Abstract
The molecular mechanism(s) by which deep-sea bacteria grow optimally under high hydrostatic pressure at low temperatures is poorly understood. To gain further insight into the mechanism(s), a previous study screened transposon mutant libraries of the deep-sea bacterium Photobacterium profundum SS9 and identified mutants which exhibited alterations in growth at high pressure relative to that of the parent strain. Two of these mutants, FL23 (PBPRA3229::mini-Tn10) and FL28 (PBPRA1039::mini-Tn10), were found to have high-pressure sensitivity and enhanced-growth phenotypes, respectively. The PBPRA3229 and PBPRA1039 genes encode proteins which are highly similar to Escherichia coli DiaA, a positive regulator, and SeqA, a negative regulator, respectively, of the initiation of DNA replication. In this study, we investigated the hypothesis that PBPRA3229 and PBPRA1039 encode DiaA and SeqA homologs, respectively. Consistent with this, we determined that the plasmid-carried PBPRA3229 and PBPRA1039 genes restored synchrony to the initiation of DNA replication in E. coli mutants lacking DiaA and SeqA, respectively. Additionally, PBPRA3229 restored the cold sensitivity phenotype of an E. coli dnaA(Cs) diaA double mutant whereas PBPRA1039 suppressed the cold sensitivity phenotype of an E. coli dnaA(Cs) single mutant. Taken together, these findings show that the genes disrupted in FL23 and FL28 encode DiaA and SeqA homologs, respectively. Consequently, our findings add support to a model whereby high pressure affects the initiation of DNA replication in P. profundum SS9 and either the presence of a positive regulator (DiaA) or the removal of a negative regulator (SeqA) promotes growth under these conditions.
Despite the fact that more than 70% of the earth's surface is covered by oceans, which have an average temperature of 3°C and exert an average hydrostatic pressure of 38 MPa (atmospheric pressure is ∼0.1 MPa), little is understood about the molecular basis of cold- and high-pressure-adapted deep-ocean life. The discovery and isolation of the pyschrotolerant facultative piezophile (high-pressure-loving organism) Photobacterium profundum SS9 (8) have made it possible to more readily address the mechanisms of piezophilic growth at cold temperatures (for a recent review, see reference 3). P. profundum SS9 is a gammaproteobacterium originally isolated from an amphipod homogenate obtained from the Sulu Sea in the Philippines at a depth of 2.5 km and a temperature of 9°C (8). Although it grows optimally at 28 MPa and 15°C, P. profundum SS9 can also grow over a wide range of pressures (0.1 to 90 MPa) and temperatures (2 to 20°C). The ability to grow at atmospheric pressure has made P. profundum SS9 more amenable to genetic manipulation than obligate piezophiles. The P. profundum SS9 genome has been sequenced and annotated (26) and consists of two chromosomes and an 80-kb plasmid. It was determined that the 80-kb plasmid is nonessential for the piezophilic growth of P. profundum SS9 (26).
To gain insights into the genetic basis of high-pressure-adapted growth, transposon mutant libraries of P. profundum SS9R (a rifampin [rifampicin]-resistant derivative of SS9) were screened in liquid culture for mutants with defects in the ability to grow at high pressure (45 MPa, 15°C) (19). One of the putative high-pressure-sensitive mutants (FL23) isolated from these screens had a mini-Tn10 insertion in the gene PBPRA3229, which encodes a protein with 75% identity (85% similarity) to Escherichia coli DiaA (DnaA initiator-associating factor) (14). Although FL23 shows growth defects at 0.1 MPa (15°C) relative to the parent strain, the ratio of growth at 45 MPa to growth at 0.1 MPa and 15°C is substantially reduced compared to that of the parent strain, confirming that disruption of PBPRA3229 results in a high-pressure sensitivity growth phenotype (19).
In E. coli, DiaA is necessary to ensure the timely initiation of DNA replication (14). DiaA forms a tetramer and binds to multiple molecules of DnaA, promoting (i) the binding of DnaA to the origin of replication in E. coli (known as oriC), (ii) ATP-DnaA-specific conformational changes in the oriC complex, and (iii) the unwinding of oriC DNA (17). Consequently, E. coli DiaA acts as a positive regulator of the initiation of DNA replication. In the absence of DiaA, initiation of DNA replication is delayed and in E. coli cells with two oriC copies, it only occurs from one of these, resulting in cells with three copies of their chromosome (14). In contrast, this is an extremely rare occurrence in wild-type E. coli cells. Although disruption of diaA in E. coli results in an asynchronous DNA replication phenotype, it does not appear to affect growth or morphology at atmospheric pressure at 37°C in a genetic background with a wild-type dnaA gene. However, disruption of the diaA gene suppresses the cold sensitivity phenotype of an E. coli dnaA(Cs) mutant at 30°C.
Even though PBPRA3229 is highly similar to E. coli DiaA, it also shows 45% identity (65% similarity) to a phosphoheptose isomerase in E. coli known as GmhA (4). GmhA is involved in lipopolysaccharide (LPS) biosynthesis and catalyzes the isomerization of d-sedoheptulose 7-phosphate into d-glycero-d-manno-heptose 7-phosphate, which is the first step in the biosynthesis of ADP-glycero-manno-heptose, a subunit of the LPS inner core. The LPS forms the outermost leaflet of the outer membrane of gram-negative bacterial cells, and in E. coli K-12 strains, the LPS is composed of inner and outer sugar cores and lipid A (25). E. coli K-12 mutants lacking GmhA produce truncated LPS species relative to that of the parent strain due to the absence of the inner core, which can be easily visualized by gel electrophoresis followed by silver staining (4). Due to the high degree of sequence similarity between PBPRA3229 and GmhA, it is also possible that FL23 has an alteration in its LPS relative to that of the parent strain.
In contrast to DiaA, SeqA is a negative regulator of the initiation of DNA replication in E. coli (20). E. coli SeqA binds to hemimethylated oriC and prevents the binding of ATP-DnaA. Disruption of seqA in E. coli also results in an asynchronous-replication phenotype. However, the effect of DiaA on the timing of DNA replication initiation appears to be SeqA independent (14). Interestingly, a putative P. profundum SS9R seqA transposon insertion mutant (PBPRA1039::Tn10) was identified as having high-pressure-enhanced growth at 45 MPa and 15°C relative to its growth at atmospheric pressure (19). Therefore, this preliminary finding suggests that the removal of a negative regulator of the initiation of DNA replication could promote the growth of P. profundum SS9R at high pressure.
In this study, we investigated the hypothesis that proteins that regulate the initiation of DNA replication play a key role in the piezophilic growth of P. profundum SS9. We determined that PBPRA3229 and PBPRA1039 encode functional DiaA and SeqA homologs, respectively, and we propose a model whereby the initiation of DNA replication is sensitive to high pressure and either the production of a positive regulator (DiaA) or the removal of a negative regulator (SeqA) can promote growth under these conditions.
