Abstract
Our views of the genes that drive phenotypes have generally been built up one locus or operon at a time. However, a given phenotype, such as virulence, is a multilocus phenomenon. To gain a more comprehensive view of the genes and interactions underlying a phenotype, we propose an approach that incorporates information from comparative genomics and network biology and illustrate it by examining the virulence phenotype of Vibrio cholerae O1 El Tor N16961. We assessed the associations among the virulence-associated proteins from Vibrio cholerae and all the other proteins from this bacterium using a functional-association network map. In the context of this map, we were able to identify 262 proteins that are functionally linked to the virulence-associated genes more closely than is typical of the proteins in this strain and 240 proteins that are functionally linked to the virulence-associated proteins with a confidence score greater than 0.9. The roles of these genes were investigated using functional information from online data sources, comparative genomics, and the relationships shown by the protein association map. We also incorporated core proteome data from the family Vibrionaceae; 35% of the virulence-associated proteins have orthologs among the 1,822 orthologous groups of proteins in the core proteome, indicating that they may be dual-role virulence genes or encode functions that have value outside the human host. This approach is a valuable tool in searching for novel functional associations and in investigating the relationship between genotype and phenotype.
The advent of high-throughput approaches to biology has forced us to rethink the way we parse the components that make up an organism, leading us away from the perceived primacy of the gene and its encoded product to a new view that encompasses how the gene product interacts with other gene products in a given set of circumstances (19). One example of how this new viewpoint is changing our understanding is found in the field of pathogenesis, specifically, in how we understand virulence (47). Virulence has been defined as the ability of a pathogen to damage a host. Virulence is mediated by virulence factors, the means by which a pathogen establishes and maintains an infection and by which it ensures its transmission to another host. Virulence factors have been classified as adhesins, invasins, impedins, aggresins, and modulins, but these factors are rarely the products of a single locus in the pathogen—groups of interacting loci are responsible for the activities of the virulence factors. Virulence can be thought of as an emergent property of the multiple interactions that manifest as a phenotype, and this implies that any attempt to define a virulence factor must take into account these interactions, or network properties, of virulence. Hence, definitions of individual loci as virulence factors have moved away from one-gene-one-factor definitions, and loci implicated in virulence are now classified in a way that attempts to reflect their roles. Wassenaar and Gaastra (71), for example, conceptualize three tiers of virulence factors. The top-level virulence factors are termed true virulence factors and include aggressins, like the bacterial toxins. The second-level virulence factors are termed virulence-associated factors and include supporting factors, such as invasins, that are required for the activities of the true virulence factors. Finally, there are factors that, while required for the establishment and maintenance of the pathogen in the host, are not exclusively expressed in that environment. They are termed virulence lifestyle genes and include, for example, adhesins, like fimbriae. As the repertoire of loci implicated in virulence grows and becomes more nuanced, the identification of all the loci involved in the manifestation of the phenotype becomes an important task. Because virulence is a multilocus phenomenon, not all the loci that work together to manifest a “virulence factor” may be recognized, despite their essential roles in virulence. Methods for detecting and identifying the loci in virulence systems are required, especially methods that can account for the uncertainties involved in identifying members of these systems. Here, we present an approach to identifying potential members of virulence systems by using comparative genomics and functional-association networks, as applied to the environmental pathogen Vibrio cholerae.
V. cholerae is widely known as the causative agent of cholera, but of the roughly 200 serotypes that are ubiquitously distributed in the world's oceans, only two, O1 and O139, have been consistently linked to epidemic cholera (15). Surveys of V. cholerae strains found in the environment show that the toxigenic strains comprise a small fraction (0.8%) of the V. cholerae strains that can be detected (16). Cholera is thought to kill over 100,000 people every year, although, due to the economic penalties imposed on countries where a cholera epidemic occurs, the disease is probably underreported. V. cholerae can be found in radically different environments, including the pelagic oceans, the human intestinal tract, biofilms adhering to plankton and shellfish, and the trophozoites of amoebas (1, 62). Movement among these niches demands a high degree of physiological flexibility.
Toxigenic strains of V. cholerae have two main virulence factors: the cholera toxin, encoded by the CTX loci that are found on a prophage, CTXphi, and the toxin-coregulated pilus (TCP), found on a pathogenicity island, VPI-1. Other virulence determinants are found on a second pathogenicity island, VPI-2; on the mannose-sensitive hemagglutination pilus loci; on the RTX toxin cluster; and on the RS1phi prophage. V. cholerae El Tor strains also carry two unique genomic islands termed the Vibrio seventh pandemic islands I and II (46). Many V. cholerae isolates in the aquatic environment carry some part of the virulence-related gene complement of the O1 and O139 serotypes; Rahman et al. found that 3.9% of non-O1 non-O139 isolates from surface waters in the Dhaka district of Bangladesh carried both the CTX genes and the TCP genes. A further 3.9% carried one or the other of these so-called major virulence determinants, and all of these strains carried at least one virulence-associated gene or group of genes (50). Such a pool of strains in the environment makes the mixing and matching of virulence-related loci from V. cholerae possible in the environment (17). In order to better understand virulence, it is necessary to complete our list of virulence-related genes and to address how they might interact with each other.
Comparative genomics has been used to identify virulence factors in newly sequenced genomes from pathogens, and this approach can also be used to aid in the classification of the identified virulence factors. The set of genomes available for the Vibrionaceae includes genomes from two nonpathogenic species of Vibrionaceae, Aliivibrio (Vibrio) fischeri and Photobacterium profundum, as well as genomes from strains that have hosts and modes of pathogenicity that differ from those of V. cholerae. By establishing the set of genes found in all the genomes of all the members of the family Vibrionaceae (the pangenome) and the subset of genes shared by each member of the family (known as the core genome), we can identify virulence-related genes that are shared among pathogenic and nonpathogenic strains. The proteins encoded by these shared genes (which we refer to here as the core proteome) can be tentatively classified as virulence lifestyle-related proteins that, in V. cholerae, support pathogenicity, while V. cholerae virulence-associated proteins that are encoded by the genes in the pangenome (which we refer to here as the panproteome) may be more directly involved in virulence. Not all virulence lifestyle proteins found in V. cholerae are found in the core proteome, but the establishment of the core proteome and panproteome still serves as a significant filter for identifying true virulence proteins.
