M. smegmatis mutants revealed that the ung and M. smegmatis ugdB gene products are partially redundant, as the mutation spectra of all three mutant strains showed an increase in G·C→A·T transitions compared to the wild type. These mutations, which arise mostly during replication of genomic DNA containing uracil residues arising from cytosine deamination, accounted for one-quarter of all mutations in the wild-type strain. In contrast, in the double mutant, they accounted for two-thirds of all mutations, while the values for the single mutants were between these two extremes (Table 4 and Fig. 3A). As shown in Table 4, there were 12- to 15-fold more G·C→A·T transitions in single-gene knockout mutants and 139-fold more G·C→A·T transitions in the double-knockout mutant than in the wild type (Fig. 3B).
TABLE 4.
rpoB mutations in Rif-resistant mutants derived from the M. smegmatis wild-type strain and the Δung, Δ5031, and Δung Δ5031 mutants
| Strain | G·C→T·A
|
G·C→C·G
|
G·C→A·T
|
A·T→C·G
|
A·T→G·C
|
A·T→T·A
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of mutants with mutation/total no. of Rifr mutantsa | Mutation frequencyb | Increase compared with wild type (fold)c | No. of mutants with mutation/total no. of Rifr mutants | Mutation frequency | Increase compared with wild type (fold) | No. of mutants with mutation/total no. of Rifr mutants | Mutation frequency | Increase compared with wild type (fold) | No. of mutants with mutation/total no. of Rifr mutants | Mutation frequency | Increase compared with wild type (fold) | No. of mutants with mutation/total no. of Rifr mutants | Mutation frequency | Increase compared with wild type (fold) | No. of mutants with mutation/total no. of Rifr mutants | Mutation frequency | Increase compared with wild type (fold) | |
| Wild type | 16/80a | 5.40 × 10−9b | 9/80 | 3.04 × 10−9 | 24/80 | 8.10 × 10−9 | 8/80 | 2.70 × 10−9 | 20/80 | 6.75 × 10−9 | 3/80 | 1.01 × 10−9 | ||||||
| Δung mutant | 5/88 | 1.14 × 10−8 | 2.1c | 10/88 | 2.27 × 10−8 | 7.5 | 52/88 | 1.18 × 10−7 | 14.6 | 6/88 | 1.36 × 10−8 | 5.1 | 10/88 | 2.27 × 10−8 | 3.4 | 5/88 | 1.14 × 10−8 | 11.2 |
| Δ5031 mutant | 2/101 | 4.36 × 10−9 | 0.8 | 10/101 | 2.18 × 10−8 | 7.2 | 47/101 | 1.02 × 10−7 | 12.6 | 4/101 | 8.71 × 10−9 | 3.2 | 33/101 | 7.19 × 10−8 | 10.6 | 5/101 | 1.09 × 10−8 | 10.8 |
| Δung Δ5031 mutant | 2/93 | 3.23 × 10−8 | 6.0 | 5/93 | 8.06 × 10−8 | 26.5 | 70/93 | 1.13 × 10−6 | 139.4 | 3/93 | 4.84 × 10−8 | 17.9 | 13/93 | 2.10 × 10−7 | 31.1 | 0/93 | <1.61 × 10−8 | NDd |
Number of mutants on Middlebrook 7H10-Rif agar with the mutation indicated/total number of Rifr mutants.
The mutation frequency of the mutation indicated was calculated by comparison with the general mutation frequency of the strain.
Increase in the mutation frequency of a mutant strain compared with the wild-type mutation frequency for the mutation indicated.
ND, no mutation detected.
FIG. 3.
Mutation spectra of Rif-resistant mutants. (A) Mutation spectra of glycosylase knockout strains. The level of each type of mutation detected is expressed as a percentage of all of the mutations detected in a strain. The nucleotide that was mutated is indicated on the y axis, and the result of the mutation is indicated on the x axis. (B) Mutation frequencies for mutations in the knockout strains compared to the mutation frequencies in the wild-type strain. n.d., no mutation detected.
There were also glycosylase-dependent differences in the frequency of A·T→G·C mutations, which may arise through adenine deamination. Whereas the frequencies of these mutations were about 3-fold higher in the ung knockout mutant than in the wild type, the frequencies of these mutations in the udgB knockout mutant and in the double-knockout mutant were >10-fold and 31-fold higher that those in the wild-type strain, respectively.