MATERIALS AND METHODS
Bacterial strains and atmospheric pressure growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. All E. coli strains were grown in Luria-Bertani (LB) medium (24) or on LB agar (1.5%, wt/vol). P. profundum SS9R strains were grown anaerobically in marine broth 2216 (28 g liter−1; Difco Laboratories) buffered with HEPES (100 mM, pH 7.5) and supplemented with 20 mM glucose. Marine agar was prepared by the addition of 1.7% (wt/vol) agar. Unless stated otherwise, all P. profundum SS9R cultures were initially grown to early stationary phase (∼48 h) in marine broth at 0.1 MPa and 15°C without shaking in heat-sealed polyethylene transfer pipettes (4.5 ml) as described previously (1). For solid-agar growth experiments, early-stationary-phase cultures of P. profundum SS9R were diluted to an optical density at 600 nm (OD600) of ∼0.2 and serially diluted until 10−5, and 10-μl aliquots were spotted onto marine agar plates. The agar plates were then incubated at different temperatures at 0.1 MPa, and bacterial growth was assessed at different times. When required, unless indicated otherwise, antibiotics were added at the following concentrations: rifampin, 100 μg ml−1 for P. profundum SS9R; streptomycin, 100 μg ml−1 for E. coli and P. profundum SS9R; kanamycin, 50 μg ml−1 for E. coli and 500 μg ml−1 for P. profundum SS9R. Where stated, thymine was also added at 50 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| P. profundum | ||
| SS9 | High-pressure- and cold-loving deep-sea isolate | 8 |
| SS9R | Rifr SS9 derivative | 7 |
| FL23 | SS9R PBPRA3229::mini-Tn10 Rifr Kanr | 19 |
| FL28 | SS9R PBPRA1039::mini-Tn10 Rifr Kanr | 19 |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | 13 |
| MM294A | pro-82 thi-1 endA1 supE44 hsdR17 | 12 |
| χ705 | F−leu-4 φr Strrarg-35 T6r λ− | 4 |
| χ711 | χ705 ΔgmhA Strr | 4 |
| KH5402-1 | ilv thyA thr tyrA trpE9829 metE deo supF6 | 15 |
| NA001 | KH5402-1 dnaA(Cs) | 16 |
| NA026 | NA001 diaA::Tn5 Kanr | 14 |
| NA141 | KH5402-1 diaA::Tn5 Kanr | 14 |
| BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 2 |
| JW0674 | BW25113 ΔseqA Kanr | 2 |
| ZE0674 | KH5402-1 ΔseqA Kanr | This study |
| Plasmids | ||
| pFL190 | Broad-host-range plasmid with PBAD inducible promoter, Strr | 18 |
| pECdiaA | pFL190 with entire E. coli diaA gene and 77 bp upstream | This study |
| pPBPRA3229 | pFL190 with entire PBPRA3229 gene and 70 bp upstream | This study |
| pECseqA | pFL190 with entire E. coli seqA gene and 81 bp upstream | This study |
| pPBPRA1039 | pFL190 with entire PBPRA1039 gene and 57 bp upstream | This study |
| pRK2013 | Helper plasmid for mating, Kanr | 9 |
High-pressure growth of P. profundum SS9.
P. profundum SS9R was grown at high pressure as previously described (1), with slight modifications. In brief, polyethylene transfer pipettes containing diluted cultures (referred to earlier) were incubated in a pressure vessel (3.5-liter capacity) at the different pressures and temperatures. The vessel was pressurized with water by using a hydraulic pump equipped with quick-connect fittings for rapid decompression and recompression, and the temperature was controlled by a water jacket connected to a refrigerated circulating water bath. To assess growth at pressure on solid agar, an overlay procedure modified from a previously published method was used (21). On cooling, an autoclaved solution of marine broth 2216-HEPES (pH 7.5) was mixed with an autoclaved aqueous solution of 5% (wt/vol) low-temperature-gelling agar (LTGA; Nacalai Tesque Inc., Japan). The mixture was then supplemented with 20 mM glucose and poured into petri dishes (LTGA plates). Early-stationary-phase cultures of P. profundum SS9R were then diluted to an OD600 of ∼0.8 and serially diluted to 10−5, and 10-μl aliquots were spotted onto LTGA plates. The LTGA plates were incubated at 15°C and 0.1 MPa for 2 h and placed on ice, and an equivalent amount of LTGA was poured directly on top of the spotted cultures. The agar “sandwich” was removed from the petri dish, placed into sterile plastic pouches (The Vacuum Pouch Ltd., United Kingdom), heat sealed, and then incubated in the pressure vessel.
Cloning of E. coli and P. profundum SS9 genes into pFL190.
The E. coli diaA and seqA genes were amplified by PCR with primers EcdiaA-77F-EcoRI (5′-GTGAATTCACCGGGAATGAGGTTGAGTGGATTAAG-3′) and EcdiaA+627R-XbaI (5′-GGTCTAGAGCTGGCGATAATGCCTTCATGTATTCTCC-3′) and primers EcseqA-81F-EcoRI (5′-GTGAATTCGGCTTGACGCTATCCGCTGC-3′) and EcseqA+549R-XbaI (5′-GCTCTAGACATTGCTTTGTCCTTTGTCTGCAACG-3′), respectively. The P. profundum SS9 PBPRA3229 and PBPRA1039 genes were amplified by PCR with primers pbpra3229-70F-EcoRI (5′-GTGAATTCATGTGGTCGCAATTCAAGGCC-3′) and pbpra3229+618R-XbaI (5′-TTCTAGATACACCAATACTTGGCGTAGTAT-3′) and primers pbpra1039-57F-EcoRI (5′-GGAATTCGCTAGAGTATAACCTCGGATCA-3′) and pbpra1039+561R-XbaI (5′-GCTCTAGAGCGCATATAGCCAGTTAAATAA-3′), respectively. The amplified genes were digested with EcoRI and XbaI and ligated into pFL190 (18) under the control of the PBAD promoter (22). Since pFL190 lacks a ribosomal binding site, the primers were designed such that the amplified gene contained the predicted ribosomal binding site for each gene (http://www.tigr.org/). Positive clones were identified by restriction digestion and sequenced with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) at the University of Edinburgh School of Biological Sciences Sequencing Service. The sequences of the cloned genes were identical to the published sequences.
RNA extraction and reverse transcription (RT)-PCR.
A culture of the defined strain was grown overnight in 5 ml of the appropriate medium, diluted 1:50, and then grown to an OD600 of ∼0.4. The cell pellets were resuspended in 1 ml of stabilization buffer (0.1% [wt/vol] sodium dodecyl sulfate [SDS], 1% [vol/vol] phenol, and 19% [vol/vol] ethanol in water). The reaction mixture was incubated on ice for 30 min and then stored at −80°C. When required, cells were lysed by thawing, followed by treatment with lysozyme (40 ng ml−1) for 5 min at room temperature. The RNA was then extracted with the SV Total RNA Isolation System (Promega) by following the manufacturer's protocol. The RNA was eluted in 100 μl of nuclease-free water and stored at −80°C. One-step RT-PCR was performed with the Access RT-PCR System (Promega) by following the manufacturer's protocol with primers pbpra3227+1506F (5′-CGCAAGCTCACGTGGCAACCC-3′) and pbpra3230+209R (5′-GTAGGTAGAGCACGAACCTTCACC-3′) for P. profundum SS9 and ECyraM+1674F (5′-CGGTAGCCAGAGCGGTGCAAC-3′) and ECyraP+319R (5′-GGCCCTGACGAATCTCGTTATACAC-3′) for E. coli. Products were visualized by agarose gel electrophoresis.