As more and more high-throughput data, derived from genomic, transcriptome, and proteomic investigations, accumulate, it has become possible to infer the interactions among the proteins in a bacterium and to build, for example, functional-association networks (27). When tested against sets of known interacting proteins, these networks have proven to perform very well in identifying novel associations among proteins (31, 51), and this reliability has increased as the amount of data used to build the networks has increased. The online service Search Tool for the Retrieval of Interacting Genes/Proteins (the STRING database) uses a standard format for representing the network of associations: nodes represent proteins, and edges, the lines between the nodes, represent functional associations. The number of edges connecting a node to other nodes is known as the degree of that node. Nodes of high degree also tend to be essential proteins (6, 28, 76). This correlation makes sense intuitively, and an analysis of protein-protein interaction networks in Saccharomyces cerevisiae showed that the correlation is due to the tendency of essential proteins to form densely connected subnetworks with proteins that are functionally involved in the same biological process (77). By analyzing a functional-association network for V. cholerae, it ought to be possible to tease out more information on recognized virulence proteins and to identify other proteins that may be important in virulence. Using the virulence proteins as in silico bait proteins, we can extract the subset of proteins linked to them, a set of proteins that will be enriched for those with unrecognized roles in virulence. Examination of this subnetwork can teach us more about how pathogenesis is manifested in V. cholerae, as well as about the proteins responsible.
MATERIALS AND METHODS
Sequence data and annotation.
Genome sequences and primary annotations for V. cholerae O1 biovar El Tor strain N16961 (21) and O1 biovar classical strain O395 (NC_009456 and NC_009457), Vibrio parahaemolyticus RIMD 2210633 (43), Vibrio vulnificus CMCP6 (32), V. vulnificus YJ016 (9), Vibrio harveyi ATCC BAA-1116 (NC_009777, NC_009783, and NC_009784), Vibrio splendidus LGP32 (NC_011744 and NC_011753), A. (Vibrio) fischeri ES114 (54), A. fischeri MJ11 (NC_011184, NC_011185, and NC_011186), A. salmonicida LFI1238 (22), and P. profundum SS9 (68) were from the GenBank and J. Craig Venter Institute's Comprehensive Microbial Resource (48). Additional annotation, primarily information about other identifiers associated with each locus, was retrieved from the Database for Annotation, Visualization, and Integrated Discovery, a comprehensive bioinformatics resource (24). The expanded list of identifiers made getting comprehensive information easier. We obtained information about the enzymatic activities encoded by V. cholerae from the Braunschweig Enzyme Database (5), which specializes in comprehensive annotation of enzymes and has a more complete set of links between enzyme commission numbers, the associated functional information, and the loci of V. cholerae than other sources. Enzyme commission numbers serve as links to metabolic-pathway information in MetaCyc (8). General information about protein function and about the domains of encoded proteins was obtained from UniProt (67) and, via UniProt, from Interpro (45). Gene ontology annotation, used in functional-enrichment analyses, was also obtained from UniProt. Information on signal transduction proteins was found at the Microbial Signal Transduction database (66).
Assembly of a list of known virulence-related proteins.
We initially used a list of 165 V. cholerae virulence-related proteins from the Virulence Factor Database (VFDB) (http://zdsys.chgb.org.cn/VFs/) (74). The first results using this set of genes and the network-based method included many proteins that had already been recognized as virulence related. The literature about these first hits led us to expand our list of virulence proteins, and we realized our list would have to be more comprehensive if we hoped to identify novel virulence-related proteins. We therefore made a more systematic search for papers containing information about V. cholerae and virulence using PubMed and GoPubMed (14). We also looked for more database sources and were directed to the National Microbial Pathogen Data Resource (NMPDR) (http://www.nmpdr.org/FIG/wiki/view.cgi) (44), which annotates the genome of V. cholerae and other pathogens in a uniform way that includes tagging virulence proteins. The addition of the NMPDR list to the results of our literature search and the VFDB list gave a total of 525 proteins. Our literature sources are shown in the supplemental material. The list of 525 proteins is shown in Table S1 in the supplemental material, and the proteins are linked to the supplemental references.
Distribution of virulence-related proteins.
In order to supply an evolutionary context and as an aid to a preliminary classification of the virulence-related proteins, we defined their distribution among the 11 strains of Vibrionaceae. We used OrthoMCL (37) to detect and group the orthologous proteins in the 11 strains of Vibrionaceae. The program builds the list of orthologs by doing an all-against-all blastp search. The orthologs are clustered using the Markov cluster algorithm, working off a matrix of corrected P values. From the results, we were able to identify two set of proteins: those that were encoded by all 11 strains, that is, the core proteome of the Vibrionaceae, and those that were encoded by fewer than 11 strains, that is, the panproteome. A hierarchical functional classification of the proteins that fell into OrthoMCL groups was performed by searching against the Clusters of Orthologous Groups (COG) database (61).
Protein association data.
The V. cholerae functional-association data were obtained from the STRING database version 7.1 (27). The associations among the proteins in the data set were visualized using Cytoscape 2.6 (57). Statistics on the connectivity in the network were calculated using the NetworkAnalyzer plug-in for Cytoscape 2.6 (4). Gene ontology term enrichment of subsets of proteins was estimated with the BiNGO plug-in (42) for Cytoscape, using the hypergeometric test and the Benjamini and Hochberg false discovery rate correction, with a selected significance level of 0.05. Functional-association candidates were assessed using information from the Database for Annotation, Visualization, and Integrated Discovery; the NMPDR; the UCSC Archaeal Genome browser (56); expression data; and other information from the published literature.
RESULTS AND DISCUSSION
We first identified and classified a list of virulence-related proteins from V. cholerae by database and literature searches and by establishing the core proteome and panproteome for the family Vibrionaceae. We then mapped these virulence proteins onto a functional-association network based on data from STRING v7.1. Statistical methods were used to establish connections between the proposed virulence-related proteins and other proteins in V. cholerae in order to identify novel candidate virulence proteins and the systems in which they were found.
Virulence-related proteins in V. cholerae.