Thymidylate synthase inhibition leads to UDG-dependent cell death.
Uracil has been hypothesized to trigger cell death through chromosomal fragmentation of uracil-rich genomes (8, 14). UDGs create abasic sites in the DNA (18) that are cleaved by AP endonucleases to generate single-strand breaks (16). UDG-associated AP endonuclease-induced double-strand breaks require a high density of genomic uracil in both the newly synthesized and template strands (14). dUTP is an intermediate in thymidylate synthesis; it is converted to dUMP plus PPi by dUTPase (Dut), and the resulting dUMP is then the sole precursor for the synthesis of dTTP catalyzed by thymidylate synthase. This conversion helps the cell maintain a low dUTP/dTTP ratio and thus reduces the likelihood of dUMP incorporation into DNA. The dut gene was shown to be essential in E. coli (2); however, mutants with low Dut activity have been found (38). These strains have significantly greater dUTP pools and display elevated DNA fragmentation and increased susceptibility to 5FdUrd (13). 5FdUrd is assumed to exert its effects predominantly through its sole metabolite, 5-fluoro-dUMP (5FdUMP), which inhibits thymidylate synthase. This leads to depletion of the dTTP pool and consequently to an increase in dUMP incorporation into newly replicated DNA (9).
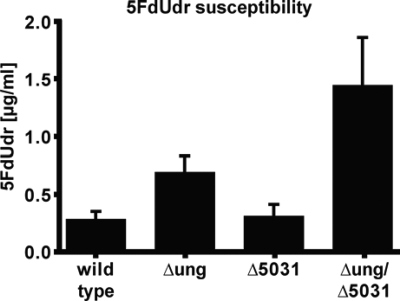
We tested the role of Ung and M. smegmatis UdgB in uracil-triggered cell death. The drug susceptibility of the mutants was studied by determining the MICs of 5FdUrd. Our results show that the ung knockout mutant tolerated this drug twofold better than the wild-type or the M. smegmatis udgB knockout mutant (Fig. 4). Loss of both UDGs decreased the sensitivity to the drug approximately fivefold compared to the wild type.
FIG. 4.
5FdUdr susceptibility. The MIC was determined in five independent experiments as described in Materials and Methods. The bars and error bars indicate the averages and standard deviations, respectively.
DISCUSSION
UdgB is conserved in hyperthermophilic archaea and in Actinomycetales.
Elevated temperatures in the environment of hyperthermophilic archaea enhance the rate of spontaneous hydrolytic deamination, especially the rate of spontaneous hydrolytic deamination of cytosine and adenine (17, 19). It is therefore hardly surprising that genomes of hyperthermophiles are protected against deamination-associated mutagenesis by more than one enzyme capable of dealing with this type of damage (17, 33). Significantly, UdgB orthologues are found not only in hyperthermophilic archaea but also (with the exception of Corynebacterium species and Tropheryma whipplei) in members of the order Actinomycetales, which have G+C-rich genomes that also require increased protection against cytosine deamination.
Mycobacteria, which are exposed to reactive oxygen or nitrogen intermediates inside host macrophages, might sustain even more DNA damage than other members of this order (39). In spite of this hostile environment, the mutation rates of mycobacteria are similar to those of other bacteria. This is even more remarkable considering that these organisms appear to lack a postreplicative mismatch repair system (35).
The fact that these organisms can effectively protect their genomes against the threat of deamination and reactive oxygen and nitrogen species is demonstrated by the finding that the general mutation rates of archaea and mycobacteria are comparable to those of E. coli (10, 35).
Both Ung and UdgB protect M. smegmatis against deamination-associated mutations.
Mutation detection systems are essential tools for determining general mutation frequencies, as well as the types of mutations resulting from specific defects in DNA metabolism. In this study, we screened for rifampin resistance, which can arise through any 1 of 69 base substitutions at 37 different sites in the rpoB gene encoding the β-subunit of RNA polymerase (11). Due to this versatility, the screen can be used to identify the substrates of DNA repair pathways of interest.