Conjugation of plasmids from E. coli into P. profundum SS9R.
The conjugation of plasmids from E. coli DH5α into P. profundum SS9R strains was performed by following a previously described method with modifications (19). Briefly, the P. profundum SS9 acceptor and E. coli donor and helper (MM294A/pRK2013) strains were grown in the appropriate media to early stationary phase and then washed in marine broth. The cultures were mixed in a 1:1:1 ratio, plated onto marine agar, and then incubated at 20°C for 40 h. The cells were then resuspended in marine broth, plated on marine agar containing streptomycin (150 μg ml−1) and rifampin, and then incubated at 0.1 MPa and 15°C for a week.
Flow cytometry analysis.
Flow cytometry experiments were performed as previously described, with modifications (17). The defined E. coli strains were grown overnight at 30°C in LB medium supplemented with thymine (50 μg ml−1) in the case of KH5402-1 derivatives. The cultures were diluted 50-fold in LB medium with l-arabinose (0.01, 0.05, or 0.2%, wt/vol) and thymine for KH5402-1 derivatives. They were incubated at 30°C with shaking until an OD600 of ∼0.2 was reached, and then cephalexin (cefalexin; 10 μg ml−1) and rifampin (300 μg ml−1) were added. The cultures were incubated as before for a further 4 h, and then the cells were pelleted by centrifugation, washed in buffer (100 mM Tris, 10 mM MgSO4, pH 7.5), fixed in 70% (vol/vol) ethanol, and stored at 4°C until required. Immediately before use, the cell pellets were washed again with buffer, the DNA was stained with SYTOX Green (2 μM prepared in buffer), and then the cells were analyzed by flow cytometry on a FACScalibur (Becton Dickinson) with the fluorescein isothiocyanate laser and filters to visualize SYTOX Green. Samples were run through the cytometer under low pressure, and the following voltage settings were used: forward scatter, E02; side scatter, 350 V; fluorescein isothiocyanate laser, 480 V. The results obtained were analyzed with the CellQuest software (Becton Dickinson) normalized with cells alone and SYTOX without cells. The chromosome number was determined by comparing the fluorescence intensity of the peaks to that of a stationary-phase culture.
LPS isolation and analysis.
The LPS was extracted from early-stationary-phase cultures (1.5 ml) by SDS lysis and resolved by SDS-polyacrylamide gel electrophoresis exactly as described previously (6). To visualize the different LPS species, the gels were fixed overnight (40% [vol/vol] ethanol, 5% [vol/vol] glacial acetic acid) and then treated with sodium meta-periodate (0.7% [wt/vol] in fixative solution) for 5 min. The gels were then stained with silver nitrate exactly as described previously (6).
Microscopy.
Microscopy was performed with a 100× oil immersion phase-contrast lens (Carl Zeiss microscope), and images were taken with an AxioCam with AxioVision version 2.0 software. Before use, the microscope slides were coated by incubation in poly-L-lysine (0.01%, wt/vol) for 5 min and then dried at 65°C for 1 h.
RESULTS
FL23 does not have an altered LPS profile.
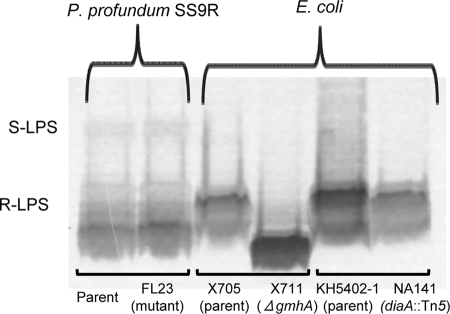
The gene disrupted in the high-pressure-sensitive P. profundum SS9R mutant FL23 encodes a protein which shows sequence similarity to both E. coli GmhA and DiaA (4, 14). Since GmhA is involved in LPS biosynthesis in E. coli (4), the LPS molecules from FL23 and the parent strain P. profundum SS9R were isolated by SDS lysis and then analyzed by gel electrophoresis-silver staining (Fig. 1). As controls, the LPS molecules were also extracted from an E. coli gmhA deletion mutant (χ711) and its parent strain (χ705) and an E. coli diaA::Tn5 mutant (NA141) and its parent strain (KH5402-1). In many bacterial species, the LPS species produced are smooth (S-LPS) and are composed of O antigen, a sugar core, and lipid A (23). However, since all of the E. coli strains used in this analysis are K-12 derivatives, they lack the O-antigen component and only produce rough LPS (R-LPS) (25). Consistent with previous studies (4), we determined that the E. coli gmhA deletion mutant produced a R-LPS species that is truncated compared to that of the parent strain (Fig. 1). In contrast, although P. profundum SS9R was able to produce a small amount of S-LPS in combination with the R-LPS, no truncations of the FL23 mutant LPS relative to that of the parent strain were observed (Fig. 1). This finding suggests that PBPRA3229 does not encode a functional GmhA homolog. In addition, since the LPS species of the E. coli diaA mutant was not truncated compared to that of the parent strain (Fig. 1), this also demonstrates that the E. coli DiaA protein is not involved in LPS modifications, despite its high degree of sequence similarity to E. coli GmhA.
FIG. 1.
The P. profundum SS9R FL23 mutant does not exhibit LPS alteration. LPS was extracted by an SDS lysis method, and the different species present in each sample were visualized by polyacrylamide gel electrophoresis-periodate-silver staining. The LPS profiles shown are representative of at least two independent experiments.
The P. profundum SS9 PBPRA3229 gene restores synchrony to E. coli DNA replication in the absence of DiaA.
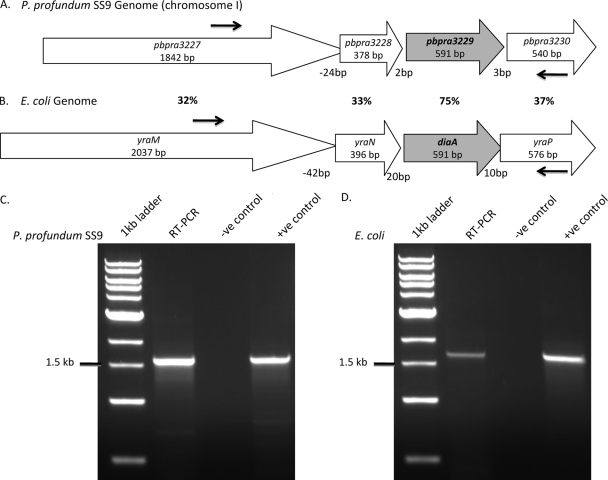
The gene disrupted in FL23 encodes a protein with 75% identity to E. coli DiaA, which promotes the timely initiation of DNA replication (14). Interestingly, the P. profundum SS9 PBPRA3229 and E. coli diaA genes appear to be similarly arranged within their respective genomes into putative operons with three other genes which encode proteins of unknown function (Fig. 2A and B). By RT-PCR, we determined that both the P. profundum SS9 PBPRA3229 and E. coli diaA genes are each cotranscribed with the other three genes and form the third gene of a four-gene operon (Fig. 2C and D, respectively). E. coli mutants lacking DiaA display asynchronous DNA replication phenotypes, which can be detected by incubation in the presence of DNA-binding dyes, followed by flow cytometry (14). However, due to technical difficulties, we were unsuccessful at using this methodology to investigate the chromosome number of either P. profundum SS9R or FL23 (data not shown). This is likely due to the complications of P. profundum SS9R having two different chromosomes (26) and producing a mixture of morphologies at atmospheric pressure (see Fig. 4C).