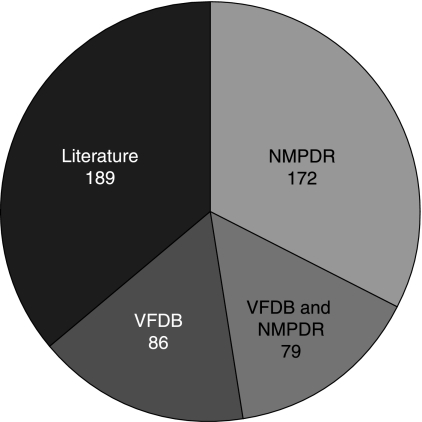
In identifying known virulence-related proteins in V. cholerae, we relied on literature searches and on two databases that list virulence proteins from V. cholerae O1 El Tor N16961, the VFDB (74) and the NMPDR (44). Together, these two databases contained 337 virulence-related proteins; only 79 of these proteins were found in both databases. We added a further 189 proteins to this list after extensive literature searching (Fig. 1). In carrying out the literature search, we looked for any protein or gene that was linked to virulence, including the level 3 loci called virulence lifestyle genes. Thus, our search results included any proteins that were known to enhance the ability of the bacterium to invade and colonize the gastrointestinal tract or to express true virulence factors. Our list also contains a number of hypothetical or uncharacterized proteins that were included, for example, because they were part of a pathogenicity island. We feel justified in using a liberal definition of a virulence-related protein for two reasons. First, the proposed virulence protein is to be viewed in the context of other proteins in the cell, as this will shed light on the function of the protein and aid in judging whether it really is a virulence protein. Second, inclusiveness is important in that even tenuous connections can point us to other, unrecognized virulence proteins and help to fill out the component lists of virulence-related subsystems. The proteins are listed in Table S1 in the supplemental material.
FIG. 1.
Visualization of the sources of the virulence-related proteins in our list.
Classifying the virulence-related proteins.
Our comparison of the gene complement of 11 strains from the Vibrionaceae established a core proteome composed of 1,882 proteins. As expected, this is significantly smaller than the single-species core genome of 2,741 genes established for V. cholerae N16961 by Keymer et al. (30). However, for a family level core proteome, it is remarkably large, even relative to genus-level core genomes for other taxa. In the genus Streptococcus, for example, Lefébure and Stanhope estimated the core genome to be 611 genes (36).
Roughly 49% of all the proteins in V. cholerae are part of the core proteome. Just over one-third (35%) of the V. cholerae N16961 proteins that have been classified as virulence or virulence-associated proteins are also found in the core proteome, that is, they are found in the avirulent strains and in strains that have other modes of infection. Thus, while it is probable that they carry out functions that are required for the establishment of an infection, these virulence-related proteins are not expressed solely in the human host environment. Indeed, it is known that certain virulence proteins can confer an advantage on V. cholerae outside the host, and these have been termed dual-role colonization factors (53, 69). For example, the TCP appears to play a role in the colonization of chitinous surfaces in the environment, as well as in colonization of the human gut (52), and a chitin binding protein, needed for the colonization of copepods, shrimp, and other chitinous surfaces in the environment, also aids in the colonization of epithelial cells (33). Thus, it is quite possible that some genes that are found in the core proteome and appear to be virulence lifestyle genes may have unrecognized roles as virulence-associated genes or even encode true virulence factors.
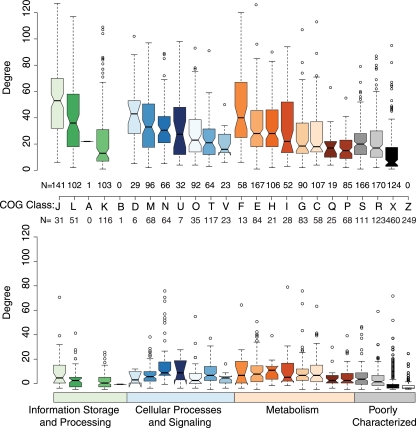
We classified the orthologous proteins according to their relationships with the COGs at NCBI. The size of each of the functional groups found in the V. cholerae N16961 proteome is shown in Fig. 2 (top), along with its degree distribution. The noncore proteins (bottom) (classified by NCBI) and the proteins that we were unable to place in a COG group (class X) have lower connectivity within the network. Proteins in COG classes J (translation, ribosomal structure, and biogenesis) and D (cell cycle control, cell division, and chromosome partitioning) had the highest mean degrees, significantly higher than those of other groups. They were followed by F (nucleotide transport and metabolism) and L (replication, recombination, and repair). This is not unexpected, given that many of these proteins are essential to the survival of the cell. The proteins in COG classes P (inorganic-ion transport and metabolism) and K (transcription) had the lowest mean degrees. This might be explained by the differences in how these sets of proteins interact with other proteins. They do not form large complexes, like the ribosome; indeed, many may be parts of specialized metabolic pathways in which they interact with a few other proteins via common substrates.
FIG. 2.
Box plots summarizing the degrees of proteins in the functional-association data set for V. cholerae N16961 as a function of their COG classes, when only associations where S is ≥0.4 are considered. (Top) Subset of proteins that are part of the core proteome. (Bottom) Subset of proteins that are not part of the core proteome. The proteins in the bottom panel were classified by NCBI. Each color and letter represents a COG class, with the exception of X (proteins that were members of an OrthoMCL group but that were not placed into a COG class) and Z (proteins that were not members of an orthologous group and could not be placed into a COG class). The plots show the range of degree values for each group of proteins; the median value is represented by the bar at the center of the notch.
The functional-association network.
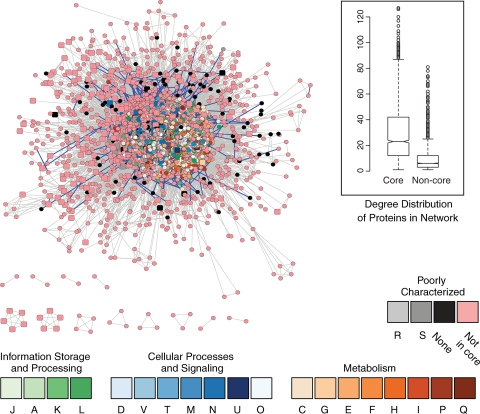
Figure 3 shows a functional-association network for V. cholerae N16961, built using data from the STRING database (27). The network includes 3,756 proteins and 159,497 associations; each association between a pair of proteins has a confidence score (S) ranging from 0.15 to 0.999 that was inferred from the evidence used to establish the association. The noncore proteins in V. cholerae and the other nodes, which are members of the core protein set for 11 strains from the family Vibrionaceae, are shown in Fig. 3. Of the 525 virulence-related proteins in our database, 3 were not connected to any other proteins in the STRING database (S ≥ 0.15) and 2 more were not connected to any other protein (S ≥ 0.4).
FIG. 3.