As shown in Table 3, the mutation frequencies of the single mutants were increased to similar extents, about sevenfold. When both ung and udgB were inactivated, the mutation frequency was increased more than 50-fold. The predominant mutation in our analysis for all strains was C→T, which results from deamination of cytosine to uracil.
The most frequently described rpoB mutations are 531TCG→TTG and 526CAC→TAC (11). To exclude the possibility that our analysis was affected by one or several such hotspots, which might have increased the observed bias in favor of cytosine alterations, we analyzed the locations of all the mutations. The nucleotide changes leading to rifampin resistance in our study were homogeneously distributed among known resistance-conferring base substitutions (data not shown).
Deamination of adenine to Hx leads to A·T→G·C mutations. The M. smegmatis udgB knockout mutant showed a >10-fold increase in A·T→G·C transitions compared to the wild type. This result supports the findings of our in vitro studies with P. aerophilum UdgB, which could be shown to remove both uracil and Hx from DNA (30).
Association of Ung with the replication fork.
Incorporation of uridine into DNA due to the use of dUTP instead of dTTP by replicative DNA polymerases takes place relatively frequently (3). It has been shown that mammalian nuclear UNG2, a homologue of bacterial Ung, associates with proliferating cell nuclear antigen, the processivity factor of replicative DNA polymerases (12). This has led to speculation that UNG2 acts on nascent DNA immediately after dUMP incorporation and is thus responsible for the immediate removal of uracil (23).
Because mammalian UNG2 was shown to largely rescue the mutator phenotype of an E. coli ung mutant, it was argued that UNG2 and Ung have similar roles (22). However, incorporation of dUMP into nascent DNA during replication is not mutagenic, as the dUMP is incorporated opposite A. Thus, the phenotypic rescue of the E. coli ung mutant by UNG2 implies only that the mammalian enzyme can process the mutagenic U·G mispairs arising through cytosine deamination in bacteria, but this provides little information concerning the role of the bacterial enzyme in the removal of uracil from replication-associated U·A pairs.
We wanted to search for evidence that might implicate the M. smegmatis Ung and UdgB enzymes in the processing of replication-associated uracil. Because the fluoropyrimidine 5FdUrd boosts cellular dUTP pools by inhibiting thymidylate synthase (9), more dUMP (and likely also more FdUMP) is incorporated into nascent DNA. Under these conditions, removal of uracil and fluorouracil by a uracil glycosylase frequently leads to cell death, because the repair synthesis requires dTTP, which is scarce in the cell. Instead, the repair polymerase might incorporate a dUMP or FdUMP residue that would be subject to renewed processing by the uracil glycosylase. When single-stranded gaps resulting from incomplete repair synthesis reach the replication fork, they give rise to cytotoxic double-strand breaks. Our results (Fig. 4) show that inactivation of the ung gene results in a 2.5-fold decrease in 5FdUrd toxicity, which confirms the hypothesis described above. Inactivation of M. smegmatis udgB did not have a noticeable effect on cell killing by the fluoropyrimidine, which suggests that the glycosylase encoded by this gene does not contribute to the cytotoxic processing of uracil and fluorouracil in 5FdUrd-treated cells, as long as Ung is present. However, the resistance of the double mutant to the drug was fivefold higher. This suggests that UdgB in the double mutant cells can and does contribute to the cytotoxic processing of incorporated uridine, albeit only when Ung is absent.
In this paper we provide key insights into the biological roles of uracil glycosylases in the model organism M. smegmatis. Phylogenic analysis showed that genes encoding UdgB are prevalent in genomes that have an elevated risk of base deamination. That the UdgB enzyme indeed counteracts the mutagenic threat of this spontaneous hydrolytic process became evident from the results of experiments described above. We found that the general mutation frequencies and mutation spectra of strains defective in Ung and M. smegmatis UdgB are very similar, which suggests that there is substantial functional redundancy, particularly in the prevention of G·C→A·T transition mutations that arise predominantly through cytosine deamination.
Differences could be detected in the abilities of the two deletion strains to prevent A·T→G·C mutations, which were repaired better when M. smegmatis UdgB was active. Because these mutations arise largely through the deamination of adenine, our results imply that UdgB removes from DNA not only uracil but also Hx, as shown in our in vitro studies with the P. aerophilum UdgB enzyme (30).