FIG. 2.
Genetic organization of the P. profundum SS9 PBPRA3229 and E. coli diaA genes. Organization of the PBPRA3229 (A) and diaA (B) genes within the P. profundum SS9 (26) and E. coli K-12 genomes (accession no. NC_000913). The percentages shown indicate the amino acid identities of the gene products. RT-PCR results obtained with total RNAs extracted from P. profundum SS9R (C) and E. coli MG1655 (D) are shown. The binding sites for the primers used are shown by the arrows in panels A and B. The RT-PCRs gave products sizes of 1.6 and 1.7 kb for P. profundum SS9 and E. coli, respectively, indicating that the four genes are transcribed together. The negative control lacks reverse transcriptase, confirming that the RNA sample is free from contaminating genomic DNA, whereas the positive control contains only genomic DNA.
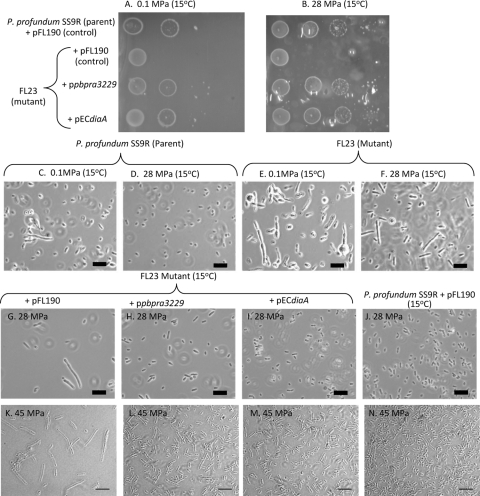
FIG. 4.
The PBPRA3229 gene and the E. coli diaA gene can both complement the growth and morphological alterations of the P. profundum SSR FL23 mutant. (A and B) Cultures of the strains shown were grown in marine broth supplemented with streptomycin, diluted, and spotted onto low-gelling-temperature marine agar overlay plates supplemented with l-arabinose (0.01%, wt/vol) but not streptomycin. The data sets shown are representative of at least three independent experiments where similar growth patterns were observed. Due to the fragile nature of the overlay plates after incubation at high pressure, the plates could only be photographed in the plastic pouches. The imperfections in the photographs are due to deformations of the plastic pouches. (C to J) Cultures were diluted 1:500 in either marine broth alone (C to F) or with streptomycin and 0.01% (wt/vol) l-arabinose (G to N) and grown for ∼40 h under the conditions shown. The images shown are representative fields of view from at least two independent experiments. Scale bars = 10 μm.
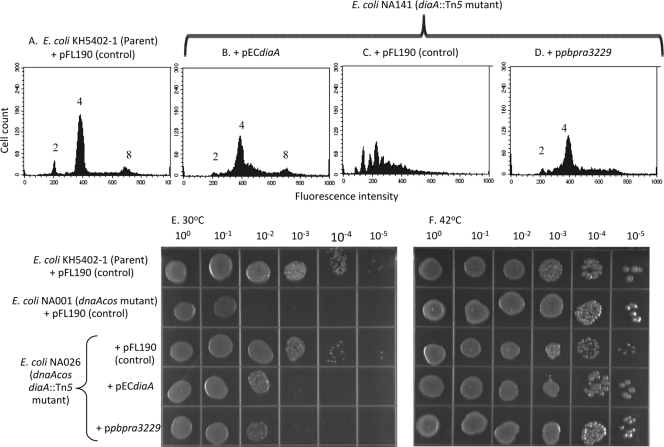
To test the hypothesis that PBPRA3229 encodes a functional DiaA homolog, we examined its ability to complement the initiation of DNA replication defect of an E. coli mutant lacking DiaA by using flow cytometry in combination with the DNA binding dye SYTOX Green. To enable us to assess this, the PBPRA3229 gene was cloned into pFL190 (pPBPRA3229) under the control of an l-arabinose-inducible promoter (22) and transformed into an E. coli mutant lacking DiaA (NA141). As a control, the strain was also transformed with either the vector alone or with the E. coli diaA gene cloned into pFL190 (pECdiaA). The genes were cloned into pFL90 since this is a broad-host-range plasmid which also replicates in P. profundum SS9 (18).
We determined under our experimental conditions that the parent strain with pFL190 and the E. coli diaA mutant with pECdiaA showed three peaks of fluorescence by flow cytometry after incubation in the presence of SYTOX Green (Fig. 3A and B, respectively). These results are consistent with previous studies (14) whereby the peaks correspond to cells with two, four, and eight copies of their chromosome, respectively. In sharp contrast, cells of the E. coli diaA mutant with the control plasmid pFL190 showed a range of fluorescence intensities under the same experimental conditions (Fig. 3C). These findings are consistent with previous studies in which, in the absence of DiaA in E. coli, there is a mistiming of the initiation of DNA replication, resulting in odd numbers of chromosomes per cell (14). We found that transformation of the E. coli diaA mutant with pPBPRA3229 restored the major fluorescence peak, corresponding to four chromosomal copies, observed for the parent strain with pFL190, as well as with the E. coli diaA mutant with pECdiaA (Fig. 3D, A, and B, respectively). Additionally, the presence of pPBPRA3229 in the E. coli diaA mutant significantly reduced the range of fluorescence intensities observed for the same mutant with control plasmid (Fig. 3D and C, respectively). Combined, these findings provide evidence that the P. profundum SS9 PBPRA3229 gene can restore synchrony to the initiation of DNA replication in E. coli in the absence of DiaA.
FIG. 3.
The P. profundum SS9 PBPRA3229 gene is functionally interchangeable with diaA in E. coli. (A to D) Exponential-phase cells of the E. coli strains shown grown in the presence of streptomycin, thymine, and 0.2% (wt/vol) l-arabinose were fixed, and the chromosome number was determined with SYTOX Green in combination with flow cytometry. The profiles shown are representative of two independent experiments. Similar results were also obtained with 0.01% (wt/vol) l-arabinose (data not shown). (E and F) The E. coli strains shown were grown overnight in LB medium (42°C) supplemented with streptomycin and thymine. Cultures were then diluted and spotted onto LB agar supplemented with thymine, streptomycin, and 0.01% (wt/vol) l-arabinose. Plates were then incubated overnight at the temperatures shown. The data set shown is representative of at least two independent experiments where similar growth patterns were observed.