Visualization of the functional-association network of V. cholerae N16961 based on data from the STRING database. The nodes that are part of the core proteome are colored according to their COG classifications, as shown below the network. Nodes that are not part of the core proteome are colored pink. The square nodes are virulence-associated proteins. There are over 159,000 associations proposed among the 3,756 proteins in the map, and those associations with an S of <0.9 (from 0.15 to 0.899) are shown in gray. The heavy blue edges represent associations with an S of ≥0.9. The inset shows the distribution of degrees (connectivities) for the core and noncore proteins as box plots. The plots show the range of degree values for each group of proteins; the median value is represented by the bar at the center of the notch.
The appearance of noncore loci at the periphery of the functional association network, shown in Fig. 3, indicates that on average the noncore proteins are less highly connected than the core proteins. When we looked at the degree distribution for these two groups, this indeed proved to be the case; the core proteins had, on average, a higher degree of connectivity within the network, and there is good statistical support for the observed difference (Fig. 3, inset). The core protein set includes many proteins essential to the viability of the cell, proteins involved in central metabolism, translation, transcription, and so on. The difference in connectivity between the two groups cannot be attributed to differences in depth of annotation, as even classifiable noncore proteins show lower connectivity (Fig. 2, bottom); furthermore, 715 (38%) of the core proteins found in V. cholerae N16961 are annotated in GenBank as hypothetical, putative, or probable. We assert that the lower degree of association observed in the noncore proteins is due, at least in part, to the peripheral roles some of them play in the cell.
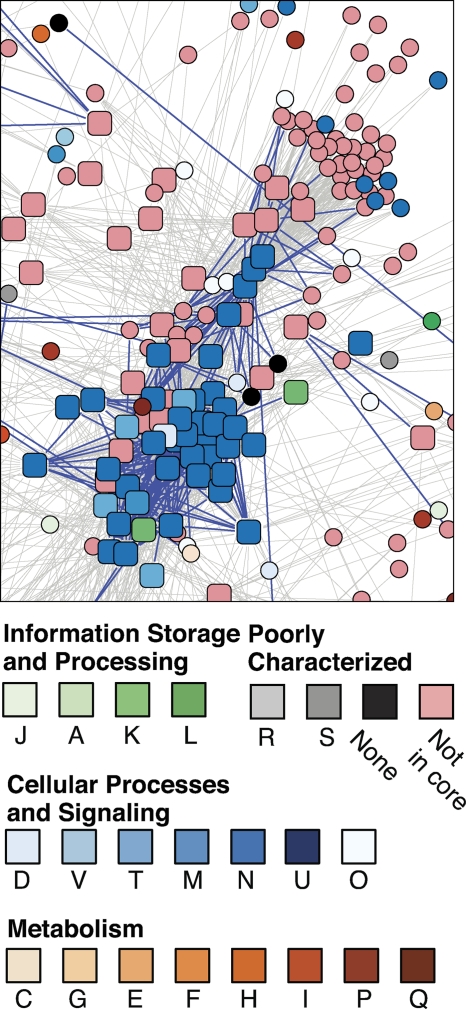
Three subsets of the association data set were extracted using scripts: (i) the set of proteins that are associated with an S of ≥0.4, which included 3,734 proteins and 36,769 associations (99% of the proteins and 23% of the edges in the main data set); (ii) the set of proteins that are associated with at least one virulence-related protein at any confidence score (3,193 proteins [85%] and 33,914 associations [21%]), which contained 523 virulence-related proteins; and (iii) the set of proteins that are associated with at least one virulence related protein at an S of ≥0.4 (2,220 proteins [59%] and 7,183 associations [5%]), which contained 521 virulence proteins. Use of the third set eliminated the edges among the non-virulence-related proteins, which made it easier to clarify which non-virulence-related proteins were associated with more than one virulence-related protein. Figure S1 in the supplemental material shows this virulence association subnetwork. There are 1,699 other V. cholerae proteins linked to the virulence proteins in this subset, and of these, 66% are in the core protein set. Several clearly defined clusters can be seen in this representation (see Fig. S1 in the supplemental material). The most highly populated cluster is primarily made up of chemotaxis- and motility-related proteins. This subnetwork is shown in Fig. 4. Since motility is an important factor for successful colonization of the intestine, nearly all the flagellar components and control elements are shown as virulence proteins. Of course, the flagellar proteins are also required for survival outside the human host. A recent report by Liu et al. indicated that the flagellar protein encoded by flgM has a more direct role in virulence; the flagella are shed when the bacterium invades the intestinal mucosa, and the cell detects the FlgM proteins, which initiates a regulatory chain that derepresses virulence gene expression (40). The chemotaxis receptor/transducer proteins are also part of this cluster, but as discussed below, their roles are not always related to chemotaxis. Many of these receptor/transducer proteins are not members of the core protein set and are not associated with the motility operon, perhaps reflecting the diverse requirements for such proteins for bacteria growing in different environments (see below).
FIG. 4.
Motility/chemotaxis subnetwork of associations (S ≥ 0.4) from Fig. 3 that involves some of the 525 virulence proteins in V. cholerae. The symbol shapes are as in Fig. 3. This cluster contains many MCPs that are not recognized as virulence-related proteins but that are associated with the nonmotility che operons, which have been linked to virulence in V. cholerae.
The associations illustrated in Fig. 3 and 4 can provide information on cellular systems involved in virulence. Furthermore, if we consider that the function of a protein has been observed to be related to the functions of the proteins with which it interacts, it should be possible to identify previously unrecognized virulence-related proteins by analyzing the associations included in Fig. 3 (and simplified in Fig. 4). Here, we discuss how association analysis can be used to narrow the search area for new virulence determinants and to help understand the roles of the implicated gene products in the cell.
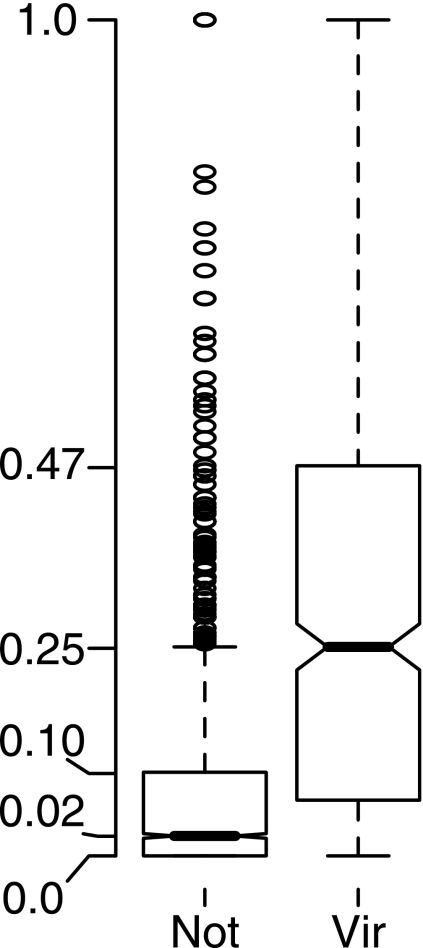
Connecting candidates to virulence-associated proteins. (i) Disproportionately connected proteins.