We also demonstrate that both Ung and UdgB can process uracil in newly synthesized DNA, even though in this case the functional redundancy is apparent only in the double mutant; in the presence of Ung, the M. smegmatis udgB strain had no detectable phenotype as measured by sensitivity to 5FdUrd. These findings suggest that Ung is highly active in the processing of U·A pairs arising during replication, whereas UdgB may act predominantly to process the mutagenic U·G and, importantly, Hx·T mispairs.
Acknowledgments
We thank Peter Sander for technical assistance, as well as for critical reading of the manuscript.
This work was supported by the Swiss National Science Foundation (D.C. and J.J.) and by the European Community (grant LSH-2003-1.1.5-5 to J.J. and grants LSHP-CT-2007-037235 and LSHM-CT-2005-518152 to B.S. and E.C.B.).
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Duncan, B. K., and J. H. Miller. 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287:560-561. [DOI] [PubMed] [Google Scholar]
- 2.el-Hajj, H. H., H. Zhang, and B. Weiss. 1988. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 170:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillet, M., and S. Boiteux. 2003. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:8386-8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guthlein, C., R. M. Wanner, P. Sander, E. O. Davis, M. Bosshard, J. Jiricny, E. C. Bottger, and B. Springer. 2009. Characterization of the mycobacterial NER system reveals novel functions of the uvrD1 helicase. J. Bacteriol. 191:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haushalter, K. A., M. W. Todd Stukenberg, M. W. Kirschner, and G. L. Verdine. 1999. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 9:174-185. [DOI] [PubMed] [Google Scholar]
- 6.Hinks, J. A., M. C. Evans, Y. De Miguel, A. A. Sartori, J. Jiricny, and L. H. Pearl. 2002. An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 277:16936-16940. [DOI] [PubMed] [Google Scholar]
- 7.Hoseki, J., A. Okamoto, R. Masui, T. Shibata, Y. Inoue, S. Yokoyama, and S. Kuramitsu. 2003. Crystal structure of a family 4 uracil-DNA glycosylase from Thermus thermophilus HB8. J. Mol. Biol. 333:515-526. [DOI] [PubMed] [Google Scholar]
- 8.Ingraham, H. A., L. Dickey, and M. Goulian. 1986. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry 25:3225-3230. [DOI] [PubMed] [Google Scholar]
- 9.Ingraham, H. A., B. Y. Tseng, and M. Goulian. 1982. Nucleotide levels and incorporation of 5-fluorouracil and uracil into DNA of cells treated with 5-fluorodeoxyuridine. Mol. Pharmacol. 21:211-216. [PubMed] [Google Scholar]
- 10.Jacobs, K. L., and D. W. Grogan. 1997. Rates of spontaneous mutation in an archaeon from geothermal environments. J. Bacteriol. 179:3298-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, R., E. M. Streicher, G. E. Louw, R. M. Warren, P. D. van Helden, and T. C. Victor. 2006. Drug resistance in Mycobacterium tuberculosis. Curr. Issues Mol. Biol. 8:97-111. [PubMed] [Google Scholar]
- 12.Ko, R., and S. E. Bennett. 2005. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair 4:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koehler, S. E., and R. D. Ladner. 2004. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol. Pharmacol. 66:620-626. [DOI] [PubMed] [Google Scholar]
- 14.Kouzminova, E. A., and A. Kuzminov. 2006. Fragmentation of replicating chromosomes triggered by uracil in DNA. J. Mol. Biol. 355:20-33. [DOI] [PubMed] [Google Scholar]
- 15.Kouzminova, E. A., and A. Kuzminov. 2008. Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-stranded breaks. Mol. Microbiol. 68:202-215. [DOI] [PubMed] [Google Scholar]
- 16.Levin, J. D., and B. Demple. 1990. Analysis of class II (hydrolytic) and class I (beta-lyase) apurinic/apyrimidinic endonucleases with a synthetic DNA substrate. Nucleic Acids Res. 18:5069-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl, T., S. Ljungquist, W. Siegert, B. Nyberg, and B. Sperens. 1977. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 252:3286-3294. [PubMed] [Google Scholar]