The P. profundum SS9 PBPRA3229 gene restores cold sensitivity to an E. coli dnaA(Cs) diaA double mutant.
It has been shown previously that, despite the importance of DiaA in the initiation of DNA replication, an E. coli mutant lacking DiaA does not display growth defects in liquid culture (14). Additionally, we determined that both the E. coli parent strain (KH5402-1) and the E. coli diaA::Tn5 mutant grew as rod-shaped cells at 0.1 MPa but grew as filaments at 28 MPa and 37°C in LB medium (data not shown), suggesting that both strains were equally affected in their growth at high pressure. Since it was shown previously that disruption of diaA suppresses the cold sensitivity phenotype of an E. coli dnaA(Cs) mutant and transformation of the double mutant strain with a plasmid-carried E. coli diaA gene restores the cold sensitivity phenotype (14), this provided us with another means to confirm that the P. profundum SS9 PBPRA3229 gene encodes a functional DiaA homolog. To investigate this, the E. coli dnaA(Cs) diaA double mutant was transformed with pFL190, pECdiaA, or pPBPRA3229 and then its growth on LB agar at 30 and 42°C was assessed (Fig. 3E and F, respectively). As controls, the growth of the parent strain and that of the dnaA(Cs) diaA double mutant with pFL190 were also assessed (Fig. 3E and F); the growth of the E. coli diaA mutant with pFL190 was unaffected relative to that of the parent strain under these conditions (data not shown). We determined that the E. coli dnaA(Cs) diaA double mutant transformed with either pPBPRA3229 or pECdiaA results in a cold sensitivity phenotype at 30°C whereby the cultures only grew down to the 10−2 dilution. In contrast, cultures of the E. coli dnaA(Cs) diaA double mutant transformed with pFL190 or the parent strain with pFL190 grew reproducibly down to the 10−4 dilution at 30°C after the same time period (Fig. 3E). All of the strains grew to the 10−5 dilution at 42°C (Fig. 3F). Therefore, these data show that the P. profundum SS9 PBPRA3229 gene can restore cold sensitivity to an E. coli dnaA(Cs) diaA double mutant, and combined with our flow cytometry analysis, these findings provide further evidence that PBPRA3229 encodes a functional DiaA homolog.
Complementation of the P. profundum SS9R FL23 mutant with either PBPRA3229 or E. coli diaA.
Previously, it was demonstrated that the FL23 mutant exhibits a high-pressure sensitivity phenotype at 45 MPa relative to its growth at atmospheric pressure (19). To further characterize the effect of pressure on FL23, we investigated the growth of FL23 with pFL190 compared to that of parent strain P. profundum SS9R with pFL190 on marine agar overlay plates at 0.1 and 28 MPa and 15°C (Fig. 4A and B, respectively). To enable growth of P. profundum SS9R at high pressure, we modified a previously published agar overlay method (21). Although the agar overlay method reduced the growth of P. profundum SS9R at atmospheric pressure and 15°C (data not shown), the parent strain with pFL190 grew significantly better in the agar overlay at high pressure (28 MPa) than at 0.1 MPa (Fig. 4B and A, respectively). In contrast, the growth of FL23 with pFL190 was substantially reduced at both 0.1 and 28 MPa at 15°C relative to that of the parent strain on the agar overlay (Fig. 4A and B, respectively). Additionally, since the growth of FL23 with pFL190 on the agar overlay incubated at either 0.1 or 28 MPa was the same (Fig. 4B and A, respectively), this shows that, in the absence of DiaA, P. profundum SS9R has lost the ability to grow optimally at high pressure.
Since the P. profundum SS9 PBPRA3229 gene is the third gene of a four-gene operon (Fig. 2A), there existed the possibility that the m-Tn10 insertion in PBPRA3229 could be exerting a polar effect on the downstream gene PBPRA3230. To confirm that the phenotypes observed for the FL23 mutant were due to disruption of PBPRA3229, pPBPRA3229 was conjugated into FL23 and its growth was investigated. Additionally, to determine whether the E. coli diaA gene could also complement the growth phenotypes of FL3, pECdiaA was also mated into FL23. We determined that the P. profundum SS9 PBPRA3229 and E. coli diaA genes were both able to complement the growth defects of FL23 at 0.1 and 28 MPa at 15°C on the agar overlay plates and restored growth to a level similar to that of the parent strain with the control plasmid (pFL190) (Fig. 4A and B, respectively). Combined, these data show that the growth defects of the FL3 mutant on solid agar are due to disruption of the PBPRA3229 gene and they also demonstrate that the E. coli DiaA protein can functionally compensate for the absence of PBPRA3229 in P. profundum SS9R at both atmospheric pressure and high pressure on solid agar.
When P. profundum SS9R is grown in liquid culture at 0.1 MPa and 15°C, it grows as a mixture of both rod-shaped and filamentous cells (Fig. 4C). However, when grown under optimal growth conditions (28 MPa, 15°C), the culture only contains rod-shaped cells, which are typically 2 to 3 μm in length (Fig. 4D). This finding is consistent with P. profundum SS9 being a high-pressure-adapted bacterium which perceives atmospheric pressure as a stress (26). In contrast, the FL23 mutant exhibited a morphology in liquid culture at both 0.1 and 28 MPa at 15°C (Fig. 4E and F, respectively) that was aberrant relative to that of the parent strain under the same conditions (Fig. 4C and D, respectively). Since FL23 fails to recover its morphology at high pressure, this finding provides further evidence that this mutant is defective in high-pressure-adapted growth. Additionally, FL23 containing pFL190 also showed aberrant morphology at 28 MPa (Fig. 4G) whereas the morphology of the FL23 mutant with either pPBPRA3229 or pECdiaA (Fig. 4H and I, respectively) appeared similar to that of the parent strain with pFL190 at 28 MPa (Fig. 4J). We also determined that pPBPRA3229 and pECdiaA could complement the altered morphology of the FL23 mutant at 45 MPa and 15°C (Fig. 4L and M, respectively). Taken together, these findings provide further evidence that disruption of PBPRA3229 is responsible for the high-pressure growth defect of FL23 and that the E. coli DiaA protein is also functionally interchangeable with the P. profundum SS9 DiaA protein at high pressure in liquid culture.
The P. profundum SS9 PBPRA1039 gene partially complements the DNA replication defect of an E. coli seqA mutant.