We first discuss the category of non-virulence-related proteins that, while not necessarily associated with any one virulence-related protein at a high confidence level, have a disproportionately large number of associations with virulence-related proteins. We calculated the ratio of associations with virulence-related proteins to associations with non-virulence-related proteins for each protein in the proteome. Figure 5 shows box plots of the ratios for the virulence-associated proteins in our database and for the non-virulence-associated proteins. Less than 8% of the non-virulence-associated proteins had a proportion of their associations with virulence-associated proteins that was greater than or equal to the median value for the virulence-associated proteins (0.25). These are the outliers in Fig. 5. It is reasonable to characterize these proteins as disproportionately connected, relative to the bulk of the proteins, and to propose that this disproportionately connected set contains proteins that play roles in the virulence of V. cholerae. There are 240 proteins in the set, and they constitute about 14% of the proteins that interact directly with the virulence-related proteins (see Fig. S1 in the supplemental material); 62 of them (26%) are core proteome proteins. These disproportionately connected proteins are listed in Table S2 in the supplemental material.
FIG. 5.
Box plot of the proportion of connections from each protein that are to virulence-associated proteins for two sets of proteins: Vir, the virulence-associated proteins, and Not, the proteins that are not identified as virulence associated. The plots show the range of proportions for each set of proteins; the median value is represented by the bar at the center of the notch.
Of the disproportionately connected proteins, 141 have a functional assignment from the genome annotation and 123 have gene ontology (GO) terms relating to biological processes associated with them. In all, 50 of the proteins have no functional information whatsoever linked to them.
The 123 disproportionately connected proteins that have GO biological-process terms associated with them are significantly enriched for the term “signal transduction.” The expression of virulence-related genes is driven by environmental cues, and these signals must be transmitted to the cell if it is to flourish in a new environment. Thirty-seven of these proteins are annotated as methyl-accepting chemotaxis proteins (MCPs). There are 45 such proteins in V. cholerae (compared to 5 in Escherichia coli) (66), and 8 of them are in our list of virulence-related proteins. The paradigmatic role of these chemoreceptors is in chemotaxis and motility, where they act in concert with the che genes to control bacterial movement toward or away from concentrations of extracellular molecules (7). E. coli has only one set of che genes, and they are essential for chemotaxis. In V. cholerae, three che operons are found, but only one of them is essential for chemotaxis (70). The motility-related operon is located in the region of cheY and cheZ, loci that encode the CheY (VC2065) and CheZ (VC2064) proteins. There is only one CheZ homolog in V. cholerae, and although there are four homologs of the soluble response regulator CheY in the V. cholerae genome, VC2065 is the only CheY homolog with a convincing FliM-binding motif, indicating that it is the only CheY homolog involved in chemotaxis. This implies that the other two operons, with which the bulk of the MCPs are associated, mediate bacterial responses other than chemotaxis (25). Figure S2 in the supplemental material shows the associations among all the MCPs and the various CheA, -W, -Z, and -Y proteins. Only 3 of the 45 MCPs in V. cholerae N16961 are associated with Che proteins from the motility operon. Two of these three MCPs (VC0098 and VCA1092) are classified as 36H MCPs, similar to the E. coli chemotaxis/motility MCPs (66). In fact, these are the only two 36H MCPs in the V. cholerae genome, and both carry the C-terminal pentapeptide motif, unique to the Proteobacteria, which is thought to aid in binding of the CheB and CheR proteins, two proteins that play key roles in the chemotactic adaptation response (3, 72). The third associated MCP, VCA1088, remains unclassified. The chromosomal locations and structural classifications of the MCPs in E. coli and V. cholerae are shown in Fig. S3 in the supplemental material, along with the locations of the various che genes. The MCPs in V. cholerae are much more structurally diverse than those seen in E. coli. They may have one, two, or no transmembrane domains and have HAMP input modules, as is seen in E. coli, or may have Cache, PAS, or no recognized input modules. The structural diversity of the MCPs in V. cholerae (2) also supports the notion that the remaining 42 MCPs in V. cholerae could be involved in the regulation of other processes.
Eight of the 45 MCPs are already classified as virulence-related genes, as they are known to be (i) involved with the TCP (VC0825 and VC0840) (see Fig. S3 in the supplemental material), (ii) implicated in the expression of a hemolysin (VCA0220), (iii) encoded on the second Vibrio-specific pathogenesis island (VC0512 and VC0514) (see Fig. S3 in the supplemental material), or (iv) expressed only during infection of a human host (VC0216, VCA1056, and VCA0176) (20). Four other MCPs that are not currently classified as virulence-related proteins, VC0449, VC1403, VCA0906, and VCA1034, are associated with recognized virulence-related proteins (S ≥ 0.4). VC0449 is associated with two phage-related replication proteins, RstA1 (VC1454) and RstA2 (VC1463), and is known to be induced by N-acetylglucosamine, the chitin polymer subunit. This is notable because chitin induces competence in V. cholerae and has been implicated in the transfer of the CTXphi prophage among toxigenic strains (65), suggesting a role for the VC0449 MCP in regulating this process. Another MCP in this group, VCA1034, also appears to be involved in chitin-induced regulation. VCA1034 is cotranscribed with, and thought to interact with, an extracellular N-acetylglucosamine binding protein, VCA1033. It is also linked to the vibriobactin outer membrane binding protein (VC2211), the RTX toxin (VC1451), and a CheY-like response regulator (VCA1086). A third MCP in this group, VCA0906, is associated with HutZ (VCA0907). Finally, the fourth member of this group, VC1403, is associated with a single virulence-related protein, VC1817. This protein is annotated as a sigma-54-dependent transcriptional regulator. Such proteins regulate the expression of genes whose promoters are specifically recognized by the sigma-54 subunit of RNA polymerase. The set of genes regulated by VC1817 is unknown, but genes transcribed by the sigma-54 RNA polymerase include iron uptake-related genes, the immunogenic protein VCA0144, and genes required for motility (60). Elimination of the sigma-54 subunit results in attenuation of virulence in V. cholerae, and this attenuation is not entirely due to the loss of motility (29, 34).