- 19.Lindahl, T., and B. Nyberg. 1974. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13:3405-3410. [DOI] [PubMed] [Google Scholar]
- 20.Mosbaugh, D. W., and S. E. Bennett. 1994. Uracil-excision DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 48:315-370. [DOI] [PubMed] [Google Scholar]
- 21.Neddermann, P., and J. Jiricny. 1993. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 268:21218-21224. [PubMed] [Google Scholar]
- 22.Olsen, L. C., R. Aasland, H. E. Krokan, and D. E. Helland. 1991. Human uracil-DNA glycosylase complements E. coli ung mutants. Nucleic Acids Res. 19:4473-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otterlei, M., E. Warbrick, T. A. Nagelhus, T. Haug, G. Slupphaug, M. Akbari, P. A. Aas, K. Steinsbekk, O. Bakke, and H. E. Krokan. 1999. Post-replicative base excision repair in replication foci. EMBO J. 18:3834-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagni, M., V. Ioannidis, L. Cerutti, M. Zahn-Zabal, C. V. Jongeneel, and L. Falquet. 2004. MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 32:W332-W335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearl, L. H. 2000. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 460:165-181. [DOI] [PubMed] [Google Scholar]
- 26.Pfister, P., S. Hobbie, Q. Vicens, E. C. Bottger, and E. Westhof. 2003. The molecular basis for A-site mutations conferring aminoglycoside resistance: relationship between ribosomal susceptibility and X-ray crystal structures. Chembiochem 4:1078-1088. [DOI] [PubMed] [Google Scholar]
- 27.Sander, P., A. Meier, and E. C. Bottger. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 16:991-1000. [DOI] [PubMed] [Google Scholar]
- 28.Sandigursky, M., and W. A. Franklin. 1999. Thermostable uracil-DNA glycosylase from Thermotoga maritima, a member of a novel class of DNA repair enzymes. Curr. Biol. 9:531-534. [DOI] [PubMed] [Google Scholar]
- 29.Sandigursky, M., and W. A. Franklin. 2000. Uracil-DNA glycosylase in the extreme thermophile Archaeoglobus fulgidus. J. Biol. Chem. 275:19146-19149. [DOI] [PubMed] [Google Scholar]
- 30.Sartori, A. A., S. Fitz-Gibbon, H. Yang, J. H. Miller, and J. Jiricny. 2002. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 21:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro, R., and S. H. Pohl. 1968. The reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry 7:448-455. [DOI] [PubMed] [Google Scholar]
- 32.Slupphaug, G., I. Eftedal, B. Kavli, S. Bharati, N. M. Helle, T. Haug, D. W. Levine, and H. E. Krokan. 1995. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry 34:128-138. [DOI] [PubMed] [Google Scholar]
- 33.Smith, K. C. 1992. Spontaneous mutagenesis: experimental, genetic and other factors. Mutat. Res. 277:139-162. [DOI] [PubMed] [Google Scholar]
- 34.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 35.Springer, B., P. Sander, L. Sedlacek, W. D. Hardt, V. Mizrahi, P. Schar, and E. C. Bottger. 2004. Lack of mismatch correction facilitates genome evolution in mycobacteria. Mol. Microbiol. 53:1601-1609. [DOI] [PubMed] [Google Scholar]
- 36.Srinath, T., S. K. Bharti, and U. Varshney. 2007. Substrate specificities and functional characterization of a thermo-tolerant uracil DNA glycosylase (UdgB) from Mycobacterium tuberculosis. DNA Repair 6:1517-1528. [DOI] [PubMed] [Google Scholar]
- 37.Starkuviene, V., and H. J. Fritz. 2002. A novel type of uracil-DNA glycosylase mediating repair of hydrolytic DNA damage in the extremely thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 30:2097-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, A. F., and B. Weiss. 1982. Role of exonuclease III in the base excision repair of uracil-containing DNA. J. Bacteriol. 151:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner, D. F., and V. Mizrahi. 2006. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin. Microbiol. Rev. 19:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner, H. R., B. K. Duncan, C. Garrett, and J. Neuhard. 1981. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J. Bacteriol. 145:687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]