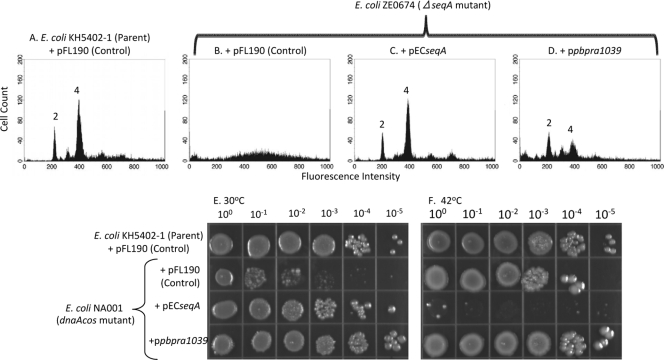
The FL28 mutant was previously isolated from a screen for high-pressure-enhanced transposon mutants of P. profundum SS9R (19). FL28 has a mini-Tn10 insertion in the PBPRA1039 gene, which encodes a product with 55% identity to the E. coli SeqA protein. In contrast to DiaA, E. coli SeqA is a negative regulator of the initiation of DNA replication (20). To investigate whether the P. profundum SS9 PBPRA1039 gene encodes a SeqA homolog, we transduced the seqA::Kan deletion from E. coli JW0674 (Keio collection) (2) into KH5402-1 to create ZE0674. The PBPRA1039 gene was cloned into pFL190 (pPBPRA1039) and transformed into the E. coli ZE0674 ΔseqA::Kan mutant. As a control, the E. coli seqA gene was also cloned into pFL190 (pECseqA) and transformed into the E. coli mutant lacking SeqA. Consistent with previous observations, and in contrast to the parent strain, which showed two defined peaks corresponding to two and four copies of its chromosome, we observed that an E. coli mutant lacking SeqA with the control plasmid pFL190 showed no defined peaks by flow cytometry after incubation in the presence of SYTOX Green (Fig. 5A and B, respectively). We found that pECseqA fully restored chromosome replication in the E. coli ΔseqA::Kan mutant to a level similar to that of the parent strain with pFL190 (Fig. 5C and A, respectively), whereas introducing pPBPRA1039 resulted in a mixed phenotype, showing smaller peaks corresponding to two and four chromosomes, as well as some of the noise signal observed with pFL190 alone (Fig. 5D and B, respectively). Therefore, these findings show that the E. coli seqA gene can complement for the absence of SeqA in E. coli when expressed from pFL190. However, they also provide evidence that the P. profundum SS9 PBPRA1039 gene can restore a degree of synchrony to the initiation of DNA replication defect in E. coli in the absence of SeqA.
FIG. 5.
The P. profundum SS9 PBPRA1039 gene partially complements the DNA replication defect of an E. coli mutant lacking SeqA and suppresses the cold sensitivity phenotype of an E. coli dnaA(Cs) mutant. (A to D) Exponential-phase cells of the strains shown were grown in the presence of streptomycin, thymine, and 0.2% (wt/vol) l-arabinose and fixed, and the chromosome number was determined with SYTOX Green in combination with flow cytometry. The profiles shown are representative of two independent experiments. (E and F) The strains shown were grown overnight in LB medium at 42°C supplemented with streptomycin and thymine but without l-arabinose. Cultures were then diluted; spotted onto LB agar supplemented with thymine, streptomycin, and 0.2% (wt/vol) l-arabinose; and then incubated under the conditions shown for 40 h. The data set shown is representative of at least two independent experiments where similar growth patterns were observed.
It was shown previously that overexpression of E. coli seqA in an E. coli dnaA(Cs) mutant suppressed the cold sensitivity phenotype at 30°C (20). To provide further evidence that the P. profundum SS9 PBPRA1039 gene encodes a SeqA homolog, pECseqA and pPBPRA1039 were transformed into the E. coli dnaA(Cs) mutant and their effect on growth at 30 and 42°C was determined (Fig. 5E and F, respectively). We found that both pECseqA and pPBPRA1039, but not the control vector pFL190, suppressed the cold sensitivity phenotype of the E. coli dnaA(Cs) mutant at 30°C to similar extents (Fig. 5E). Therefore, combined with our flow cytometry analysis, our findings provide evidence that pPBPRA1039 encodes a SeqA homolog. In addition, as determined previously (20), we found that the overexpression of E. coli seqA, but not the P. profundum SS9 PBPRA1039 gene, conferred lethality on the E. coli dnaA(Cs) mutant at 42°C (Fig. 5F). Taken together, these data provide evidence that the P. profundum SS9 PBPRA1039 gene encodes a functional SeqA homolog. They also demonstrate that there are temperature-dependent differences between the plasmid-carried E. coli and P. profundum SS9 seqA genes in E. coli.
The P. profundum SS9 PBPRA1039 gene complements the growth defect of FL28 on solid agar at atmospheric pressure.
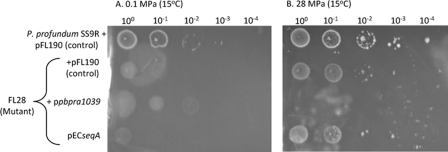
It was shown previously that the FL28 mutant shows high-pressure-enhanced growth at 45 MPa relative to its growth at 0.1 MPa (19). Consistent with this observation, although we found that the growth of FL28 with pFL190 was significantly reduced relative to that of the parent strain with pFL190 at both 0.1 and 28 MPa at 15°C on agar overlay plates (Fig. 6A and B), it grew substantially better after incubation at 28 MPa than at 0.1 MPa at 15°C (Fig. 6B and A), respectively. Transferring pPBPRA1039 into FL28 improved its growth at 0.1 MPa and 15°C on agar overlay plates, but no growth of this strain was observed at 28 MPa (Fig. 6A and B, respectively). This finding showed that the plasmid-carried P. profundum SS9 PBPRA1039 gene complements the growth defect of FL28 at atmospheric pressure but is deleterious to growth at high pressure. In contrast, mating pECseqA with FL28 had no effect on its growth at either 0.1 or 28 MPa at 15°C (Fig. 6A and B, respectively). Additionally, we found that increasing the l-arabinose concentration in the growth medium from 0.01 to 0.2% (wt/vol) had no further effect on the growth of any of the strains at either 0.1 or 28 MPa (data not shown). Thus, the plasmid-carried E. coli seqA gene is unable to complement the growth defects of the FL28 mutant at either atmospheric or high pressure.
FIG. 6.
The P. profundum SS9 PBPRA1039 gene complements the P. profundum FL28 mutant at 0.1 MPa but is lethal at 28 MPa. Cultures of the strains shown were grown in marine broth and streptomycin until early stationary phase, and then 10-μl aliquots were spotted onto low-gelling-temperature marine agar supplemented with 0.01% (wt/vol) l-arabinose. The data sets shown are representative of at least two independent experiments where similar growth patterns were observed. Due to the fragile nature of the overlay plates after incubation at high pressure, the plates could only be photographed in the plastic pouches. The imperfections in the photographs are due to deformations of the plastic pouches.
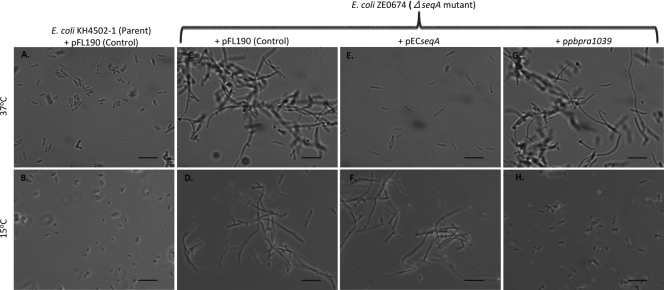
The ability of the plasmid-carried P. profundum SS9 PBPRA1039 and E. coli seqA genes to complement the morphological alteration of the E. coli seqA mutant is dependent upon temperature.