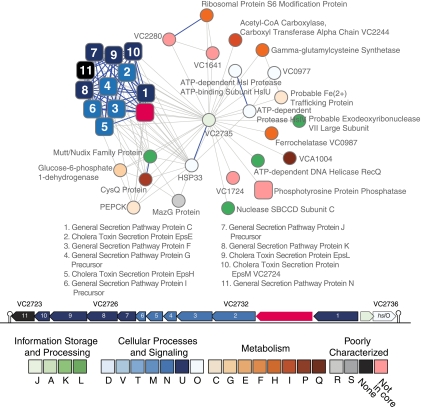
Of the 117 disproportionately associated proteins with no GO biological-process annotation, 86 are annotated as “hypothetical proteins,” and 74 of these have no GO annotation at all. Some of these 74 proteins are candidates for functional assignment. For example, VC2735 is encoded upstream of the eps operon and is thought to be cotranscribed with VC2736 as part of an operon that is divergently transcribed from the eps operon (49, 56). The eps operon plays a central role in virulence. It encodes the type II secretion system (T2SS) (and not, as implied by the genome annotation, the general secretion pathway [13]), a set of proteins that facilitates the export of the cholera toxin and a hemagglutinin/protease protein and that is also involved in the secretion of the filamentous phage that encodes the cholera toxin (12, 55). This diversity of substrates is an unusual feature of the V. cholerae T2SS (11). Figure 6 shows the proteins that interact with VC2735 and the chromosomal arrangement of the genes around it. As shown in Fig. 6, the divergently transcribed eps genes encode proteins that form a tight cluster. With the exception of VC2733, all the proteins in this cluster have orthologs in the other 11 genomes. The location of the gene encoding VC2735 is often occupied by genes that modulate some aspect of protein secretion in other species (18), but it is not a homolog of any of these proteins. VC2735 has an S4 RNA-binding motif that indicates it may play a role in translational regulation. The gene downstream of VC2735 in the putative operon encodes a redox-sensitive chaperone that is similarly disproportionately connected to virulence proteins. Chaperones similar to VC2736 are activated in response to oxidative stress and elevated temperature; they are very efficient chaperones (26). Chaperones are also commonly required to aid in the translocation and assembly of secreted proteins. We speculate that VC2735 and VC2736 are involved in translational regulation and protein stabilization under the changing conditions faced by V. cholerae, possibly when the cells enter the human host. Under these conditions, the role of the T2SS changes from involvement in filamentous phage production to secretion of cholera toxin. Presumably, deletion of these two genes would lead to a decrease in the production of cholera toxin under infective conditions.
FIG. 6.
Virulence-associated proteins that interact with VC2735. The edge styles are as in Fig. 3. The gene neighborhood is also shown, with putative transcriptional terminators indicated as stylized stems and loops. VC2733 is shown in red, as its functionality is unclear. It encodes the EpsD protein, an outer membrane protein that forms a channel through which secreted proteins and phage reach the exterior of the cell. This gene, in V. cholerae O1 biovar El Tor strain N16961, has a frameshift mutation near the N terminus and has therefore been annotated as nonfunctional. Nonetheless, the strain is able to excrete the cholera toxin and to produce viable filamentous CTXphi, so either the encoded protein is functional or a viable substitute exists and the frameshift mutation does not disrupt expression of the eps operon genes.
(ii) Proteins with high-confidence associations.
One very promising group of potential virulence-related proteins is composed of non-virulence-related proteins that are associated at a high confidence score (S ≥ 0.9) with recognized virulence proteins. We extracted a list of these proteins from the data represented in Fig. 3, and the 262 non-virulence-related proteins in this set are listed in Table S3 in the supplemental material. Associations among the virulence-related proteins and this set are seen in Fig. S1 in the supplemental material, especially near the virulence proteins that form modules involved in iron uptake, chemotaxis, pilin formation, and so on. Unlike the set of disproportionately connected proteins, most of these proteins (77%) are members of the core proteome. There are 28 proteins that are annotated as “hypothetical proteins” in this set. Two hundred eleven of the proteins have GO biological-process annotations. In contrast to the group of disproportionately connected non-virulence-related proteins, which are enriched for only a single GO term, this group is enriched for a diversity of GO terms, including “glycolysis,” “NAD biosynthetic process,” and “serine biosynthetic process”; overall, the enriched terms fall into the “metabolic process” category, which is significantly overrepresented in this group of loci. This probably reflects links between metabolism and virulence; sugar transport has been linked to the regulation of biofilm formation in V. cholerae (23), and the ability to synthesize 2,3-butanediol has been credited with the ability of El Tor strains to survive the acidity of the human stomach, thereby enhancing the virulence of these strains (75). Research looking at in vivo gene expression profiles also indicates that there are strong regulatory links between metabolism and virulence (41, 73). Recently, such a regulatory connection has been elucidated in group A Streptococcus strains (58, 59).
Some of the proteins in this set are clearly examples of virulence-related proteins that have been overlooked due to differences in annotation. For example, the genes encoding VC0244 and VC0247 are part of the operon made up of genes needed to synthesize the O antigen component of the lipopolysaccharide but were not included in our list of virulence-related proteins because, unlike the other genes in this operon, they were not designated rfb genes in the annotation.
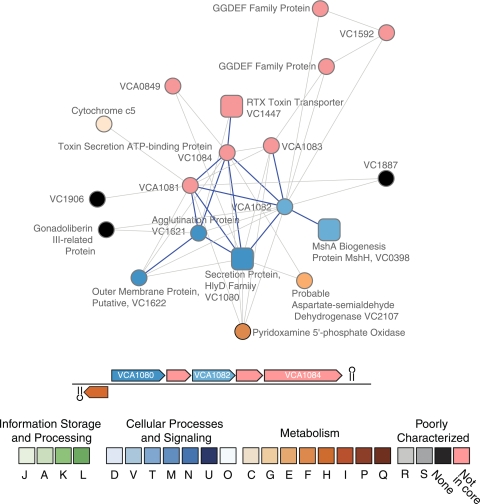
Other candidate virulence-related proteins detected include three proteins that are members of a putative six-gene operon found on chromosome 2 (Fig. 7). One of these proteins, VCA1084, is annotated as a toxin secretion ATP-binding protein. The second, VCA1082, has GGDEF and EAL domains. GGDEF/EAL proteins modulate levels of bis-(3′-5′)-cyclic di-GMP (c-di-GMP) (10). This compound is a second messenger implicated in the regulation of biofilm formation (63), motility (38), and virulence (39, 64) in V. cholerae. GGDEF domains are involved in the synthesis of c-di-GMP, while EAL domains encode phosphodiesterase activity, which breaks down c-di-GMP. The importance of c-di-GMP in the regulation of V. cholerae is underscored by the presence of 62 GGDEF and/or EAL domains in the proteome. VCA1083, which is encoded on this putative operon but not associated with high confidence with any virulence proteins, also has GGDEF and EAL domains. Interestingly, in other strains of V. cholerae, VCA1082 and VCA1083 appear to be fused into a single protein (44). The third protein is a predicted periplasmic protein of unknown function (35). The fifth protein encoded in the operon, VCA1080, is on our list of virulence-related proteins. NMPDR assigned it to its virulence protein collection on the basis of its homology with ABC-type protease exporter proteins in other taxa. This type I secretion protein has been designated a putative RTX transport protein in other species of Vibrio (35). Figure 7 reveals that these three proteins are linked to one another, as well as to other proteins involved in virulence, including VC1447, an RTX transporter protein; VC0398, which is encoded by the first gene in the msh operon, and is another GGDEF/EAL protein; and VC1622, a putative outer membrane protein that has an OmpA protein domain.
FIG. 7.
Proteins that interact with VCA1081, VCA1082, and VCA1084. Edge styles and gene neighborhood conventions are as in Fig. 6.
Conclusion.
By assembling a list of virulence-related proteins for V. cholerae N16961 and using these proteins as in silico bait proteins in a computationally generated functional-association network, we were able to generate a list of 463 proteins that are candidates for roles in virulence systems in the pathogen. This list includes proteins that are obviously involved in virulence but that were overlooked because of the annotation, as well as proteins that require follow-up to confirm their roles in virulence. This group of candidate proteins was significantly enriched for proteins involved in chemotaxis, cell communication, and signal transduction and, to a lesser degree, for proteins involved in the regulation of cellular processes and a variety of metabolic processes. This is consistent with the notion that virulence depends on the actions of a large number of proteins, many of which control the pathogen's behavior, rather than on a few proteins acting directly on the host. The associations shown in Fig. 3 and 4 hint that evolutionarily driven changes in any one protein must have far-reaching effects and suggest that studying the evolution of systems will aid greatly in understanding how pathogenesis emerges.
Supplementary Material
Acknowledgments
This work is supported by NIH grant 1R21AI067543 to T. G. Lilburn and Y. Wang, NIH grants SC1GM081068 and SC1AI080579 to Y. Wang, and the PSC-CUNY Research Award PSCREG-39-497 and CUNY Summer Research Award to J. Gu.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences, National Institute of Allergy and Infectious Diseases; the National Institutes of Health; or ATCC.
Footnotes
Published ahead of print on 7 August 2009.
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Abd, H., A. Saeed, A. Weintraub, G. B. Nair, and G. Sandström. 2007. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 60:33-39. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 104:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alon, U., M. G. Surette, N. Barkai, and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature 397:168-171. [DOI] [PubMed] [Google Scholar]
- 4.Assenov, Y., F. Ramírez, S. E. Schelhorn, T. Lengauer, and M. Albrecht. 2008. Computing topological parameters of biological networks. Bioinformatics 24:282-284. [DOI] [PubMed] [Google Scholar]
- 5.Barthelmes, J., C. Ebeling, A. Chang, I. Schomburg, and D. Schomburg. 2007. BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 35:D511-D514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batada, N. N., and L. D. Hurst. 2007. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat. Genet. 39:945-949. [DOI] [PubMed] [Google Scholar]
- 7.Butler, S. M., and A. Camilli. 2005. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspi, R., H. Foerster, C. A. Fulcher, P. Kaipa, M. Krummenacker, M. Latendresse, S. Paley, S. Y. Rhee, A. G. Shearer, C. Tissier, T. C. Walk, P. Zhang, and P. D. Karp. 2008. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 36:D623-D631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, P. A., and S. Stibitz. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17-23. [DOI] [PubMed] [Google Scholar]
- 11.Davis, B. M., E. H. Lawson, M. Sandkvist, A. Ali, S. Sozhamannan, and M. K. Waldor. 2000. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science 288:333-335. [DOI] [PubMed] [Google Scholar]
- 12.Davis, B. M., and M. K. Waldor. 2000. CTXphi contains a hybrid genome derived from tandemly integrated elements. Proc. Natl. Acad. Sci. USA 97:8572-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desvaux, M., N. J. Parham, A. Scott-Tucker, and I. R. Henderson. 2004. The general secretory pathway: a general misnomer? Trends Microbiol. 12:306-309. [DOI] [PubMed] [Google Scholar]
- 14.Doms, A., and M. Schroeder. 2005. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 33:W783-W786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmer, J. J., J. M. Janda, F. W. Brenner, D. N. Cameron, and K. M. Birkhead. 2005. Genus I. Vibrio Pacini 1854, 411AL, p. 494-546. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, vol. 2, part B. Springer, New York, NY. [Google Scholar]
- 16.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, M. Dziejman, M. H. Rahman, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. USA 101:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruque, S. M., and G. B. Nair. 2002. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 46:59-66. [DOI] [PubMed] [Google Scholar]
- 18.Filloux, A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694:163-179. [DOI] [PubMed] [Google Scholar]
- 19.Fraser, A. G., and E. M. Marcotte. 2004. A probabilistic view of gene function. Nat. Genet. 36:559-564. [DOI] [PubMed] [Google Scholar]
- 20.Hang, L., M. John, M. Asaduzzaman, E. A. Bridges, C. Vanderspurt, T. J. Kirn, R. K. Taylor, J. D. Hillman, A. Progulske-Fox, M. Handfield, E. T. Ryan, and S. B. Calderwood. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. USA 100:8508-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hjerde, E., M. S. Lorentzen, M. T. Holden, K. Seeger, S. Paulsen, N. C. Bason, C. Churcher, D. Harris, H. Norbertczak, M. A. Quail, S. Sanders, S. Thurston, J. Parkhill, N. P. Willassen, and N. Thomson. 2008. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houot, L., and P. I. Watnick. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190:311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, D. W., B. Sherman, Q. Tan, J. Collins, W. G. Alvord, J. Roayaei, R. Stephens, M. Baseler, H. C. Lane, and R. Lempicki. 2007. DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyakutake, A., M. Homma, M. J. Austin, M. A. Boin, C. C. Hase, and I. Kawagishi. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J. Bacteriol. 187:8403-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakob, U., W. Muse, M. Eser, and J. C. Bardwell. 1999. Chaperone activity with a redox switch. Cell 96:341-352. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, L. J., M. Kuhn, M. Stark, S. Chaffron, C. Creevey, J. Muller, T. Doerks, P. Julien, A. Roth, M. Simonovic, P. Bork, and C. von Mering. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412-D416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong, H., S. P. Mason, A. L. Barabasi, and Z. N. Oltvai. 2001. Lethality and centrality in protein networks. Nature 411:41-42. [DOI] [PubMed] [Google Scholar]