To investigate whether the inability of pECseqA to complement the growth phenotypes of the FL28 mutant at 15°C was due to the temperature, we investigated the effect of temperature on the ability of either pECseqA or pPBPRA1039 to complement an E. coli ΔseqA::Kan mutant. Since it was shown previously that an E. coli seqA mutant forms filaments in LB medium at atmospheric pressure (20), we used this phenotype to assess complementation. We found that, unlike the parent strain, an E. coli ΔseqA::Kan mutant with the control plasmid pFL190 formed filaments at both 37 and 15°C at atmospheric pressure (Fig. 7A to D). Additionally, we found that pECseqA complemented the filamentation defect of the E. coli ΔseqA::Kan mutant at 37°C but not at 15°C (Fig. 7E and F, respectively). Conversely, we determined that pPBPRA1039 complemented the filamentation defect of the E. coli ΔseqA::Kan mutant at 15°C but not at 37°C (Fig. 7H and G, respectively). Combined, these findings show that the ability of the plasmid-carried PBPRA1039 and seqA genes to complement the filamentation phenotype of an E. coli seqA mutant is dependent upon the growth temperature. Additionally, we also found that both the E. coli parent and seqA mutant strains formed filaments at high pressure (28 MPa, 37°C) (data not shown), providing evidence that SeqA does not affect the growth of E. coli at high pressure.
FIG. 7.
The ability of the P. profundum SS9 PBPRA1039 and E. coli seqA genes to complement the morphological defect of an E. coli seqA mutant is dependent upon temperature. Cultures of the strains shown were diluted 1:500 in LB broth with thymine, streptomycin, and 0.01% (wt/vol) l-arabinose and grown to mid-exponential phase (OD600, ∼0.4) at the temperatures shown. The images shown are representative fields of view from at least two independent experiments. Scale bars = 10 μm.
DISCUSSION
Our data clearly show that the gene disrupted in the high-pressure-sensitive P. profundum SS9R FL23 mutant, PBPRA3229, encodes a protein which is functionally interchangeable with the E. coli DiaA protein. We found that DiaA is important for the growth of P. profundum SS9R at atmospheric pressure on solid agar but also plays a key role in adaptation to high pressure under these conditions. Additionally, consistent with the previously reported growth defect of the P. profundum SS9R diaA mutant at high pressure in liquid culture (19), we found that DiaA is essential for P. profundum SS9R to have a rod-shaped, uniform morphology under these conditions. In contrast, despite showing an asynchronous DNA replication phenotype (14), the E. coli diaA::Tn5 mutant did not show altered growth relative to that of the parent strain at either atmospheric or high pressure. The differences in the behavior of the E. coli and P. profundum SS9R diaA mutants is unlikely due to differences in the function of the DiaA proteins per se in these two bacterial species, since we found that the E. coli and P. profundum SS9 DiaA proteins were functionally interchangeable at both atmospheric pressure and high pressure. Additionally, since E. coli is a mesophile, they also suggest that the P. profundum SS9 DiaA protein is not specifically adapted over the E. coli DiaA protein to function at high pressure. Instead, the importance of DiaA for the growth of P. profundum SS9 but not for E. coli growth is likely due to other differences in the DNA replication machinery of these two bacterial species.
Unlike the genome of E. coli, which has one chromosome, that of P. profundum SS9 is similar to those of other members of the family Vibrionaceae and consists of two chromosomes (26). In Vibrio cholerae, the replication of both chromosomes occurs through two different mechanisms (11). For example, V. cholerae chromosome I replication is dependent upon DnaA and is thought to use a mechanism similar to that used by E. coli (11). In contrast, the replication of V. cholerae chromosome II involves a DnaA-independent mechanism (11). However, although these replication processes involve different mechanisms, the replication processes of both chromosomes are synchronous and this is essential to ensure cell survival (10). Therefore, the growth defects we observed for the P. profundum SS9R mutant lacking DiaA could be due to alterations in DnaA-dependent chromosome I replication, leading to a loss of synchrony between chromosome I replication and chromosome II replication. Such a model could rationalize why loss of DiaA in E. coli, which only has one chromosome, would not affect growth.
Since the growth phenotypes of the P. profundum SS9R mutant lacking DiaA were exacerbated at high pressure and previous microarray studies determined that the transcription of the P. profundum SS9 diaA gene did not appear to be regulated in response to pressure changes (5, 26), high pressure may exert a greater inhibitory effect on the initiation of DNA replication in P. profundum SS9R and the presence of DiaA may be essential to overcome this block. However, due to technical difficulties in measuring chromosome number, we were unfortunately unable to determine the effect of DiaA on the initiation of chromosome replication in P. profundum SS9R. As mentioned earlier, this could be due to the fact that P. profundum SS9R has two different chromosomes and exhibits a mixture of morphologies at atmospheric pressure. Further studies are necessary to develop novel methodologies to investigate the effect of pressure changes and DiaA on the initiation of chromosome replication in P. profundum SS9.
Interestingly, consistent with our proposed model, another P. profundum SS9R mutant (FL31) isolated from the high-pressure sensitivity screen was determined to have a transposon insertion in a putative rctB gene (19). In V. cholerae, RctB is an essential protein involved in the initiation of chromosome II replication (11). Since the transposon insertion in FL31 is inserted near the 3′ end of the gene, it was proposed that this could give rise to a partially functional RctB protein (19). Although, further work is necessary to determine whether the gene disrupted in FL31 encodes an RctB homolog, this finding provides further support for our model whereby disruption of the replication of one chromosome, this time chromosome II, could affect the ability of P. profundum SS9R to grow optimally at high pressure.
In addition to our findings on DiaA, we also found that the gene disrupted in a high-pressure-enhanced transposon mutant of P. profundum SS9R (FL28) (19) appears to encode a SeqA homolog. We found that the P. profundum SS9 putative seqA gene (PBPRA1039) could suppress the cold sensitivity phenotype of an E. coli dnaA(Cs) mutant and could restore a degree of synchrony to the initiation of DNA replication in an E. coli mutant lacking SeqA. Although we found that the P. profundum SS9R seqA mutant (FL28) had a growth defect at atmospheric pressure on solid agar relative to the parent strain, we found, consistent with previous findings in liquid culture (19), that its growth was greatly improved at high pressure under these conditions. Since SeqA is a negative regulator of the initiation of DNA replication and prevents the binding of DnaA-ATP to oriC in E. coli (20), these findings suggest that the loss of a negative regulator of the initiation aids the growth of P. profundum SS9R at high pressure. Therefore, this adds further support for a model whereby the initiation of chromosome replication in P. profundum SS9R is susceptible to high pressure and either the presence of a positive regulator (DiaA) or the absence of a negative regulator (SeqA) is necessary to enable optimum growth under these conditions. However, as with DiaA, further studies are necessary to determine the precise role of SeqA in regulating the initiation of chromosome replication in P. profundum SS9R at atmospheric pressure and high pressure.