- 29.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keymer, D. P., M. C. Miller, G. K. Schoolnik, and A. B. Boehm. 2007. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Appl. Environ. Microbiol. 73:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. M., P. M. Bowers, D. Pal, M. Strong, T. C. Terwilliger, M. Kaufmann, and D. Eisenberg. 2007. Functional linkages can reveal protein complexes for structure determination. Structure 15:1079-1089. [DOI] [PubMed] [Google Scholar]
- 32.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirn, T. J., B. A. Jude, and R. K. Taylor. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863-866. [DOI] [PubMed] [Google Scholar]
- 34.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 35.Krishnamurthy, N., D. P. Brown, D. Kirshner, and K. Sjölander. 2006. PhyloFacts: an online structural phylogenomic encyclopedia for protein functional and structural classification. Genome Biol. 7:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefébure, T., and M. J. Stanhope. 2007. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 39.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Z., T. Miyashiro, A. Tsou, A. Hsiao, M. Goulian, and J. Zhu. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. USA 105:9769-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombardo, M., J. Michalski, H. F. Martinez-Wilson, C. Morin, T. Hilton, C. Osorio, J. Nataro, C. Tacket, A. Camilli, and J. Kaper. 2007. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. USA 104:18229-18234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maere, S., K. Heymans, and M. Kuiper. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448-3449. [DOI] [PubMed] [Google Scholar]
- 43.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 44.McNeil, L. K., C. Reich, R. K. Aziz, D. Bartels, M. Cohoon, T. Disz, R. A. Edwards, S. Gerdes, K. Hwang, M. Kubal, G. R. Margaryan, F. Meyer, W. Mihalo, G. J. Olsen, R. Olson, A. Osterman, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, D. A. Rodionov, X. Shi, O. Vassieva, V. Vonstein, O. Zagnitko, F. Xia, J. Zinner, R. Overbeek, and R. Stevens. 2007. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 35:D347-D353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bork, V. Buillard, L. Cerutti, R. Copley, E. Courcelle, U. Das, L. Daugherty, M. Dibley, R. Finn, W. Fleischmann, J. Gough, D. Haft, N. Hulo, S. Hunter, D. Kahn, A. Kanapin, A. Kejariwal, A. Labarga, P. S. Langendijk-Genevaux, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, C. McAnulla, J. McDowall, J. Mistry, A. Mitchell, A. N. Nikolskaya, S. Orchard, C. Orengo, R. Petryszak, J. D. Selengut, C. J. A. Sigrist, P. D. Thomas, F. Valentin, D. Wilson, C. H. Wu, and C. Yeats. 2007. New developments in the InterPro database. Nucleic Acids Res. 35:D224-D228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Shea, Y. A., F. J. Reen, A. M. Quirke, and E. F. Boyd. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 42:4657-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pallen, M. J., and B. W. Wren. 2007. Bacterial pathogenomics. Nature 449:835-842. [DOI] [PubMed] [Google Scholar]
- 48.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price, M., K. H. Huang, E. Alm, and A. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahman, M., K. Biswas, M. A. Hossain, R. B. Sack, J. Mekalanos, and S. M. Faruque. 2008. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 27:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajagopala, S. V., J. Goll, N. D. Gowda, K. C. Sunil, B. Titz, A. Mukherjee, S. S. Mary, N. Raviswaran, C. S. Poojari, S. Ramachandra, S. Shtivelband, S. M. Blazie, J. Hofmann, and P. Uetz. 2008. MPI-LIT: a literature-curated dataset of microbial binary protein-protein interactions. Bioinformatics 24:2622-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reguera, G., and R. Kolter. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187:3551-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 54.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, V. J. DiRita, and M. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider, K. L., K. S. Pollard, R. Baertsch, A. Pohl, and T. M. Lowe. 2006. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 34:D407-D410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shannon, P., A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski, and T. Ideker. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shelburne, S., M. Davenport, D. Keith, and J. Musser. 2008. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 16:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shelburne, S. A., D. Keith, N. Horstmann, P. Sumby, M. T. Davenport, E. A. Graviss, R. G. Brennan, and J. M. Musser. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 105:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, F. L., T. Iida, and J. Swings. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Udden, S. M., M. S. Zahid, K. Biswas, Q. S. Ahmad, A. Cravioto, G. B. Nair, J. J. Mekalanos, and S. M. Faruque. 2008. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc. Natl. Acad. Sci. USA 105:11951-11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ulrich, L. E., and I. B. Zhulin. 2007. MiST: a microbial signal transduction database. Nucleic Acids Res. 35:D386-D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.UniProt Consortium. 2008. The universal protein resource (UniProt). Nucleic Acids Res. 36:D190-D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 69.Vezzulli, L., C. A. Guzmán, R. Colwell, and C. Pruzzo. 2008. Dual role colonization factors connecting Vibrio cholerae's lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr. Opin. Biotechnol. 19:254-259. [DOI] [PubMed] [Google Scholar]
- 70.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 71.Wassenaar, T. M., and W. Gaastra. 2001. Bacterial virulence: can we draw the line? FEMS Microbiol. Lett. 201:1-7. [DOI] [PubMed] [Google Scholar]
- 72.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 73.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, J., L. Chen, L. Sun, J. Yu, and Q. Jin. 2008. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 36:D539-D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon, S. S., and J. J. Mekalanos. 2006. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect. Immun. 74:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu, H., P. M. Kim, E. Sprecher, V. Trifonov, and M. Gerstein. 2007. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 3:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zotenko, E., J. Mestre, D. P. O'Leary, and T. M. Przytycka. 2008. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput. Biol. 4:e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.