It should be noted that although the plasmid-carried P. profundum SS9R seqA gene could complement the growth defect of the P. profundum SS9R seqA mutant (FL28) at atmospheric pressure, no growth of this strain was observed at high pressure. Consistent with our model, it is interesting to speculate that the overexpression of the P. profundum SS9 SeqA protein, which is a negative regulator of the initiation of DNA replication, may be lethal to P. profundum SS9 at high pressure.
Although we did observe some functional complementation of the E. coli seqA mutant at 15 and 30°C by the plasmid-carried P. profundum SS9 seqA gene, unlike the plasmid-carried E. coli seqA gene, there was no effect on the growth of the E. coli dnaA(Cs) mutant at 42°C. This could be due to differences in amount and/or activity between the P. profundum SS9 SeqA and E. coli SeqA proteins within E. coli. Since P. profundum SS9 is a cold-loving bacterium which can only grow at up to 20°C (8), it is possible that the P. profundum SS9 SeqA protein is heat labile and/or has reduced activity compared to the E. coli SeqA protein at 42°C. However, further studies are necessary to investigate this possibility.
Additionally, we also found that, unlike the plasmid-carried P. profundum SS9 seqA gene, the plasmid-carried E. coli seqA gene was unable to complement the growth defects of the P. profundum SS9 seqA mutant at 15°C. The reason for this difference is unknown, but it could be due to differences in the amounts and/or activity of the P. profundum SS9 and E. coli SeqA proteins within P. profundum SS9. Since the same plasmid-carried E. coli seqA gene was able to prevent filamentation of an E. coli seqA deletion mutant at 37°C but not at 15°C, this provides evidence that the inability of the plasmid-carried E. coli seqA gene to complement the P. profundum SS9 seqA mutant is likely due to the growth temperature. The temperature effect appears to be a feature of the plasmid-carried E. coli seqA gene, since the E. coli parent strain did not form filaments at 15°C, suggesting that the chromosome-carried seqA gene produces a functional SeqA protein at this temperature. However, further studies are necessary to determine whether the filamentation phenotypes observed are due to reduced/elevated levels or reduced/elevated activity of the different SeqA proteins at different temperatures.
In conclusion, these findings provide evidence that high pressure exerts an effect at the level of the initiation of DNA replication and that proteins that regulate this process play a key role in the high-pressure-adapted growth of P. profundum SS9. Based on our findings on DiaA, they also suggest that for P. profundum SS9 to grow optimally at 28 MPa, its individual proteins do not all need to be more high pressure adapted than their counterparts in nonpiezophilic bacterial species.
Acknowledgments
This research was supported by a NERC New Investigator Grant (NE/D000203/1) and a Royal Society Grant awarded to G.P.F. BBSRC (BB/D000564/1) and MRC New Investigator (G0501107) grants awarded to G.P.F. also supported this work. The Ph.D. studentships of Z.H. and D.A. were funded by the Darwin and Leverhulme Trusts, respectively. While at the University of Edinburgh, the lectureship of G.P.F. was funded by the Leverhulme Trust, and start-up grants at the Universities of Edinburgh and Aberdeen also helped support this project. D.H.B. gratefully acknowledges funding from the National Science Foundation (Microbial Interactions and Processes NSF0801793 and Microbial Genome Sequencing Program NSF0827051).
We thank Miguel Valvano for the E. coli gmhA mutant and Tsutomu Katayama for the E. coli diaA and dnaA(Cs) mutants. Thanks also to Andrew Hall for the use of his pressure vessels, to the ocean laboratory at the University of Aberdeen for donating pressure vessels and equipment, and to Linda Duncan for help with the flow cytometry.
Footnotes
Published ahead of print on 21 August 2009.
REFERENCES
- 1.Allen, E. E., D. Facciotti, and D. H. Bartlett. 1999. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 65:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 21 February 2006, posting date. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, D. H., G. P. Ferguson, and G. Valle (ed.). 2008. Adaptations of the psychrotolerant piezophile Photobacterium profundum strain SS9, p. 319-337. In C. Michiels, D. H. Bartlett, and A. Aertsen (ed.), High-pressure microbiology. ASM Press Washington, DC.
- 4.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 5.Campanaro, S., A. Vezzi, N. Vitulo, F. M. Lauro, M. D'Angelo, F. Simonato, A. Cestaro, G. Malacrida, G. Bertoloni, G. Valle, and D. H. Bartlett. 2005. Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics 6:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, G. R. O., L. A. Sharypova, H. Scheidle, K. M. Jones, K. Niehaus, A. Becker, and G. C. Walker. 2003. Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J. Bacteriol. 185:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, E., and D. H. Bartlett. 1993. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain SS9. J. Bacteriol. 175:7533-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong, E. F. 1986. Adaptations of deep-sea bacteria to the abyssal environment. Ph.D. dissertation. University of California, San Diego.
- 9.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X.-W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 10.Duigou, S., K. G. Knudsen, O. Skovgaard, E. S. Egan, A. Lobner-Olesen, and M. K. Waldor. 2006. Independent control of replication initiation of the two Vibrio cholerae chromosomes by DnaA and RctB. J. Bacteriol. 188:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan, E. S., and M. K. Waldor. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521-530. [DOI] [PubMed] [Google Scholar]
- 12.Finan, T. M., B. Kunkel, G. F. de Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Ishida, T., N. Akimitsu, T. Kashioka, M. Hatano, T. Kubota, Y. Ogata, K. Sekimizu, and T. Katayama. 2004. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 279:45546-45555. [DOI] [PubMed] [Google Scholar]
- 15.Katayama, T. 1994. The mutant DnaAcos protein which overinitiates replication of the Escherichia coli chromosome is inert to negative regulation for initiation. J. Biol. Chem. 269:22075-22079. [PubMed] [Google Scholar]
- 16.Kellenberger-Gujer, G., A. J. Podhajska, and L. Caro. 1978. A cold sensitive dnaA mutant of E. coli which overinitiates chromosome replication at low temperature. Mol. Gen. Genet. 162:9-16. [DOI] [PubMed] [Google Scholar]
- 17.Keyamura, K., N. Fujikawa, T. Ishida, S. Ozaki, M. Su'etsugu, K. Fujimitsu, W. Kagawa, S. Yokoyama, H. Kurumizaka, and T. Katayama. 2007. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21:2083-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauro, F. M., E. A. Eloe, N. Liverani, G. Bertoloni, and D. H. Bartlett. 2005. Conjugal vectors for cloning, expression, and insertional mutagenesis in gram-negative bacteria. BioTechniques 38:708-712. [DOI] [PubMed] [Google Scholar]
- 19.Lauro, F. M., K. Tran, A. Vezzi, N. Vitulo, G. Valle, and D. H. Bartlett. 2008. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J. Bacteriol. 190:1699-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 21.Masui, N., and C. Kato. 1999. New method of screening for pressure-sensitive mutants at high hydrostatic pressure. Biosci. Biotechnol. Biochem. 63:235-237. [DOI] [PubMed] [Google Scholar]
- 22.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the -l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 23.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1982. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Stevenson, G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. Reeves. 1994. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]